Abstract
Numerous environmental and endogenous signals control the highly orchestrated and intricate process of plant senescence. Ethylene, a well-known inducer of senescence, has long been considered a key endogenous regulator of leaf and flower senescence, but the molecular mechanism of ethylene-induced ovule senescence has not yet been elucidated. In this study, we found that blockage of fertilization caused ovule abortion in the pear cultivar ‘1913’. According to transcriptome and phytohormone content data, ethylene biosynthesis was activated by pollination. At the same time, ethylene overaccumulated in ovules, where cells were sensitive to ethylene signals in the absence of fertilization. We identified a transcription factor in the ethylene signal response, ethylene-insensitive 3-like (EIL1), as a likely participant in ovule senescence. Overexpression of PbEIL1 in tomato caused precocious onset of ovule senescence. We further found that EIL1 could directly bind to the promoter of the SENESCENCE-ASSOCIATED CYSTEINE PROTEINASE 1 (PbCysp1) gene and act upstream of senescence. Yeast one-hybrid and dual-luciferase assays revealed the interaction of the transcription factor and the promoter DNA sequence and demonstrated that PbEIL1 enhanced the action of PbCysp1. Collectively, our results provide new insights into how ethylene promotes the progression of unfertilized ovule senescence.
Similar content being viewed by others
Introduction
Seedlessness, a very desirable trait that improves ease of consumption, has been bred into many fruit species, including grape1, litchi2, citrus3, and watermelon4. Previous studies have reported that phytohormones (e.g., gibberellins [GAs], auxin, and cytokinins), fertilization failure, female and male sterility, embryo abortion, and various other factors can induce the formation of seedlessness4,5,6,7,8,9.
In angiosperms, pollination and fertilization are necessary for ovule and fruit development10. After pollination, the pollen tube enters the embryo sac and releases two sperm cells. One sperm cell fuses with the egg cell to form a diploid embryo, while the other sperm cell fuses with the two polar nuclei to form a triploid endosperm. A normal seed is composed of three genetically distinct tissues: the embryo, endosperm, and seed coat11,12. The seed coat, including the inner and outer integuments, is necessary for ovule development. Multiple pathways are involved in ovule and fruit development after pollination, and a number of independent and possibly redundant hormone pathways regulate development at the early process13. In orchids, ovary and gametophyte development are coordinately regulated by auxin and ethylene after pollination. In the presence of auxin, ethylene regulates the initiation of ovary development and indirectly promotes subsequent ovule differentiation14. In Arabidopsis thaliana, GA regulates integument development by interfering with the transcription factor ABERRANT TESTA SHAPE15. In tomato, auxin and GA rapidly accumulate after pollination and act as positive regulatory signals during early fruit development16.
As a key hormone, ethylene regulates many aspects of the plant life cycle, such as fertilization, seed germination, flower development, sex determination, senescence, fruit ripening, and responses to biotic and abiotic stresses17. In early maize development, ethylene mediates programmed cell death (PCD) during endosperm development and regulates carpel senescence18,19. Ethylene also regulates carpel senescence in Pisum sativum19. Furthermore, ethylene controls both petal and unfertilized whole-pistil senescence20. In the ethylene biosynthesis and signal transduction pathways, methionine is first converted to S-AdoMet by S-AdoMet synthetase. S-AdoMet is then transformed into 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS). Under the catalytic action of ACC oxidase (ACO), ACC is finally converted to ethylene. The formation of ACC is the rate-limiting step in this pathway17. In ethylene signal transduction, ethylene-insensitive 3 (Ein3) and Ein3-like 1 (EIL1) control most ethylene responses and regulate the vast majority of downstream target genes21,22,23,24. Previous studies have reported that ethylene plays a critical role in preventing the attraction of a second pollen tube and that activation of the Ein3-dependent ethylene response pathway is necessary for pollen tube attraction25. Furthermore, overaccumulation of Ein3 in the micropyle blocks pollen tube attraction26. Ein3 also regulates ovule senescence to modulate GA-mediated fruit set in Arabidopsis27. Although these findings highlight the role of ethylene in early ovule development and the involvement of this hormone in ovule senescence, little is known about the mechanism by which ethylene controls ovule senescence.
In plants, PCD occurs during: sex determination; anther development; tracheary element differentiation; degeneration of the nucellus, endosperm, and endothelium; leaf, carpel, and petal senescence; and early plant senescence18,19,27,28,29,30,31,32,33. In plants, most enzymes in the large cysteine protease (Cysp) family belong to the papain-like, metacaspase, and legumain subfamilies34. Cysp enzymes are ubiquitously involved in cell degeneration and senescence in plants and are induced during processes such as the differentiation of tracheary elements35. In Arabidopsis, senescence-associated gene 12 (SAG12), SAG2, XYLEM CYSTEINE PROTEASE 1 (XCP1), and XCP2 are closely connected to cell senescence36,37,38. AtSAG12 is a senescence marker gene induced during leaf senescence36,39. In Brassica napus, BnCysp1 is an ovule integument-specific cysteine proteinase associated with PCD of the inner integument33. Despite these findings, a relationship between ethylene and Cysp genes has not been reported.
In this study, we aimed to characterize the involvement of ethylene in the initiation and progression of ovule senescence in unfertilized pear. To achieve this goal, we analyzed the expression patterns and functions of ethylene-related genes as well as senescence-related target genes acting downstream of the ethylene signal. Our analysis revealed that the PbEIL1-dependent regulation of PbCysp1 controls the senescence of ovules experiencing fertilization blockage.
Results
Fertilization blockage leads to ovule abortion in the seedless pear cultivar ‘1913’
To verify the cause of seedlessness in ‘1913’ pear, we carried out field experiments, including artificial emasculation followed by bagging (nonpollination), self-pollination with bagging, artificial pollination of inactivated pollen followed by bagging, and cross-pollination. According to our results, fruit development and set occurred 12 days after pollination (DAP) in pear subjected to cross-pollination, whereas the fruit of the pear plants subjected to the other treatments fell off (Fig. 1a). Cross-pollination is thus necessary for fruit set in ‘1913’ pear. We next monitored the development of early ovules subjected to cross-pollination treatment. We observed that the ovules began to turn brown at 7 DAP and were completely brown at 12 DAP (Fig. 1b). In addition, the seeds in mature fruits were shriveled (Fig. 1c). When we stained ovules with Alcian blue and nuclear fast red after pollination, we found that most integument cells of ‘1913’ were stained deeply blue at 7 DAP, and a notable amount of intercellular space was observed in the integument area (Supplementary Fig. S1). In contrast, ovule cells of the seeded variety ‘Dangshansu’, used as a control, remained intact and were barely stained blue (Supplementary Fig. S1). These results demonstrate that integument cells of ‘1913’ begin to die and degenerate at 7 DAP.
a Developing ‘1913’ fruitlets subjected to different treatments and then observed at 12 days after treatment. UP, unpollinated; SP, self-pollinated; IP, pollinated with inactivated pollen; P, cross-pollinated. b, c Development of ‘1913’ ovules and fruits at early (b) and mature (c) stages. At 7 DAP, the ovules began to abort. d Paraffin section-based observation of the fertilization process in ‘1913’ prior to ovule death. EC, egg cell; SC, sperm cell; PN, polar nucleus. e Embryo sac and f ovule development at 12 DAP. In both images, ‘Dangshansu’ and ‘1913’ are on the left and right, respectively. A zygotic embryo was formed in ‘Dangshansu’, whereas the embryo sac of ‘1913’ was empty. The different tissues were stained with safranin and fast green for histocytological observation. DAP, days after pollination
To determine the reason for ovule abortion, we collected ovules of ‘1913’ every 24 h after pollination to check for ovule abortion. We found that sperm cells entered the embryo sac at 3 DAP but did not fuse with the egg cell or polar nucleus cells until 7 DAP—when the ovule began to abort (Fig. 1b, d). At 12 DAP, an empty embryo sac was present, and the ovule cells had died (Fig. 1e, f). In ‘Dangshansu’, in contrast, a developing zygotic embryo formed, and ovule cells remained intact (Fig. 1e, f). All these results indicate that fertilization failure is the fundamental cause of ovule abortion in ‘1913’.
Participation of ethylene in the abortion of unfertilized ovules in ‘1913’ pear
To determine the mechanism of ovule abortion in ‘1913’, we collected ovules of ‘1913’ and ‘Dangshansu’ at 0, 3, and 7 DAP for transcriptome analysis. To verify the RNA-seq results, quantitative real-time PCR (qRT-PCR) was conducted on candidate genes. The results of the qRT-PCR analysis suggest that the RNA-seq data from this study are reliable (Supplementary Fig. S2). Venn diagrams were constructed to visualize the distributions of genes differentially expressed between ‘1913’ and ‘Dangshansu’ during the three ovule developmental periods (0, 3, and 7 DAP) (Supplementary Fig. S3a). Overlapping genes in these comparisons were used to analyze the molecular mechanism underlying ovule development after pollination. A Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicated that differentially expressed genes (DEGs) were significantly enriched in 20 pathways, especially those related to biosynthesis of amino acids, carbon metabolism, and plant hormone signal transduction (Supplementary Fig. S3b and Supplementary Table S2). These analyses revealed that multiple physiological mechanisms are involved in ovule development after pollination and fertilization. In subsequent analyses, we focused on significantly enriched DEGs involved in plant hormone signal transduction.
Because heatmaps revealed that pollination activated auxin, GA, and ethylene biosynthesis and signal transduction pathways (Supplementary Fig. S3c and Supplementary Table S3), we investigated whether ovule abortion was due to significant differences in hormone content between seeded and seedless varieties. We thus compared active GA, auxin, and ACC levels among ‘1913’ and two-seeded varieties, ‘Dangshansu’ and ‘Starkrimson’. We found that ACC, the precursor of ethylene and thus a marker of endogenous ethylene content, was the only hormone whose content was significantly different in ‘1913’ vs. the two-seeded varieties. This result suggests that ethylene is related to the death of unfertilized ovules. To verify this inference, we designed an experiment in which normal ovules were treated with exogenous ACC in vitro. In this experiment, most ACC-treated ovules, but few control ovules, turned dark at 4 days after treatment, thus implying that ACC may promote ovule senescence in vitro (Fig. 2b). Trypan blue staining revealed that ACC treatment resulted in premature ovule cell death. Taken together, these results indicate that ethylene participates in the degradation of abnormal ovules of ‘1913’. In our next analysis, we focused on how ethylene contributes to unfertilized ovule death.
a Levels of active gibberellins (GA1, GA3, and GA4), IAA, and ACC in the ovules of ‘1913’ and the seeded cultivars ‘Dangshansu’ and ‘Starkrimson’. b Observations of ovule development in vitro after different treatments. ACC, exogenous ACC treatment; Control, solvent treatment. c Ovule senescence was observed by trypan blue staining. Control and ACC-treated ovules are on the right and left, respectively. DAT, days after treatment
Role of PbEIL1 in unfertilized ovule senescence
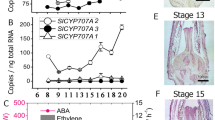
In this portion of the study, we first examined the expression patterns of ethylene synthesis-related genes after pollination in the ovules of seedless and seeded pear. We collected ovules of ‘1913’, ‘Dangshansu’, and ‘Starkrimson’ at 0, 3, and 7 DAP for analysis. We found that PbACS and PbACO family genes were significantly upregulated in ‘1913’, ‘Dangshansu’, and ‘Starkrimson’ at 7 DAP compared with 0 and 3 DAP. At 7 DAP, the two classes of genes were significantly more upregulated in seedless pear than in seeded pear (Fig. 3a and Supplementary Fig. S4). These results demonstrate that pollination activates ethylene biosynthesis and that ethylene is synthesized at higher levels in seedless pear than in seeded pear. Ein3, a transcription factor that can activate ethylene responses, may play a key role in the relationship between ethylene overaccumulation and ovule abortion. Taking into account that AtEin3 is expressed in ovules at the early stage in Arabidopsis26, we searched for the closest homolog of AtEin3 in pear and found it to be PbEIL1 (61% identity). Analysis of integrated transcriptomic data indicated that PbEIL1 was upregulated at 7 DAP in ‘1913’ (Supplementary Table S3). In light of these results, we used PbEIL1 to further ascertain the relationship between ethylene signaling and senescence.
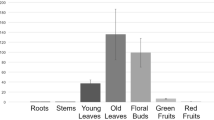
a PbACS and PbACO expression patterns at different stages in seeded pear cultivars and ‘1913’. b Tissue-specific and spatiotemporal expression of PbEIL1. c Identification of positive transgenic plants by GUS staining. The black arrows indicate the longitudinal sections of the ovules. The staining revealed the expression of PbEIL1 under the control of the PeFBP7 promoter in the ovule integument. d Seed development of 20-day-old fruits. The black arrows point to areas of significant seed senescence. e Paraffin sections of developing integument cells stained with Alcian blue and nuclear fast red. The black arrows indicate regions where the structures of transgenic and WT cells were significantly different. f Observation of seed death by trypan blue staining. The inner integuments of transgenic seeds were obviously stained (red arrows). In (a) and (b), the expression levels of each gene in ‘Starkrimson’ at 0 DAP and the expression of PbEIL1 in petals were normalized to 1.0. The data are mean ± SDs of three biological replicates. Different letters between bars indicate significant differences at P < 0.05 (Duncan’s multiple range test)
We analyzed the tissue expression specificity of PbEIL1 and found that PbEIL1 was more highly expressed in ovules than in petals, sepals, hypanthia, stamens, and stigmas (Fig. 3b). In a qRT-PCR assay, PbEIL1 was significantly upregulated at 7 DAP in ‘1913’, ‘Dangshansu’, and ‘Starkrimson’, with the highest expression observed in ‘1913’ (Fig. 3b). These results suggest that PbEIL1 is associated with early ovule development and indicate that the high level of PbEIL1 in ‘1913’ unfertilized ovules may be involved in the senescence of abnormal ovules.
To verify the relationship between PbEIL1 and unfertilized ovule abortion, we expressed PbEIL1 in tomato under the control of the ovule-specific PeFBP7 promoter. By monitoring GUS signals, we found that PbEIL1 was expressed in the ovule integument of transgenic tomato (Line 6 and Line 9) (Fig. 3c). When we observed the developing seeds at 20 DAP, we found that the transgenic seeds were yellower than wild-type seeds and had some black areas (Fig. 3d). This finding indicates that PbEIL1 accelerates seed senescence. Observation of paraffin sections and trypan blue staining confirmed the senescence of the transgenic seeds (Fig. 3e, f). The structures of inner integument cells were incomplete in transgenic seeds (Fig. 3e), and their integument was strongly stained by trypan blue as a result of senescence. These results demonstrate that PbEIL1 participates in the senescence process of unfertilized ovules in ‘1913’.
Screening of BnCysp1 homologs in pear
In angiosperms, the integument is an important structure in ovule development. In ‘1913’, ovule abortion was associated with apparent death of the integument (Fig. 1e, f). Previous studies have shown that BnCysp1 and ATSAG12 specifically regulate integument and leaf death, respectively, and that the expression levels of these genes are elevated when the corresponding tissues undergo senescence33,40. These findings implicate senescence-specific cysteine proteases in the senescence of different tissues. In light of these results, we analyzed transcriptomic data for ‘1913’ to identify senescence-specific DEGs possibly involved in ovule death. A heatmap visualizing differences in the expression levels of these genes is shown in Fig. 4a. An evolutionary analysis of homologous proteins indicated that PbCysp1 (LOC103928267) and PbSAG39-like (LOC103945424) were more closely related to BnCysp1 than to ATSAG12 (Fig. 4b). We collected different floral organs at 7 DAP and mature leaves at 153 DAP and performed a qRT-PCR assay, which revealed that PbCysp1 was highly expressed in ovules but only weakly expressed in mature leaves and other floral organs. The expression of PbSAG39-like was highest in leaves, with lower expression in stigmas, ovules, and stamens (Fig. 4c). PbCysp1 and PbSAG39-like were significantly upregulated at 3 DAP and 7 DAP compared with 0 DAP in ‘1913’, whereas no expression changes were detected in ‘Dangshansu’. In ‘Starkrimson’, however, PbCysp1 was upregulated and PbSAG39-like was downregulated at 7 DAP relative to 0 DAP and 3 DAP, respectively (Fig. 4d). These results suggest that PbCysp1 is an ovule-specific senescence-related gene, and that PbSAG39-like is primarily associated with leaf senescence. Consequently, PbCysp1 is more similar to BnCysp1 than PbSAG39-like, and the accumulation of PbCysp1 is related to ovule abortion in ‘1913’. We thus selected PbCysp1 for further study.
a Heatmap showing the differential expression of all PbCysp genes in the ‘1913’ transcriptome. b Phylogenetic tree of amino acid sequences of Cysp proteins in pear and senescence-related Cysp proteins in Arabidopsis thaliana and Brassica napus. c Tissue-specific expression of PbCysp1 and PbSAG39-like. d PbCysp1 and PbSAG39-like expression patterns at different stages in ‘1913’, ‘Dangshansu’, and ‘Starkrimson’. The expression levels of each gene in ‘Starkrimson’ at 0 DAP and those of PbCysp1 and PbSAG39-like in petals were normalized to 1.0. The data are mean ± SDs of three biological replicates. Different letters between bars indicate significant differences at P < 0.05 (Duncan’s multiple range test)
Direct binding of PbEIL1 to the promoter of PbCysp1 leads to ovule senescence
According to previous studies, Ein3 family members can bind to the DNA sequence A(C/T)G(A/T)A(C/T)CT41,42. We analyzed the PbCysp1 promoter sequence and found Ein3 binding sites. In addition, we observed that the phenotype of PbEIL1-transgenic tomato was similar to a phenotype in B. napus attributed to BnCysp. Taking all of these results into consideration, we hypothesized that PbEIL1 and PbCysp1 coregulate the death of ‘1913’ ovules. When we compared the expression patterns of PbEIL1 and PbCysp1, we found that both genes were highly expressed in ovules and significantly upregulated at 7 DAP in ‘1913’ (Fig. 3b–d). In addition, PbEIL1 and PbCysp1 were both upregulated in ACC-treated ovules (Fig. 5a). In general, PbEIL1 and PbCysp1 had similar expression patterns and were regulated by ethylene. In a yeast one-hybrid assay, PbEIL1 was able to bind to the PbCysp1 promoter (Fig. 5b). Moreover, dual-luciferase assays revealed that PbEIL1 could promote the activity of PbCysp1 (Fig. 5c). These results demonstrate that PbEIL1 is upstream of PbCysp1 and positively regulates the expression of PbCysp1. Collectively, our observations indicate that overaccumulation of PbEIL1 in ovules activates PbCysp1, resulting in ovule senescence.
a Promotion of PbEIL1 and PbCysp1 expression by ACC treatment. b Yeast one-hybrid assay results showing the interaction of PbEIL1 with the PbCysp1 promoter. c Validation of the activation effect of PbEIL1 on the PbCysp1 promoter based on a dual-luciferase assay. Left: schematic diagrams of effector and reporter constructs used in the transient dual-luciferase assay; right: image revealing that LUC relative activity was significantly increased when PbEIL1 was overexpressed. Relative promoter activity is represented by the ratio of the activity of the structural gene luciferase (LUC) to that of 35S Renilla (REN). An empty vector was used as a reference. Asterisks indicate significantly elevated LUC activity compared with that in the negative control (**P < 0.01, Student’s t-test)
Discussion
In this study, we identified and characterized a previously unknown molecular interaction between the ethylene pathway and ovule senescence, a discovery that enhances our understanding of the gene regulatory networks governing ovule abortion. In summary, we have demonstrated that PbEIL1 binds to the promoter of PbCysp1 to participate in unfertilized ovule abortion at the early stage. Previous studies have reported that auxin, GA, and ethylene increase in ovules after pollination and participate in the regulation of early ovule development17,43,44,45. In the present study, we similarly found that auxin, GA, and ethylene biosynthesis-associated genes are activated after pollination in pear ovules, with significant increases observed at 7 DAP (Fig. 3a and Supplementary Figs. S1, S4). When we compared the levels of these hormones between the seedless pear cultivar ‘1913’ and the seeded pear cultivars ‘Dangshansu’ and ‘Starkrimson’, we found that ACC was the only hormone whose content was significantly higher in seedless than in seeded pear; no consistent differences in auxin and GA levels were observed among the three varieties (Fig. 2a). This result thus suggests a prominent role for ethylene in ovule abortion in ‘1913’ and indicates that auxin and GA do not play key roles in this process. In this study, the selection of two different pear varieties as controls helped us to exclude the influences of genetic background differences. Nevertheless, our interpretations using data derived from different cultivars should be treated with some degree of caution.
In a previous study, Carbonell-Bejerano et al.45 reported that ethylene biosynthesis and response genes were activated upon unfertilized ovule senescence, a finding similar to our observations that ethylene is associated with ovule senescence. In our in vitro assay, spraying with ACC accelerated the senescence of pollinated ovules of ‘Dangshansu’ (Fig. 2b), which further verified the involvement of ethylene in ovule cell senescence. In this in vitro experiment, ovules at 30 DAP were more useful than early ovules for drawing conclusions because they were more resistant to mechanical damage inflicted by the experiment.
In the ethylene signaling pathway, Ein3 and EIL1 regulate the vast majority of downstream target genes22,23,24. Although ethylene is involved in the senescence of many different tissues in plants, most studies examining the role of ethylene in senescence have focused on leaves and petals22,23,24,46. Ethylene is known to participate in ovule senescence27, but no direct evidence exists to prove how ethylene causes ovule death. In our study, PbEIL1 was significantly increased in the ovules of ‘1913’ at 7 DAP, when the ovules began to abort (Fig. 3b). Ectopic expression of PbEIL1 in tomato ovules caused early ovule senescence, thus demonstrating that the observed overaccumulation of PbEIL1 in ovules was indeed related to ovule senescence. Unlike in ‘1913’, however, ovule senescence was not obvious in tomato at the early stage of development. We speculated that this latter result was due to the rapid development of ovules after fertilization, which may have weakened the effect of the ethylene signal, as developing ovules are insensitive to ethylene. A previous study has reported that ethylene signaling is activated at a suitable point in development. At this time, ethylene receptor levels are adjusted in tissues fated to die, and these tissues become appropriately sensitive to the generated ethylene signals47. Furthermore, ethylene does not readily cause young leaves to undergo senescence, but it can induce senescence in leaves that have reached a defined age48. These pieces of evidence indicate that ethylene is not the basic cause of ovule senescence but instead acts more like an executor. For example, ethylene plays an indispensable role in the senescence of degenerated ‘1913’ ovules, which are themselves the products of unsuccessful fertilization.
A recent study has demonstrated that overaccumulation of Ein3 in synergid cells prevents pollen tube attraction, with SAG29 activated by Ein3 playing a role in this function of synergid cells26. After fertilization, the Ein3-dependent ethylene response pathway is necessary for synergid cell PCD25. These results indicate that Ein3 participates in many early developmental processes. Because the mechanism by which Ein3 regulates ovule senescence is still unclear, we focused on the downstream genes of Ein3 in the present study. Wan et al.33 identified an integument-specific gene, Cysp1, associated with PCD in the inner integument in B. napus. In our current investigation, we confirmed that PbCysp1 is very closely related to BnCysp1 and that PbEIL1 can promote the expression of PbCysp1 (Figs. 3b, 4c, d, and 5b, c). This regulatory mechanism is a novel discovery in pear and provides a reference for future research on early ovule development. In addition, our genomic sequence analysis uncovered no differences in amino acid sequences among PbACS, PbACO, PbEIL1, and PbCysp, but we found six single-nucleotide polymorphisms (SNPs) in the PbEIL1 promoter sequence of ‘1913’ that differed from those in ‘Starkrimson’ and ‘Dangshansu’ pear (Supplementary Figs. S5 and S6). An analysis of cis-acting elements allowed us to exclude the influences of these sites on the ethylene signal response, but other potential functions of these six SNPs need to be further studied.
Taken together, our data indicate that pollination activates ethylene biosynthesis in pear ovules and that overaccumulation of ethylene in unfertilized ovules is involved in ovule abortion. Taking into account the transcriptomic data, the results of expression pattern analysis, and the results of interaction and activation assays on candidate genes, we conclude that ethylene signaling enhances the transcription of PbEIL1, after which PbEIL1 promotes the expression of PbCysp1 for participation in unfertilized ovule senescence (Fig. 6).
Materials and methods
Plant materials and growth conditions
The pear samples used in this study were collected from a pear orchard in Meixian, Shaanxi, China, in 2018 and 2019. All selected materials were bagged 2 days before flowering to prevent pollination. The following materials were used: the seedless pear cultivar ‘1913’ (Pyrus communis L.), and the seeded pear cultivars ‘Dangshansu’ (Pyrus bretschneideri Rehd.) and ‘Starkrimson’ (Pyrus communis). The pear cultivar ‘1913’ is a hybrid selected from a cross of ‘Bartlett’ (Pyrus communis L.) with a male parent of ‘Zaosu’, a hybrid of ‘Pingguoli’ (Pyrus bretschneideri), and ‘Shenbuzhi’ (Pyrus communis). The seedless characteristic of ‘1913’ has proven stable over many years of observation. The sun-exposed surfaces of ‘1913’ fruits are bright red, and the fruits need to be softened after maturity, which is a well-known characteristic of European pear fruits. All samples were randomly screened to ensure uniform size and the absence of mechanical damage. Several samples were used for observation of ovules, and some samples were fixed in FAA fixative solution to observe histocytes of ovules. The remaining samples were immediately frozen in liquid nitrogen and stored at −80 °C for total RNA extraction.
Tomato (Solanum lycopersicum L. ‘Micro-Tom’) was used as the material for the transgenic experiment. For tomato germination and growth, the seeds were first imbibed in 50 °C water for 4–5 h and then bagged in wet gauze. After germination for 48 h, the seedlings were transferred to pots (120 × 100 mm) containing a mixture of peat:vermiculite (1:1, v/v) and cultured in a growth chamber (25 °C, relative air humidity of 70–80%, 16-h/8-h day/night photoperiod and photon flux of 115 µmol/m2/s). The seedlings were irrigated daily with Hoagland’s solution, and natural light was supplemented with Osram lamps (Powerstar HQI-BT, 400 W) to provide 16 h of daylight49.
Transcriptome analysis
Ovules collected at 0 DAP, 3 DAP, and 7 DAP were used for RNA sequencing. The ovules were collected from ‘1913’ and ‘Dangshansu’ pear. Total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. RNA contamination and degradation were monitored on 0.8% agarose gels, and RNA purity was detected on a Tecan MultiskanGO full-wavelength multifunctional enzyme labeling instrument (Thermo, Waltham, MA, USA). A total of 1 µg of RNA per sample was used as input material for RNA sample preparation. Sequencing libraries were prepared by random fragmentation of the cDNA sample and used for Illumina HiSeq sequencing. After constructing a reference genome index in Bowtie v2.2.3, paired-end clean reads were aligned to the reference genome using TopHat v2.0.12. The reads mapped to each gene were counted in HTSeq v0.6.1. The RNA-sequencing data from ovules at 0 DAP were used as controls. A false discovery rate <0.001 was used as the threshold for the significance of DEGs. Genes were annotated using the ‘Dangshansu’ pear database (http://www.ncbi.nlm.nih.gov/genome/?term=pyrus) as a reference. Three independent biological replications were sequenced and analyzed.
Histological and microscopic examination
Ovules of ‘1913’ and ‘Dangshansu’ were collected, fixed in FAA for 24 h, dehydrated through an ethanol series, and embedded in paraffin wax. For safranin and fast green staining, tissues were cut into longitudinal sections (5 μm thickness), dewaxed with xylene, hydrated with an ethanol series and stained with safranin and fast green before microscopic examination (BX51 + PD72 + IX71, Olympus, Japan) and imaging. For Alcian blue staining, the tissues were prepared and sectioned in the same manner, and then the tissues were stained with Alcian blue and 1% nuclear fast red before microscopic examination and imaging50.
Phytohormone analysis
Ovules collected at 7 DAP were used as the materials for phytohormone analysis. Samples (0.1 g) were ground in liquid nitrogen and placed in 2 mL centrifuge tubes. Then, 1 mL of ethyl acetate was added, and the samples were shaken at 2000 rpm for 10 min. Next, the suspensions were centrifuged at 12,000 rpm for 10 min, and the supernatant was withdrawn. The supernatant was dried under nitrogen gas, dissolved in 200 µL of methanol and filtered through a 0.22-µm filter membrane before testing. GA1, GA3, GA4, IAA, and ACC levels were determined by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) (AB SCIEX TripleTOF 5600+, Darmstadt, IN, USA). UPLC-MS/MS was performed using an ACQUITY UPLC HSS T3 (1.8 mm, Waters, USA) column (2.1 × 100 mm). The mobile phase solvent was the same as that described by Balcke et al.51, and the injection volume was 2 µL. The mass spectrometry conditions were as follows: a spray voltage of 4500 V and air curtain, nebulizer and auxiliary gas pressures of 15, 65, and 70 psi, respectively. The atomizing temperature was 400 °C. Each sample consisted of three replicates from independent experiments.
ACC spraying assay
The fruit of ‘Dangshansu’ was collected at 30 DAP, and then the ovules were separated and precultured in a Petri dish with phosphate-buffered solution (PBS)-moistened filter paper at 16 °C in the dark for 12 h. The ovules with black surfaces were removed, and the rest of the ovules were treated with water (as controls) or 10 µM ACC52. Then, the development of seeds was observed, and the samples were stained with trypan blue to verify cell death.
Trypan blue staining
Trypan blue staining was performed as described previously, with minor modifications53. For staining, the samples were immersed in boiled lactophenol (glycerol:lactic acid:liquid phenol:distilled water, 1:1:1:1) with 0.25 mg/mL trypan blue for 20 s. Next, the samples were destained with destaining buffer (ethanol:liquid phenol:lactic acid, 2:1:1) at 65 °C for 1 h and washed three times with 75% (v/v) ethanol. Then, the samples were photographed under a microscope (MZ10F, Leica, Germany).
Quantitative real-time PCR (qRT-PCR) assay
Total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, Beijing, China). The RNA concentration and quality were assessed by UV spectrophotometry and on a 0.8% agar ethidium bromide-stained gel, respectively. Next, 1 µg of total RNA was reverse-transcribed into cDNA using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China). qRT-PCR amplifications were performed on an ABI instrument (Thermo Fisher Scientific, Massachusetts, USA) using a SYBR Premix Ex Taq Kit (Takara). The primers used for qRT-PCR are listed in Supplementary Table S1. Three biological replicates were used in the assay, and PbActin7 (LOC103926846) was used as a reference gene. For data analysis, the relative expression level of each gene was calculated using the cycle threshold (Ct) 2−ΔΔCt method54.
In this study, we collected ovules at 0 DAP, 3 DAP, and 7 DAP from ‘1913’, ‘Dangshansu’, and ‘Starkrimson’ to analyze the differential expression of candidate genes. In the tissue-specific expression assay, we collected petals, sepals, hypanthium, stamens, stigmas, and ovules at 7 DAP and leaves at 153 DAP from ‘1913’ for analysis.
Production of transgenic tomato plants
The complete coding sequence (CDS) of PbEIL1 was fused to the PeFBP7 promoter of a pBI121 binary vector. The PeFBP7 promoter sequence was cloned from petunia DNA. PeFBP7 is an ovule-specific expression gene. The primers are listed in Supplementary Table S1. The pFBP7::PbEIL1 construct was transformed into tomato using Agrobacterium tumefaciens strain EHA105. For transformation, the unfolded cotyledons were first cut off before the true leaves emerged. Then, the cotyledons were cultured for 1 day in the dark and placed on wet filter paper in Petri dishes containing solidified preculture (PC) medium (MS salts supplemented with vitamins, 3% [w/v] sucrose, 100 mg/L myo-inositol, 1 mg/L kinetin, and 0.7% [w/v] agar). Next, they were immersed in bacterial suspensions (OD600nm = 0.4) containing 200 µM acetosyringone for 10 min. After that, the bacterial suspension on the explants was removed with filter paper, and the explants were then cultured in the dark on PC medium for 2 days. Then, the explants were transferred to PC medium containing 2 mg/L zeatin, 300 mg/L cefotaxime, and 100 mg/L kanamycin. Explants that developed resistant calli produced shoots, which were excised and placed on rooting medium (MS salts, 2% [w/v] sucrose, 100 mg/L myo-inositol, 1 mg/L thiamine, 1 mg/L IAA, and 0.7% [w/v] agar). The rooted explants were cultured in pots containing vermiculite, watered with Hoagland’s solution, and conditioned in a growth chamber before being transferred to a greenhouse.
GUS staining assay
For GUS staining, 20-day-old tomato fruit was stained with 5-bromo-4-chloro-3-indolyl glucuronide at 37 °C for 12 h as described by Fillatti et al.55.
Homologous evolution analysis
The amino acid sequences of senescence-specific cysteine protease genes in pear, A. thaliana and B. napus were aligned using ClustalW56. A phylogenetic tree was constructed using MEGA 5.10 with the neighbor-joining statistical method. In addition, 1000 bootstrap replications were performed to test the phylogeny.
The genes included were as follows: PbSAG101 (Pyrus bretschneideri, LOC103928442), PbSAG101-like (Pyrus bretschneideri, LOC103928444), PbSAG101-like (Pyrus bretschneideri, LOC103939636), PbSAG101-like (Pyrus bretschneideri, LOC103939637), PbZingipain2-like (Pyrus bretschneideri, LOC103944110), PbSAG39-like (Pyrus bretschneideri, LOC103945424), PbCysp1 (Pyrus bretschneideri, LOC103928267), BnCysp1 (Brassica napus, AF448505), and ATSAG12 (Arabidopsis thaliana, AT5G45890).
Yeast one-hybrid assay
A yeast one-hybrid assay was performed following the manufacturer’s instructions for a Matchmaker Gold Yeast One-Hybrid System Kit (Clontech, Mountain View, CA, USA). The full-length CDS of PbEIL1 was amplified and inserted into the MCS of pGADT7 AD, and the approximately 500 bp promoter of PbCysp1 was inserted into pAbAi bait vectors. The pAbAi bait vectors were linearized and transformed into Y1HGold separately. Then, the colonies were selected on a plate without uracil for selective glucose synthesis. By colony PCR analysis (Matchmaker Insert Check PCR Mix 1; Clontech), we confirmed the correct integration of the plasmids into the genome of Y1HGold. Then, the bait yeast strains were cultured on SD/–Ura medium with different concentrations of aureobasidin A (AbA) to select the minimum inhibitory concentration. Next, the AD-prey vectors were transformed into the bait yeast strains and selected on an SD/-Leu/AbA plate. All transformations and screenings were performed three times. The related primers are listed in Supplementary Table S1.
Dual-luciferase assay
The promoter sequence of PbCysp1 cloned from ‘1913’ genomic DNA using PrimeSTAR Max Premix (Takara) was inserted into the dual-LUC plasmid pGreenII 0800-LUC (reporter vector). The full-length CDS of PbEIL1 was cloned into the MCS region of a pGreenII 0029 62-SK binary vector (effector vector)57. The effector and reporter vector constructs are shown in Fig. 5c.
Each recombinant plasmid was transferred into Agrobacterium strain GV3101. Agrobacterium cells containing PbEIL1-62sk and pPbCysp1-LUC were mixed at a 1:1 ratio before infiltration into 4-week-old Nicotiana benthamiana leaves. The controls received mixed injections of Agrobacterium cells with an empty 62sk vector and PbCysp1-LUC, and the control and treatment plants were injected on the left and right sides of the same leaf. Then, the injected plants were grown in darkness at room temperature for 1 day and in light for 2 days. Next, the leaves were collected in PBS for the dual-luciferase assay. The ratio of firefly luciferase enzyme activity to Renilla luciferase enzyme activity was analyzed using a dual-luciferase reporter assay system (Promega, Madison, WI, USA) on a Tecan Infinite M200 PRO full-wavelength multifunctional enzyme labeling instrument (Tecan, Hombrechtikon, Switzerland). Five independent biological replicates were analyzed. The related primers are listed in Supplementary Table S1.
Statistical analysis
The data were subjected to analysis of variance and tested for significant (*P < 0.05, **P < 0.01) treatment differences using Duncan’s test and Student’s t-test. The results are presented as mean ± standard deviation (SDs) of three replicate samples.
References
Lo’ay, A. A. & El-Boray, M. S. Improving fruit cluster quality attributes of ‘Flame Seedless’ grapes using preharvest application of ascorbic and salicylic acid. Sci. Hortic. 233, 339–348 (2018).
Chu, Y. C., Lin, T. S. & Chang, J. C. Pollen effects on fruit set, seed weight, and shriveling of ‘73-S-20’ Litchi-with special reference to artificial induction of parthenocarpy. Hortsciece 50, 369–373 (2015).
Ma, Y. W., Li, Q. L., Hu, G. B. & Qin, Y. H. Comparative transcriptional survey between self-incompatibility and self-compatibility in Citrus reticulata Blanco. Gene 609, 52–61 (2017).
Wijesinghe, S. A. E. C., Evans, L. J., Kirkland, L. & Rader, R. A global review of watermelon pollination biology and ecology: the increasing importance of seedless cultivars. Sci. Hortic. 271, 109493 (2020).
Cong, L. et al. 2,4-D-induced parthenocarpy in pear is mediated by enhancement of GA4 biosynthesis. Physiol. Plant 166, 812–820 (2019).
Zhang, S. J. et al. Comparative transcriptome analysis during early fruit development between three seedy citrus genotypes and their seedless mutants. Hortic. Res. 4, 17041 (2017).
Zeng, Y. X., Hu, C. Y., Lu, Y. G., Li, J. Q. & Liu, X. D. Abnormalities occurring during female gametophyte development result in the diversity of abnormal embryo sacs and leads to abnormal fertilization inindica/japonica hybrids in Rice. J. Integr. Plant Biol. 51, 3–12 (2009).
Cong, L. et al. CPPU may induce gibberellin-independent parthenocarpy associated with PbRR9 in ‘Dangshansu’ pear. Hortic. Res. 7, 68 (2020).
Liu, L. L. et al. Histological, hormonal and transcriptomic reveal the changes upon gibberellin-induced parthenocarpy in pear fruit. Hortic. Res. 5, 1 (2018).
Llop-Tous, I., Barry, C. S. & Donald, G. Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol. 123, 971–978 (2015).
Robert, H. S. et al. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4, 548–553 (2018).
Figueiredo, D. D. & Köhler, C. C. Auxin: a molecular trigger of seed development. Genes Dev. 32, 479–490 (2018).
Chiwocha, S. D. S. et al. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42, 35–48 (2005).
Zhang, X. S. & ONelll, S. D. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant cell 5, 403–418 (1993).
Gomez, M. D., Ventimilla, D., Sacristan, R. & Perez-Amador, M. A. gibberellins regulate ovule integument development by interfering with the transcription factor ATS. Plant Physiol. 172, 2403–2415 (2016).
Sjut, V. & Bangerth, F. Induced parthenocarpy—a way of changing the levels of endogenous hormones in tomato fruits (Lycopersicon esculentum Mill.) 1. Extractable hormones. Plant Growth Regul. 1, 243–251 (1982).
Lin, Z. F., Zhong, S. L. & Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336 (2009).
Young, T. E., Callie, D. R. & Demason, D. A. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 115, 737–751 (1997).
Orzaez, D. & Granell, A. DNA fragmentation is regulated by ethylene during carpel senescence in Pisum sativum. Plant J. 11, 137–144 (1997).
Orzaez, D., Blay, R. & Granell, A. Programme of senescence in petals and carpels of Pisum sativum L. fowers and its control by ethylene. Planta 208, 220–226 (1999).
Chang, K. N. et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2, e00675 (2013).
Qiu, K. et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11, e1005399 (2015).
Kim, H. J. et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ethylene-insensitive2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036 (2014).
Li, Z. H., Peng, J. Y., Wen, X. & Guo, H. W. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant cell 25, 3311–3328 (2013).
Volz, R., Heydlauff, J., Ripper, D., Lyncker, L. V. & Gross-Hardt, R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev. Cell 25, 310–316 (2013).
Zhang, C. et al. Ethylene signaling is critical for synergid cell functional specification and pollen tube attraction. Plant J. 96, 176–187 (2018).
Carbonell-Bejerano, P., Urbez, C., Granell, A., Carbonell, J. & Perez-Amador, M. A. Ethylene is involved in pistil fate by modulating the onset of ovule senescence and the GA-mediated fruit set in Arabidopsis. BMC Plant Biol. 11, 84 (2011).
Gunawardena, A. H. L. A. N., Greenwood, J. S. & Dengler, N. G. Programmed cell death remodels lace plant leaf shape during development. Plant cell 16, 60–73 (2004).
Hiratsuka, R., Yamada, Y. & Terasaka, O. Programmed cell death of Pinus nucellus in response to pollen tube penetration. J. Plant Res 115, 141–148 (2001).
Domínguez, F., Moreno, J. & Cejudo, F. J. The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213, 352–360 (2001).
Fath, A., Bethke, P., Lonsdale, J., Meza-Romero, R. & Jones, R. Programmed cell death in cereal aleurone. Plant Mol. Biol. 44, 255–266 (2000).
Wang, M. et al. Apoptosis in developing anthers and the role of ABA in this process during androgenesis in Hordeum vulgare L. Plant Mol. Biol. 39, 489–501 (1999).
Wan, L. L., Xia, Q., Qiu, X. & Selvaraj, G. Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 30, 1–10 (2002).
Zhang, D. D. et al. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant cell 26, 2939–2961 (2014).
Fukuda, H. Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 44, 245–253 (2000).
Otegui, M. S. et al. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41, 831–844 (2005).
Funk, V., Kositsup, B., Zhao, C. & Beers, E. P. The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128, 84–94 (2002).
Ahmed, S. U. et al. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149, 1335–1344 (2000).
Gan, S. S. & Amasino, R. M. Making sense of senescence molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 113, 313–319 (1997).
Lohman, K. N., Gan, S., John, M. C. & Amasino, R. M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant 92, 322–328 (1994).
Kosugi, S. & Ohashi, Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28, 960–967 (2000).
Yamasaki, K. et al. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. Plant Mol. Biol. 348, 253–264 (2005).
McAtee, P., Karim, S. K., Schaffer, R. & David, K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci. 4, 79 (2013).
Jones, M. L. & Woodson, W. R. Pollination-induced ehylene in carnation: role of stylar ethylene in corolla senescence. Plant Physiol. 115, 205–212 (1997).
Carbonell-Bejerano, P., Urbez, C., Carbonell, J., Granell, A. & Perez-Amador, M. A. A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol. 154, 163–172 (2010).
Ma, N. et al. Petal senescence: a hormone view. J. Exp. Bot. 69, 719–732 (2018).
Trobacher, C. P. Ethylene and programmed cell death in plants. Botany 87, 757–769 (2009).
Jing, H. C., Schippers, J. H. M., Hille, J. & Dijkwel, P. P. Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 56, 2915–2923 (2005).
Martinez-Bello, L., Moritz, T. & Lopez-Diaz, I. Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 66, 5897–5910 (2015).
Crawford, B. C. W., Ditta, G. & Yanofsky, M. F. The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 17, 1101–1108 (2007).
Balcke, G. U. et al. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8, 47 (2012).
Vanderstraeten, L., Depaepe, T., Bertrand, S. & Straeten, V. D. D. The ethylene pecursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 10, 1591 (2019).
Zhou, J. G. et al. Proteolytic processing of SERK3/BAK1 regulates plant immunity, development, and cell death. Plant Physiol. 180, 543–558 (2019).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Fillatti, J. J., Kiser, J., Rose, R. & Comai, L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730 (1987).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Acknowledgements
This research was funded by the National Key R&D Program of China (2019YFD1001400) and the China Agriculture Research System (CARS 28-45).
Author information
Authors and Affiliations
Contributions
L.X., Z.W., and H.W. designed the experiments. H.W., H.Z., F.L., L.C., L.S., and X.L. performed the experiments. H.W. analyzed the data. H.W., R.Z., C.Y., Z.W., F.M., and L.X. wrote and revised the manuscript. All authors participated in this research and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, H., Zhang, H., Liang, F. et al. PbEIL1 acts upstream of PbCysp1 to regulate ovule senescence in seedless pear. Hortic Res 8, 59 (2021). https://doi.org/10.1038/s41438-021-00491-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-021-00491-5