Abstract
The structurally robust biopolymer sporopollenin is the major constituent of the exine layer of pollen wall and plays a vital role in plant reproductive success. The sporopollenin precursors are synthesized through an ancient polyketide biosynthetic pathway consisting of a series of anther-specific enzymes that are widely present in all land plant lineages. Tetraketide α-pyrone reductase 1 (TKPR1) and TKPR2 are two reductases catalyzing the final reduction of the carbonyl group of the polyketide synthase-synthesized tetraketide intermediates to hydroxylated α-pyrone compounds, important precursors of sporopollenin. In contrast to the functional conservation of many sporopollenin biosynthesis associated genes confirmed in diverse plant species, TKPR2’s role has been addressed only in Arabidopsis, where it plays a minor role in sporopollenin biosynthesis. We identified in gerbera two non-anther-specific orthologues of AtTKPR2, Gerbera reductase 1 (GRED1) and GRED2. Their dramatically expanded expression pattern implies involvement in pathways outside of the sporopollenin pathway. In this study, we show that GRED1 and GRED2 are still involved in sporopollenin biosynthesis with a similar secondary role as AtTKPR2 in Arabidopsis. We further show that this secondary role does not relate to the promoter of the gene, AtTKPR2 cannot rescue pollen development in Arabidopsis even when controlled by the AtTKPR1 promoter. We also identified the gerbera orthologue of AtTKPR1, GTKPR1, and characterized its crucial role in gerbera pollen development. GTKPR1 is the predominant TKPR in gerbera pollen wall formation, in contrast to the minor roles GRED1 and GRED2. GTKPR1 is in fact an excellent target for engineering male-sterile gerbera cultivars in horticultural plant breeding.
Similar content being viewed by others
Introduction
Pollen grains of land plants are surrounded by a sculpted pollen wall, which plays critical roles in protecting male gametophytes against various biotic and abiotic stresses and, therefore, in plant sexual reproduction1. Pollen wall commonly comprises of a pectocellulosic inner intine layer and a tough outer exine layer mainly composed of sporopollenin1,2,3. Exhibiting remarkable chemical stability and physical strength, sporopollenin has been considered as the most resistant of natural biopolymers, and it is regarded as the major component enabling the resistance of pollen walls to various stresses3,4. Sporopollenin has been widely reported in various extant and fossil species across the plant kingdom, including Bryophytes where it is part of the spore wall. Production of sporopollenin is considered to have been a key prerequisite for plants to spread on land3,4.
Despite the importance, the chemical composition of sporopollenin has remained obscure due to its biochemically and physically extremely resistant nature. Decades’ attempts by applying multiple degradation studies and nuclear magnetic resonance (NMR) methods have demonstrated that sporopollenin is a complex biopolymer consisting of highly cross-linked hydroxylated aliphatic, aromatic, and phenylpropanoid-derived moieties2,5,6. Recent investigations by using thioacidolysis degradation in combination with NMR analysis further revealed aliphatic polyketide-derived polyvinyl alcohol units as the major structural components of sporopollenin and that phenylpropanoid derivatives are essential components that exist as covalently linked structural units7,8. Investigations of Arabidopsis mutants with defective pollen exine have facilitated the understanding of sporopollenin composition through genetic and biochemical studies9,10,11,12,13,14. Aliphatic polyhydroxylated tetraketide α-pyrones were revealed to be the important sporopollenin building blocks in these studies12. A series of genes encoding key enzymes involved in sporopollenin precursor biosynthesis was identified and characterized in Arabidopsis9,10,11,12,13,14, and a pathway of sporopollenin biosynthesis therefore has been proposed12,15 (Fig. 1). In the proposed pathway, medium- to long-chain fatty acids undergo hydroxylation catalyzed by cytochrome P450-enzymes CYP703A2 and CYP704B1, and are then esterified to CoA by a fatty acyl-CoA synthetase (ACOS5), the generated hydroxyl fatty acyl-CoA thioesters being subsequently condensed with malonyl-CoA by the chalcone synthase-like polyketide synthase A (PKSA) and PKSB to yield tetraketide α-pyrones, which are further reduced by tetraketide α-pyrone reductase 1 (TKPR1) and TKPR2 (Fig. 1). The resulting polyhydroxylated tetraketide products function as the building blocks for sporopollenin8,12 (Fig. 1). These enzymes have been shown to be specifically expressed in the tapetum of Arabidopsis anthers and tightly co-regulated during pollen development, and their close orthologues have been found widespread in land plants ranging from mosses to angiosperms with conserved male-organ-specific expression patterns9,10,11,12,13,14.
Medium- to long-chain fatty acids undergo hydroxylation catalyzed by cytochrome P450-enzymes CYP703A2 and CYP704B1, and are then esterified to CoA thioesters by the fatty acyl-CoA synthetase ACOS5. The generated hydroxyl fatty acyl-CoA esters are subsequently condensed with malonyl-CoA by the chalcone synthase-like polyketide synthases PKSA and PKSB to yield tetraketide α-pyrones, which are further reduced by tetraketide α-pyrone reductases TKPR1 and TKPR2. The resulting polyhydroxylated tetraketide compounds then function as building blocks for sporopollenin
In contrast to Arabidopsis ACOS5, CYP703A2, CYP704B2, PKSA, PKSB, and TKPR1, whose orthologues have been characterized to play conserved functions in sporopollenin biosynthesis in many species including rice16, tobacco16, and rapeseed17, AtTKPR2’s role has been addressed only in Arabidopsis12. Both AtTKPR1 and AtTKPR2 were shown to catalyze the same enzymatic reaction in vitro; however, AtTKPR2 seems to play a minor role in sporopollenin biosynthesis12. While the tkpr1 mutant in Arabidopsis makes completely sterile pollen grains with severely damaged exine, the tkpr2 mutant produces fertile pollen with only mildly impaired exine12. Unlike the other sporopollenin biosynthesis associated enzymes that were demonstrated to constitute an endoplasmic reticulum localized metabolon, AtTKPR2 was shown to preferentially localize in the cytoplasm12,18.
The ornamental plant Gerbera hybrida, from the sunflower family Asteraceae, is one of the economically most important cut-flower crops globally19. Gerbera has been worked into a model plant of Asteraceae, and has contributed to flower development and secondary metabolite biosynthesis studies for decades19. The gerbera type III polyketide synthases include a small family of the archetypical chalcone synthases20,21, but we also identified the first plant pyrone polyketide synthase, gerbera 2-pyrone synthase 1 (G2PS1), that utilizes acetyl-CoA as a starter to initiate the synthesis of two antimicrobial polyketide derivatives, gerberin and parasorboside22. Later, we identified two homologues of G2PS1 (G2PS2 and G2PS3) that use the same substrates but catalyze a longer elongation in the biosynthesis of another defense-related polyketide, 4-hydroxy-5-methylcoumarin23. Orthologues of Arabidopsis PKSA and PKSB, the gerbera anther-specific chalcone synthase-like 1 (GASCL1) and GASCL2, display the conserved tapetum-specific expression pattern and catalyze the conserved sporopollenin precursor biosynthesis with medium- to long-chain fatty CoA esters as starters24. During the exploration of accessory reductases in gerberin and parasorboside biosynthesis, we identified a reductase gene that displays a similar wide expression pattern as G2PS1, Gerbera reductase 1 (GRED1), and its close paralogue GRED2. Both GRED1 and GRED2 are orthologues of AtTKPR2.
The dramatically expanded expression pattern of GRED1 and GRED2 implies involvement in pathways outside the anthers. In this study, we show that GRED1 and GRED2 are nevertheless involved in sporopollenin biosynthesis with a similar secondary role as AtTKPR2 in Arabidopsis. We further show that this secondary role does not relate to the promoter of the gene, AtTKPR2 cannot rescue pollen development in Arabidopsis even when controlled by the AtTKPR1 promoter. We also identified the gerbera orthologue of AtTKPR1, GTKPR1, and characterized its crucial role in gerbera pollen development. GTKPR1 is the predominant TKPR in gerbera pollen wall formation, in contrast to the minor roles of GRED1 and GRED2. GTKPR1 is in fact an excellent target for engineering male-sterile gerbera cultivars in horticultural plant breeding.
Results
Identification and analysis of orthologous genes for AtTKPR1 and AtTKPR2 in gerbera
A single orthologue of AtTKPR1 was identified in gerbera by a BLAST search25 of the gerbera transcriptome using AtTKPR1 as the query sequence and named GTKPR1. GTKPR1 was recovered in full length from gerbera anther cDNA. The cDNA contains a 990 bp open reading frame (ORF) showing 68% identity with AtTKPR1 at nucleotide sequence level. GRED1 and GRED2 are two orthologues of AtTKPR2 that were identified and cloned during our exploration of the accessory reductases in the biosynthesis of the defense-related polyketides gerberin and parasorboside. Both GRED1 and GRED2 contain a 960 bp ORF and they share 85% nucleotide sequence identity with each other, and 68% and 66% identity with AtTKPR2, respectively (Table S2). Further BLAST searches of the gerbera transcriptome did not uncover any additional AtTKPR2-like transcripts in gerbera. Pairwise comparisons of the gerbera and Arabidopsis TKPR1 and TKPR2 encoded amino acid sequences showed that GTKPR1 and AtTKPR1 showed 73% identity in amino acid sequence, GRED1, GRED2, and AtTKPR2 shared 67% and 65% identity, respectively, while AtTKPR1 and GTKPR1 showed no more than 50% identity with AtTKPR2 or GRED1 or GRED2 (Table S2).
AtTKPR1 and AtTKPR2 orthologues from selected species ranging from mosses to eudicots were extracted from the GenBank database. We found that there was only a single AtTKPR1 orthologue in all species we analyzed (except in soybean), while some species, like gerbera, had two AtTKPR2 orthologues. In phylogenetic analysis, the reductases clustered in two clades. GTKPR1, AtTKPR1, and other AtTKPR1 orthologues clustered the TKPR1 clade while AtTKPR2, GRED1, GRED2, and the other AtTKPR2 orthologues formed the TKPR2 clade (Fig. 2).
Full amino acid sequences from Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Helianthus annuus (Ha), Glycine max (Gm), Populus trichocarpa (Pt), Vitis vinifera (Vv), Arabidopsis thaliana (At), Gerbera hybrida (G) were aligned and then back-translated to the corresponding original nucleotide sequences. In the unrooted maxiumum-likelihood phylogenic tree TKPR1- and TKPR2-like genes form separate clades. Bootstrap support values are indicated at the nodes. The scale indicates branch lengths measured in the number of substitutions per site
GTKPR1 shows tapetum-specific expression, while GRED1 and GRED2 are universally expressed in various gerbera organs but in the anther, they are tapetum-specific
To investigate the expression patterns of GRED1, GRED2, and GTKPR1 in gerbera, we analyzed their expression profiles in 15 gerbera tissues by RT-PCR. Our data shows that GRED1 is highly expressed in various gerbera organs but only moderately expressed in anthers; GRED2 is preferentially expressed in the disc flowers, anthers, receptacle, scape, and roots (Fig. 3a). Neither of them displayed the anther-specific expression pattern like the Arabidopsis AtTKPR2. Unlike the universally expressed GRED1 and GRED2, GTKPR1 was specifically expressed in disc flowers at inflorescence stage 6 with developing anthers (Fig. 3a), consistent with the reported expression pattern of Arabidopsis AtTKPR1, tobacco NtTKPR1, and rice OsTKPR112,16.
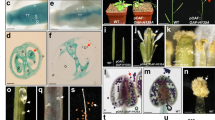
a Semi-quantitative RT-PCR analysis of expression levels of GTKPR1, GRED1, GRED2, and GMYB80 in a series of gerbera tissues. Stage 6 disc flowers have active tapetal cells, the anther sample is a mixture of stages 6, 8, and 10. The constitutively expressed gerbera GAPDH gene was used as control. b RNA in situ hybridization of GTKPR1, GRED1, and GRED2 in developing anthers of stage 6 disc flowers. Sense and anti-sense probes of GASCL1 were applied as negative and positive controls. MMC microspore mother cells, Tds tetrads, T tapetum
To further investigate the precise locations of GTKPR1, GRED1, and GRED2 expression in gerbera anthers, we conducted RNA in situ hybridization experiments with gene-specific antisense probes in sections containing anthers at various developmental stages. An antisense probe for GASCL1, the gerbera tapetum-specific orthologue of AtPKSA in sporopollenin biosynthesis, was used as a positive control, and its sense probe was used as a negative control. Hybridization signals of the GTKPR1 probe were exclusively detected in the tapetum cells, starting from the meiosis stage when the tapetum was first visible until the microspore stage when the tapetum degenerated. The strongest signals were observed at the microspore stage, which was consistent with the in situ localization of AtTKPR1 in Arabidopsis anthers and NtTKPR1 in tobacco anthers12,16 (Fig. 3b). The hybridization signals of GRED1 and GRED2 were also exclusively observed in the tapetum cells of the anthers; however, they were only observed at the microspore stage when the microspores were released from the tetrads, and the GRED1 probe displayed much weaker signals compared with the GTKPR1 or GRED2 probes (Fig. 3b). This observation is consistent with the reported AtTKPR2 expression, which was specifically expressed in tapetum but was more restricted temporally compared with AtTKPR112.
Sporopollenin biosynthesis genes are under common transcriptional regulation in Arabidopsis and particularly the transcription factor MYB80 (MS188) is an upstream regulator of all known biosynthetic genes, including AtTKPR226. We investigated if the expanded patterns of GRED1 and GRED2 expression in gerbera would be the result of expanded expression of the gerbera MYB80 homologue GMYB80. Expression of GMYB80 follows closely the pattern of GTKPR1, not GRED1 or GRED2 (Fig. 3a) indicating that GMYB80 is not responsible for their expression in vegetative tissues.
Downregulation of GTKPR1, GRED1, and GRED2 differentially affect gerbera pollen development
To investigate the physiological functions of GTKPR1 in gerbera pollen grain development, we applied a double-stranded RNA interference (RNAi) vector to produce GTKPR1 downregulated gerbera transgenic lines. We characterized several lines that showed specific suppression of GTKPR1 (Fig. 4a) and observed that their inflorescences showed visible phenotypes (Fig. S1). Compared with the wild-type inflorescences displaying healthy anthers and large amounts of released pollen, the severely downregulated lines (GTKPR1_RNAi TR17 and TR20) and the moderately downregulated line (GTKPR1_RNAi TR8) showed shrunken anthers without any released pollen, and the mildly downregulated line (GTKPR1_RNAi TR11) showed normal anthers and a small amount of released pollen grains in the spring (Fig. S1) but not in the winter (not shown in figure). We characterized four lines (GRED1/2 RNAi TR3, TR8, TR9, and TR12) that showed cross-suppression of both GRED1 and GRED2 (Fig. 4b). All four lines grew similar inflorescences as the GTKPR1 mild suppression line that showed normal anthers and small amounts of release pollen grains in the spring (Fig. S1).
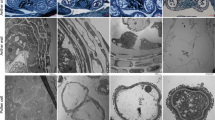
a, b Expression analysis of severely, moderately, and mildly repressed GTKPR1 RNAi lines and GRED1/2 RNAi lines. qRT-PCR shows the expression levels of the corresponding genes in disc flowers of inflorescence development stage 6. c Statistics for the normal and dead pollen grains in gerbera wild-type and transgenic lines. d Alexander staining and SEM observation of pollen grains made by gerbera wild-type and transgenic lines, arrows refer to damaged exine. Error bars (± SE) are calculated from three biological replicates. Scale bars are 30 μm
We further applied Alexander staining to examine the viability of pollen grains produced by the transgenic lines and applied SEM to examine the morphology of the pollen grains in detail. The results show that all pollen grains produced by the GTKPR1 severe suppression lines (GTKPR1_RNAi TR17 and TR20) were severely distorted with damaged exine and showed no viability at all (Fig. 4c, d). More than 80% of the pollen grains produced by the GTKPR1 moderate suppression line (GTKPR1_RNAi TR8) were dead and these pollen grains showed a degree of distortion in shape and damage in exine (Fig. 4c, d). The mild suppression line (GTKPR1_RNAi TR6) produced around 50% of dead pollen grains based on the staining, and SEM showed that these pollen grains were not distorted but showed a mild exine damage (Fig. 4c, d). The GRED1/2 downregulation lines produced 10% to 30% dead pollen grains that also showed mild exine damage (Fig. 4c, d), a similar phenotype as the pollen grains in the Arabidopsis tkpr2 mutant12. The pollen exine damage was relatively heavier in the severe suppression lines (GRED1/2 RNAi TR3, TR8), which displayed a mildly distorted shape compared with pollen in the moderated suppression lines (GRED1/2 RNAi TR9, TR12) that showed a nearly normal shape (Fig. 4c, d). As a control, the wild-type gerbera produces 95% alive pollen grains with a smooth surface and healthy exine (Fig. 4c, d).
Expression of GTKPR1 in Arabidopsis tkpr1 mutant rescues the male-sterile phenotype, while expression of AtTKPR2, GRED1, and GRED2 do not
AtTKPR1 and AtTKPR2 have been shown to catalyze the same in vitro enzymatic reactions and both were considered to catalyze the reduction of tetraketide α-pyrones in Arabidopsis sporopollenin biosynthesis12. To verify that GTKPR1 plays conserved function with AtTKPR1, and to investigate whether AtTKPR2 and its orthologues GRED1 and GRED2 can catalyze the same in vivo functions as AtTKPR1, we performed Arabidopsis mutant complementation experiments. Coding sequences of GTKPR1, AtTKPR2, GRED1, and GRED2 were independently transformed into the sterile Arabidopsis tkpr1 homozygous background under the control of the native AtTKPR1 promoter (Fig. 5a), and the AtTKPR1 coding sequence (in the same vector) was transformed as a positive control. We conducted the floral dipping transformations on the fertile flower shoots of Arabidopsis tkpr1/+ heterozygote and selected the transgenic lines by germinating seeds on hygromycin selection medium. The transgenic lines in the tkpr1 homozygous background were identified through genotyping PCR with specific primers, and RT-PCR with gene-specific primers was applied to verify the expression of the transformed genes (Fig. S2).
a Structural representation of complementation constructs for AtTKPR1, AtTKPR2, GTKPR1, GRED1, and GRED2, driven by the AtTKPR1 promoter. b–f Inflorescences, siliques, flowers, anthers, and viability staining of pollen grains in Arabidopsis nontransgenic plants (Col-0 and tkpr1 homozygous) and AtTKPR1, AtTKPR2, GTKPR1, GRED1, and GRED2 transgenic plants in tkpr1 homozygous background. Scale bars are 50 mm in c, 500 μm in d, and 50 μm in e and f
Three or more individual transgenic lines in tkpr1 homozygous background were screened from each construct. By observing the phenotypes of these transgenic lines, we found that the restoration of pollen fertility and seed formation were achieved in AtTKPR1 and GTKPR1 transgenic lines, while the AtTKPR2, GRED1, and GRED2 transgenic lines displayed a male sterile phenotype, the same as the nontransgenic tkpr1 homozygous control (Fig. 5b, c). Released pollen grains were found on the mature anther surfaces of AtTKPR1 and GTKPR1 transgenic lines in tkpr1 homozygous background, while the AtTKPR2, GRED1, and GRED2 transgenic lines in tkpr1 homozygous background did not show any released pollen grains on the anther surfaces, a similar phenotype as the tkpr1 mutant (Fig. 5c, d). However, the pollen viability assay showed tiny amounts of viable pollen grains in few of the mature anthers of AtTKPR2, GRED1, and GRED2 transgenic lines in contrast to the tkpr1 mutant that displayed practically empty anthers (Fig. 5e). This indicates that the viability of pollen grains was partially resorted in these lines.
Discussion
The structurally robust biopolymer sporopollenin is the major constituent of the exine layer of pollen wall and plays a vital role in plant reproductive success. In Arabidopsis, enzymes responsible for sporopollenin biosynthesis are encoded by a set of anther-specific genes CYP703A2/CYP704B1, ACOS, PKSA/PKSB, and TKPR1/TKPR29,10,11,12,13,14. Extensive evidence suggests that sporopollenin biosynthesis is a highly conserved polyketide biosynthetic pathway present in all land plant lineages and was probably associated with the evolution of land plants10,11,12. In contrast to the functional conservation of many sporopollenin biosynthesis associated genes having been confirmed in several plant species like rice, tobacco, and rapeseed, orthologues of AtTKPR2 have not been studied in other species than Arabidopsis. Gerbera has two orthologues of AtTKPR2, GRED1, and GRED2. They are expressed not only in anthers (like AtTKPR2), but also widely in vegetative tissues, possibly indicating recruitment to biochemical pathways outside of the anthers. In this study, we characterized that GRED1 and GRED2 have kept the conserved tapetum-specific expression pattern in anthers and in fact can and do participate in sporopollenin biosynthesis. Concerning the anther domain, GRED1 and GRED2 of gerbera are similar to AtTKPR2 of Arabidopsis. We also characterized the orthologue of AtTKPR1 in gerbera, GTKPR1, and revealed its critical function in sporopollenin biosynthesis. Our results confirm the conservation of the TKPR function in sporopollenin biosynthesis, and that TKPR1s (e.g., AtTKPR1 and GTKPR1) are the predominate enzymes. The roles of TKPR2s (e.g., AtTKPR2, GRED1, and GRED2) are minor, and the strong expression of GRED1 and GRED2 outside anthers in gerbera rises the interesting question of whether the main function of TKPR2 is in fact outside of sporopollenin biosynthesis even in Arabidopsis.
GRED1, GRED2, and GTKPR1 are involved in gerbera pollen exine formation
Gerbera GRED1 and GRED2, the two orthologues of AtTKPR2, encode two reductases showing high sequence similarities with AtTKPR2, and GTKPR1 is the orthologue of AtTKPR1 (Table S2). TKPRs catalyze the final reduction of the PKSA/PKSB-synthesized tetraketide intermediates to hydroxylated α-pyrone compounds, important precursors of sporopollenin12 (Fig. 1). Despite exhibiting the same in vitro enzymatic functions, AtTKPR2 was suggested to play a minor function in sporopollenin biosynthesis compared to AtTKPR1’s predominant role in the pathway. Homozygous tkpr2 mutant plants produce fertile pollen with mildly impaired exine while the tkpr1 mutant makes completely sterile pollen grains with a severely damaged exine12. In gerbera, although GRED1 and GRED2 were expressed in various organs, in anthers they were exclusively expressed in the tapetum at microspore stage, a precisely similar expression pattern as the one reported for AtTKPR212 (Fig. 3a, b). This suggests that GRED1 and GRED2 may still be involved in sporopollenin biosynthesis in the anthers.
An important question relating to the minor role of AtTKPR2 in sporopollenin biosynthesis is whether this is due to the promoter and expression pattern of the gene. In vitro, AtTKPR2 catalyzes the same reaction as AtTKPR112. In constructs with a 1500 bp promoter fragment of AtTKPR1 attached to the cDNA of the reductases, we could show that AtTKPR1 and GTKPR1 could fully complement the tkpr1 mutant in Arabidopsis (Fig. 5b, c). However, the AtTKPR2 construct could not rescue fertility, but still restored viability of the pollen grains to some extent, concluded from the vital staining that marks the pollen grain cytoplasm (Fig. 5b–f). This shows that AtTKPR1 and AtTKPR2 are not functionally equivalent. GRED1 and GRED2 (under the AtTKPR1 promoter) led to a similar partial restoration of pollen viability as AtTKPR2 (Fig. 5f), indicating a similar function in this context.
The gerbera GRED1/GRED2 cross-downregulated transgenic lines produced 10–30% of dead pollen grains with a mild exine damage (Fig. 4c), a similar phenotype as reported Arabidopsis tkpr2 mutant9. The gerbera GTKPR1 severely downregulated lines were however completely sterile and produced 100% of distorted pollen grains with severe exine damage, and milder suppressed lines were partially sterile producing around 50% of dead pollen grains with relative milder exine damage (Fig. 4c, d), resembling the Arabidopsis tkpr1 null mutants and partial mutants12. GTKPR1 exhibited a typical tapetum-specific expression pattern as do various reported sporopollenin biosynthesis associated genes (Fig. 3b), including gerbera GASCL1 and GASCL224. This is highly consistent with AtTKPR1 expression pattern in Arabidopsis in the tapetum of anthers, ranging from the meiosis stage to microspore stage12.
Completely male-sterile and terribly distorted pollen grains were also observed in rice Ostkpr1 mutant and tobacco NtTKPR1 downregulation lines, in which the crucial TKPR functions of OsTKPR1 and NtTKPR1 were characterized previously16. These observations suggest that all the three reductases are involved in exine formation in gerbera. Similar as AtTKPR1, GTKPR1 is the predominate TKPR in gerbera sporopollenin biosynthesis and its function is irreplaceable in pollen exine formation. Consistent with AtTKPR2, GRED1, and GRED2 are relatively less crucial for this pathway, majority of pollen grains are alive, and the plants are fertile even without their functions.
Functional conservation and differentiation of tetraketide α-pyrone reductases in land plants
Searching the latest transcriptome data of several model plants, we showed that TKPR1 genes nearly always appear as single copy with male-organ specific expression pattern while TKPR2s are present as two copies with non-anther-specific expression patterns in several species like gerbera, soybean, and poplar (Fig. 2, Table S3). This pattern suggests that while TKPR1s are highly functionally conserved in sporopollenin biosynthesis, TKPR2s may have differentiated into novel functions in the process of gene duplication during land plant evolution. The functional conservation of TKPR1s has been confirmed by the precisely consistent tapetum-specific expression pattern of AtTKPR1, OsTKPR1, NtTKPR1, and GTKPR1 and consistent severely distorted exine phenotype presented in corresponding mutant plants reported previously and in this work12,16. This is also strongly supported by our observation that GTKPR1 successfully restored the fertility of Arabidopsis tkpr1 mutant. TKPR2s have been rarely studied previously and TKPR2’s minor role in sporopollenin biosynthesis was only characterized in Arabidopsis12. Consistent male organ-specific expression pattern in most species suggests that TKPR2s would play conserved functions in sporopollenin biosynthesis. However, the dramatically expanded expression pattern of gerbera GRED1 and GRED2, and of soybean and poplar TKPR2 genes, implies that they might have acquired novel functions involved in pathways outside the anthers.
TKPR1 is a potential molecular target for male sterility in breeding
Male sterility of plants is an important trait with applications in agricultural and horticultural crop breeding, such as effective production of hybrid seeds, guarding of germplasm in commercial ornamental breeding and in preventing of undesired diffusion of seeds or pollen grains produced by genetically modified crops in the open field27. In this study, we showed that TKPR1 is one of the key genes controlling the plant’s male fertility and is highly functionally conserved in land plants. TKPR1 can be applied as a molecular target for controlling male fertility in crops through modern genetic methods such as RNAi or CRISPR-Cas928. Present as single copy, the gene is a straightforward target with a low off-target risk. In this work, we successfully made male sterile gerbera transgenic lines by suppression of GTKPR1 expression, demonstrating the application in an important ornamental plant species.
Materials and methods
Plant materials
Gerbera hybrida cultivar ‘Regina’ was obtained from Terra Nigra B.V, The Netherlands. Wild-type and transgenic gerbera plants were multiplied in vitro and grown as previously described29. Arabidopsis thaliana (ecotype Col-0) T-DNA insertion line SAIL_837_D01 seeds of a tkpr1 heterozygous plant were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). In the progeny, the wild-type, tkpr1 homozygous, and heterozygous individuals were identified by PCR using gene-specific and T-DNA-specific primers (Table S1). The identified tkpr1 heterozygous plants were used for floral dipping transformation30. For the growth of Arabidopsis, seeds were surface sterilized and stratified for 2 days at 4 °C in the dark, then germinated on MS medium (no antibiotics for control plants and supplemented with 30 mg/L hygromycin B (Sigma) for transgenic lines) at 20 °C under continuous light. Two-week-old seedlings were transplanted to soil and grown at 22 °C under long-day conditions (light/dark cycles of 16 h/8 h). Developmental stages of gerbera inflorescences and Arabidopsis flowers are described by Helariutta et al.31 and Smyth et al.32, respectively.
Identification and cloning of GTKPR1, GRED1, GRED2, and GMYB80
Using the Arabidopsis tetraketide α-pyrone reductase 1 (AtTKPR1) as the query sequence to blast against the gerbera transcriptome data from 35 gerbera tissues (unpublished data), a TKPR1-like reductase encoding transcript was identified and named as GTKPR1. The GTKPR1 sequence was recovered in full length and its full coding sequence was amplified from the gerbera cultivar ‘Regina’ anther cDNA with gene-specific primers (Table S1). GRED1 and GRED2 are two TKPR2-like genes that were identified and cloned based on GRED1 having a significantly similar expression pattern as the broadly expressed gerbera 2-pyrone synthase G2PS1 and GRED2 being similar in sequence with GRED1. GRED1 and GRED2 sequences were recovered in full length and their full coding sequences were amplified from the cultivar ‘Regina’ petal cDNA of inflorescence development stage 6 with gene-specific primers (Table. S1). GMYB80 was identified through significant similarity (84–87% identity, 100% coverage) of the encoded amino acid sequence with MYB80 transcription factors of Asteraceae, e.g., with those of Lactuca sativa and Helianthus annuus (XP_023740601 and XP_022039565, respectively). GenBank accession numbers for the nucleotide sequences for GTKPR1, GRED1, GRED2, and GMYB80 are MW842918, MW842919, MW842920, and MZ328313, respectively.
Phylogenetic analysis
To conduct phylogenetic analysis, putative TKPR1 and TKPR2 orthologues from selected plant species were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) by conducting BLASTP searches using AtTKPR1 and AtTKPR2 as queries, and their accession numbers are listed in Table S3. The full-length amino acid sequences were aligned using Clustal Omega software33 and then back-translated to the corresponding nucleotide sequences. The generated alignment was used to construct a maximum-likelihood phylogenetic tree in MEGA X34 with default parameters, and a total of 1000 bootstrap replicates. Expression patterns of TKPR1 and TKPR2 orthologues were searched from published articles, ePlant database or model plant genome databases (Table S3).
Generation of gerbera RNAi transgenic lines
For the gene silencing construct, the full-length ORFs for GTKPR1, GRED1, and GRED2 were cloned into the entry vector pDONR221 through the Gateway BP reaction35, and the corresponding RNAi constructs were then generated by LR recombination between the entry clones and the Gateway binary vector pK7GWIWG2D(II)36. For transformation, the GTKPR1, GRED1, and GRED2 RNAi constructs were electroporated into Agrobacterium tumefaciens strain C58C1(pGV2260)37, and then used for the agrobacterium-mediated transformation of the cultivar ‘Regina’, performed as described earlier38.
Arabidopsis complementation experiment
A 1.5 kb region upstream of the AtTKPR1 initiation codon was Gateway cloned from Arabidopsis genomic DNA into the entry vector pDONRP4-P1r (Invitrogen) using the primers listed in Table S1 to generate the promoter entry vector pPro_TKPR1_entry. The pEnNosT2-R2R3 vector containing a nopaline synthase 3′-terminator flanked by attR2 and attL3 sites was a gift from Ari Pekka Mähönen (University of Helsinki, Finland). Full-length coding sequences of AtTKPR1 and AtTKPR2 were PCR amplified from Arabidopsis Col-0 flower bud cDNA using gene-specific primers (Table S1) and further cloned into pDONR221. The complementation constructs for AtTKPR1, AtTKPR2, GTKPR1, GRED1, and GRED2, driven by the AtTKPR1 promoter, were constructed through Multisite Gateway LR reactions between the pPro_TKPR1_entry, entry clones of the target genes, pEnNosT2-R2R3 vector, and the destination vector pHm43GW, according to the manufacturer’s instructions (Invitrogen). The generated complementation constructs were electroporated into Agrobacterium tumefaciens strain GV3101(pMP90)39 and further transformed independently into the Arabidopsis heterozygous tkpr1/+ mutant line by floral dipping30. The harvested seeds were screened on MS medium supplemented with 30 mg/L hygromycin B (Sigma-Aldrich) to select the transgenic plants. The transgenic plants in tkpr1 homozygous background (tkpr1/tkpr1) were identified by PCR as described above.
Gene expression analysis
Semiquantitative RT-PCR was performed to analyze gene expression in gerbera tissue series and in Arabidopsis nontransgenic and transgenic lines. Quantitative real-time PCR (qRT-PCR) was performed to analyze gene expression analysis in gerbera wild-type and transgenic lines. For determination of the expression patterns of GTKPR1, GRED1, and GRED2 in gerbera, a series of 15 vegetative and reproductive tissue series were sampled from gerbera cultivar ‘Regina’ for total RNA isolation, including ray and disc flowers from inflorescence development stage 6, 8, and 10, pooled bract, carpel, anther, ovary, pappus, receptacle, and scape samples from inflorescence development stages 6, 8, and 10, pooled leaf sample from 5–8 cm, 25 cm, and 37–38 cm long leaves, and root. For verification of RNAi-efficiency in GTKPR1 RNAi and GRED1/2 RNAi gerbera transgenic lines, disc flowers at inflorescence development stage 6 were collected from wild-type, five independent GTKPR1 RNAi lines (TR8, TR11, TR17, and TR20) and four independent GRED1/2 RNAi lines (TR3, TR8, TR9, and TR12). For verification of the expression of transformed genes in Arabidopsis, flower buds at stage 9 from transgenic lines and nontransgenic controls were collected for RNA isolation. The total RNA isolation and cDNA synthesis was performed as described previously24. For RT-PCR, gene-specific primers (Table S1) were used to amplify the corresponding cDNA sequences under the following PCR program: 95 °C for 3 min, followed by 28 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min, and followed by 72 °C for 8 min. qRT-PCR was performed with gene-specific primers (Table S1) as described previously24 and relative expression was calculated by using the ΔΔCt method as described by Pfaffl40. The gerbera housekeeping gene GAPDH and Arabidopsis TUBULIN were used as reference genes for gerbera and Arabidopsis, respectively.
RNA in situ hybridization
The 1–5 outermost rings of disc flowers of the cultivar ‘Regina’ at inflorescence development stage 6 were collected for RNA in situ hybridization, performed as described by Kontturi et al.24. Gene-specific fragments for GRED1 (199 bp), GRED2 (240 bp), and GTKPR1 (212 bp) were PCR amplified from the GRED1, GRED2, and GTKPR1 entry clones with primers containing the T7 polymerase promoter sequence (Table S1), and then applied to synthesis of digoxigenin-labeled antisense probes following the manufacturer’s protocols of DIG RNA labeling kit (Roche). Gene-specific sense and antisense probes for GASCL1 synthesized previously24 were used as the positive and negative controls.
Pollen grain viability assay
Optimized Alexander staining described by Peterson et al.41 was applied to analyze the viability of pollen grains in gerbera and Arabidopsis transgenic lines. Anthers of gerbera wild-type, GTKPR1 RNAi, and GRED1/2 RNAi transgenic lines at inflorescence development stage 9, and flowers of Arabidopsis wild-type, tkpr1 homologous mutant, and transgenic lines at floral stage 14–15 were collected and fixed in 60% ethanol, 30% chloroform, and 10% glacial acetic acid (V/V) for 2–3 hours, then stained with 2–4 drops of stain solution (0.01% Malachite green, 0.05% Acid fuchsin, 0.005% Orange G, 10% ethanol, and 4% glacial acetic acid) on a slide with a moderate rate of heating until the stain solution was nearly boiling. Rinsed with water and the stained pollen grains were released by pressing slightly with a cover slip. Slides were examined and photographed using the LeitzLaborlux S Microscope (Wetzlar, Germany) equipped with a Leica DFC420 C Digital Camera. Cytoplasm of viable pollen grains is stained in purple while aborted and empty pollen grains stain blue. The alive and dead pollen grains from each gerbera sample were counted from three independent fields with 20–100 pollen grains each.
Scanning electron microscopy (SEM)
Surface morphology of gerbera pollen grains was examined by SEM. For sampling, flowers were collected from wild-type and selected transgenic lines, at similar developmental stages as compared to those used for pollen grain staining. Pollen grains were squeezed out from the anthers, and directly mounted to carbon double-sided tape supported by a sample stub. Platinum coating was done to all mounted samples by a Quorum Q150TS high-resolution coater (Quorum Technologies, UK). The samples were then imaged by a Quanta 250 SEM device (FEI, Hillsboro, OR, USA) located at the electron microscopy unit, University of Helsinki, Finland. In close-up images of individual pollen grains, background was removed in Adobe Photoshop CC in order to better illustrate the pollen structures.
References
Jiang, J., Zhang, Z. & Cao, J. Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol. 15, 249–263 (2013).
Quilichini, T. D., Grienenberger, E. & Douglas, C. J. The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113, 170–182 (2015).
Wallace, S., Fleming, A., Wellman, C. H. & Beerling, D. J. Evolutionary development of the plant spore and pollen wall. AoB Plants 11, plr027 (2011).
Brooks, J. & Shaw, G. Sporopollenin: a review of its chemistry, palaeochemistry and geochemistry. Grana 17, 91–97 (1978).
Ariizumi, T. & Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460 (2011).
Ahlers, F., Thom, I., Lambert, J., Kuckuk, R. & Wiermann, R. 1H NMR analysis of sporopollenin from Typha Angustifolia. Phytochemistry 50, 1095–1098 (1999).
Xue, J. S. et al. Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Mol. Plant 13, 1644–1653 (2020).
Li, F. S., Phyo, P., Jacobowitz, J., Hong, M. & Weng, J. K. The molecular structure of plant sporopollenin. Nat. Plants 5, 41–46 (2019).
Dobritsa, A. A. et al. CYP704B1 is a long-chain fatty acid ω-Hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151, 574–589 (2009).
De Azevedo Souza, C. et al. A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21, 507–525 (2009).
Kim, S. S. et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22, 4045–4066 (2010).
Grienenberger, E. et al. Analysis of TETRAKETIDE α-PYRONE reductase function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22, 4067–4083 (2010).
Tang, L. K., Chu, H., Yip, W. K., Yeung, E. C. & Lo, C. An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. N. Phytol. 181, 576–587 (2009).
Morant, M. et al. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19, 1473–1487 (2007).
Kim, S. S. & Douglas, C. J. Sporopollenin monomer biosynthesis in Arabidopsis. J. Plant Biol. 56, 1–6 (2013).
Wang, Y., Lin, Y.-C., So, J., Du, Y. & Lo, C. Conserved metabolic steps for sporopollenin precursor formation in tobacco and rice. Physiol. Plant 149, 13–24 (2013).
Qin, M. et al. Heterodimer formation of BnPKSA or BnPKSB with BnACOS5 constitutes a multienzyme complex in tapetal cells and is involved in male reproductive development in Brassica napus. Plant Cell Physiol. 57, 1643–1656 (2016).
Lallemand, B., Erhardt, M., Heitz, T. & Legrand, M. Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol. 162, 616–625 (2013).
Teeri, T. H., Elomaa, P., Kotilainen, M. & Albert, V. A. Mining plant diversity: Gerbera as a model system for plant developmental and biosynthetic research. Bioessays 28, 756–767 (2006).
Helariutta, Y. et al. Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae). Plant Mol. Biol. 28, 47–60 (1995).
Deng, X. et al. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. N. Phytol. 201, 1469–1483 (2014).
Eckermann, S. et al. New pathway to polyketides in plants. Nature 396, 387–390 (1998).
Pietiäinen, M. et al. Two polyketide synthases are necessary for 4-hydroxy-5-methylcoumarin biosynthesis in Gerbera hybrida. Plant J. 87, 548–558 (2016).
Kontturi, J. et al. Functional characterization and expression of GASCL1 and GASCL2, two anther-specific chalcone synthase like enzymes from Gerbera hybrida. Phytochemistry 134, 38–45 (2017).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Wang, K. et al. The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 178, 283–294 (2018).
Mitsuda, N. et al. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4, 325–332 (2006).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Ruokolainen, S. et al. Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol. 10, 1–11 (2010).
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W. & Chua, N. H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006).
Helariutta, Y., Elomaa, P., Kotilainen, M., Seppänen, P. & Teeri, T. H. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 22, 183–193 (1993).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Sievers, F. et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Katzen, F. Gateway® recombinational cloning: a biological operating system. Expert Opin. Drug Discov. 2, 571–589 (2007).
Karimi, M., Inzé, D. & Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Deblaere, R. et al. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13, 4777–4788 (1985).
Elomaa, P. & Teeri, T. H. Biotechnology in Agriculture and Forestry, 48 (ed Bajaj, Y. P. S.) (Springer, 2001).
Koncz, C. & Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396 (1986).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Peterson, R., Slovin, J. P. & Chen, C. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1, 66–69 (2010).
Acknowledgements
We would like to thank Dr Ari Pekka Mähönen for sharing pEnNosT2-R2R3 vector, our laboratory technicians Anu Rokkanen, and Eija Takala for their great technical assistance, gardener Sanna Peltola for taking care of the plants in the greenhouse, and the Electron Microscopy Unit at the Institute of Biotechnology, University of Helsinki, for providing excellent facilities. The China Scholarship Council is thanked for providing grant for PhD studies of LZ.
Author information
Authors and Affiliations
Contributions
T.H.T. and L.Z. designed the research. L.Z. performed the research, collected, and analyzed the data. T.Z. conducted the SEM observations. L.Z. and T.H.T. interpreted the data and wrote the manuscript, which all authors commented.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, L., Zhang, T. & Teeri, T.H. Tetraketide α-pyrone reductases in sporopollenin synthesis pathway in Gerbera hybrida: diversification of the minor function. Hortic Res 8, 207 (2021). https://doi.org/10.1038/s41438-021-00642-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00642-8
This article is cited by
-
Comparative proteomic analysis of papaya bud flowers reveals metabolic signatures and pathways driving hermaphrodite development
Scientific Reports (2024)
-
Cytological Analysis of Male-Sterile MS5 Japanese Cedar (Cryptomeria japonica D. Don) and Comparison with Other Male-Sterile Mutants
Journal of Plant Biology (2024)
-
A novel 7-base pair deletion at a splice site in MS-2 impairs male fertility via premature tapetum degradation in common bean (Phaseolis vulgaris L.)
Theoretical and Applied Genetics (2023)
-
Genome resequencing reveals the population structure and genetic diversity of almond in Xinjiang, China
Genetic Resources and Crop Evolution (2023)
-
De novo transcriptome assembly of Conium maculatum L. to identify candidate genes for coniine biosynthesis
Scientific Reports (2022)