Abstract
Chrysanthemum white rust disease, which is caused by the fungus Puccinia horiana Henn., severely reduces the ornamental quality and yield chrysanthemum. WRKY transcription factors function in the disease-resistance response in a variety of plants; however, it is unclear whether members of this family improve resistance to white rust disease in chrysanthemum. In this study, using PCR, we isolated a WRKY15 homologous gene, CmWRKY15-1, from the resistant chrysanthemum cultivar C029. Real-time quantitative PCR (RT-qPCR) revealed that CmWRKY15-1 exhibited differential expression patterns between the immune cultivar C029 and the susceptible cultivar Jinba upon P. horiana infection. In addition, salicylic acid (SA) treatment strongly induced CmWRKY15-1 expression. Overexpression of CmWRKY15-1 in the chrysanthemum-susceptible cultivar Jinba increased tolerance to P. horiana infection. Conversely, silencing CmWRKY15-1 via RNA interference (RNAi) in C029 increased sensitivity to P. horiana infection. We also determined that P. horiana infection increased both the endogenous SA content and the expression of salicylic acid biosynthesis genes in CmWRKY15-1-overexpressing plants, whereas CmWRKY15-1 RNAi plants exhibited the opposite effects under the same conditions. Finally, the transcript levels of pathogenesis-related (PR) genes involved in the SA pathway were positively associated with CmWRKY15-1 expression levels. Our results demonstrated that CmWRKY15-1 plays an important role in the resistance of chrysanthemum to P. horiana by influencing SA signaling.
Similar content being viewed by others
Introduction
When plants are exposed to external pathogens through stomata or wounds, the first line of defense is triggered: pathogen-associated molecular pattern-triggered immunity (PTI). PTI prevents several types of pathogens from entering cells by triggering reactive oxygen species bursts and callose deposition. Once they are activated, pattern recognition receptors on the cell membrane in turn induce the activity of related kinases and the activation of downstream signal transduction pathways1,2,3. Signals in these pathways culminate in the nucleus, where they mediate a series of resistance-related reactions, such as the transcription and translation of disease-related proteins, the expression of various transcription factors, and microRNA synthesis4. The second plant line of defense is effector-triggered immunity (ETI), which inhibits the growth and spread of pathogenic bacteria via the process of programmed cell death. WRKY transcription factors play critical roles in both PTI and ETI.
Members of the WRKY transcription factor family, one of the largest transcription factor protein families in plants, are involved in biotic stress tolerance and participate in both signal transduction pathways and gene expression regulation. WRKY proteins contain two highly conserved domains: the WRKY domain, with the amino acid motif WRKYGQK, at their N-terminus and a novel C2–H2 or C2HC type zinc-finger motif at their C-terminus5,6,7. Based on their motif arrangements, WRKY proteins can be classified into four groups8. Moreover, WRKY transcription factors can specifically bind to the W-box element (TTGACC/T) in the promoters of various downstream biotic stress-related genes to regulate their transcription and enhance plant defense9. Arabidopsis WRKY genes such as WRKY18, WRKY28, WRKY52, and WRKY33 have been shown to enhance resistance to the several pathogenic bacterial species, including Pseudomonas syringae, Sclerotinia sclerotiorum, Ralstonia solanacearum, and Botrytis cinerea10,11,12,13. Similarly, the wheat (Triticum aestivum) genes TaWRKY45 and TaWRKY70 promote resistance to stripe rust and leaf rust in the cultivar Xiaoyan 614,15. WRKY transcription factors regulate a variety of signaling networks involved in plant disease-resistance-related responses, such as networks involving abscisic acid (ABA), salicylic acid (SA), jasmonic acid/ethylene (JA/ET), mitogen-activated protein kinases (MAPKs), and histone deacetylases16,17. The expression of WRKY genes during plant defense responses closely parallels that of genes involved in these signaling pathways18. In Arabidopsis (Arabidopsis thaliana), WRKY18, WRKY38, WRKY54, and WRKY66 participate in SA signal transduction19. AtWRKY18 enhances the resistance of transgenic A. thaliana to P. syringae by activating PR gene expression in the SA pathway20. WRKY transcription factors function in conjunction with PR1-1 and PR2 in the SA signaling pathway to regulate plant resistance to anthracnose; moreover, exogenous SA application reduces the disease index of banana (Musa acuminata) infected with anthracnose21. Overexpression of CsWRKY50 in cucumber (Cucumis sativus) enhances plant resistance to the fungal pathogen Psilocybe cubensis and upregulates the transcript levels of several phytohormone-related defense genes, including SA- and JA-responsive genes and SA biosynthesis genes22.
Chrysanthemum morifolium is a popular ornamental plant species worldwide with great economic and cultural value. Chrysanthemum white rust caused by the fungus Puccinia horiana severely affects chrysanthemum cultivation. Once the disease takes hold, it tends to spread extensively, which causes economic losses and hinders production. In addition to reducing chrysanthemum ornamental quality and yield, P. horiana may even lead to plant death. Although the roles of WRKY transcription factors in the resistance mechanism of plants have been extensively studied, it is unclear whether members of the WRKY family in chrysanthemum contribute to the response to chrysanthemum white rust infection. Here, we identified a gene whose expression is significantly induced, CmWRKY15-1, from transcriptomic data collected upon P. horiana infection23. We explored the role of CmWRKY15-1 in the regulation of chrysanthemum resistance to P. horiana infection and established that this gene improves resistance to white rust disease either directly or indirectly via the SA-mediated disease-resistance signaling pathway.
Results
Isolation and Sequence Analysis of CmWRKY15-1
We isolated an 810-bp full-length cDNA encoding a predicted protein of 269 amino acids from C029 (Fig. 1a) and analyzed the sequence via BLAST searches against published sequences in GenBank. The nucleotide sequence was 98% similar to that of the previously published chrysanthemum gene WRKY15; therefore, we named this gene CmWRKY15-1. The predicted CmWRKY15-1 protein contains a typical WRKY domain that contains a WRKYGQK motif distributed between amino acids 137 and 195 and a C2–H2 zinc-finger motif, both of which are hallmarks of class II WRKY transcription factors (Fig. 1b). The protein contains 46 negatively charged residues and 40 positively charged residues. The instability coefficient of CmWRKY15-1 is 49.68, while its average hydrophobicity is −0.647. These results indicated that CmWRKY15-1 is likely to be an unstable and hydrophilic protein.
a PCR amplification product of CmWRKY15-1. b Multiple amino acid sequence alignment between CmWRKY15-1 and several related WRKY proteins from the Asteraceae. The box indicates the WRKYGQK heptapeptide sequence; the triangle indicates the zinc-finger motif. c Secondary structure of the predicted CmWRKY15-1. Blue, alpha-helix; red, folding and extending chain; green, beta-turn; purple, random coil. d Three-dimensional structure of predicted CmWRKY15-1. e Phylogenetic analysis of CmWRKY15-1. M Marker, La Lupinus angustifolius; Vr Vigna radiata var. radiata; Mn Morus notabilis; Si Sesamum indicum; Vv Vitis vinifera; Ga Gossypium aridum; Bn Brassica napus; Dc Daucus carota subsp. Sativus; Cc Cynara cardunculus L.; Ha Helianthus annuus L.; Cm Chrysanthemum morifolium.
Based on predictions of protein structure and phylogenetic analysis of CmWRKY15-1, we determined the secondary structure of CmWRKY15-1 using SOPMA (http://npsa-pbil.ibcp.fr/). Of the 269 amino acids present in CmWRKY15-1, 118 were part of alpha-helices, 37 were part of folded-form extension chains, 7 were part of beta-turns, and 107 were part of random coils, accounting for 43.9, 13.75, 2.6, and 39.8% of the protein, respectively (Fig. 1c). Figure 1d depicts the three-dimensional structure of the predicted CmWRKY15-1 protein. We next compared the structure of CmWRKY15-1 to similar protein structures reported in the Phyre database to predict its tertiary structure. The CmWRKY15-1 protein was 97% similar to CmWRKY15 and 68 and 62% similar, respectively, to other WRKY proteins from two other Asteraceae species, sunflower (Helianthus annuus) and artichoke thistle (Cynara cardunculus). CmWRKY15-1 appears to be most closely related to WRKY proteins from rapeseed (Brassica napus) and carrot (Daucus carota subsp. sativus) (Fig. 1e).
Expression Profiles of CmWRKY15-1 Under Stress Treatments
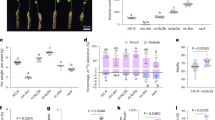
To explore the potential role of CmWRKY15-1 in chrysanthemum, we first determined the expression patterns of CmWRKY15-1 during a 72 h inoculation treatment in the resistant cultivar C029 and the susceptible cultivar Jinba by RT-qPCR. CmWRKY15-1 transcript levels were high from 24 to 48 h after inoculation (Fig. 2a). In addition, CmWRKY15-1 expression was generally much higher in the resistant cultivar C029 than in the susceptible cultivar Jinba. We also analyzed CmWRKY15-1 expression following exogenous application of phytohormones in C029 and observed an ~7.8-fold increase in CmWRKY15-1 transcript levels after 1 h of SA treatment, while JA or ET resulted in more modest increases (4.0-fold for JA and 5.3-fold for ETH), although all the tested phytohormones did increase CmWRKY15-1 transcript levels (Fig. 2b). These results suggest that, during infection with P. horiana, a temporal increase in CmWRKY15-1 expression in the leaves may be associated with modulating resistance to P. horiana and that CmWRKY15-1 expression is strongly induced by the phytohormone SA.
a CmWRKY15-1 transcript levels in chrysanthemum leaves during infection with P. horiana in C029 and Jinba. b CmWRKY15-1 expression levels after water and exogenous hormone treatments. SA, 0.1 mM salicylic acid; MeJA, 50 μM methyl jasmonate; ETH, 0.5 g L−1 ethephon (ETH) in C029. The error bars indicate the standard deviations of three replicates. The different letters mean significant differences according Duncan’s multiple range test at p < 0.05; the same scheme applies below.
Plasmid Construction and Generation of Transgenic Chrysanthemum
To investigate whether CmWRKY15-1 plays a role in controlling resistance to P. horiana infection, we generated an overexpression vector based on the pBI121 vector, which contains the selectable marker gene nptII, affording kanamycin resistance (Fig. 3a). We generated overexpression (OE) and silenced (RNA interference [RNAi]) CmWRKY15-1 chrysanthemum plants, obtaining 12 T0 clones of CmWRKY15-1-OE plants and 18 T0 clones of CmWRKY15-1-RNAi plants. We selected two representative and independent positive plants for each of the transgenic lines (OE-1, OE-2, RNAi-1, and RNAi-2) (Fig. 3b) to measure CmWRKY15-1 transcript levels via RT-qPCR. Compared with untransformed wild-type (WT) plants, the OE lines exhibited 12- and 8-fold increases in CmWRKY15-1 relative expression levels, and the RNAi plants presented reduced CmWRKY15-1 transcript levels, with downregulation ranging from 55 to 62% (Fig. 3c). We then assessed the phenotypes of these plants in terms of their growth and development but observed no obvious differences in plant height, crown diameter, or leaf number for any of the transgenic plants relative to the wild type.
a Diagram of the pBI121-CmWRKY15-1 and pRNAi-CmWRKY15-1 vectors. CaMV 35 S, promoter; NOS, nopaline synthase terminator; nptII, neomycin phosphotransferase gene; CmWRKY15-1, target fragment; LB and RB, left and right borders of the T-DNA; SacI, XbaI, BamHI, and HindIII, cloning sites; A1, antisense fragment; S1, sense fragment; PDK, intron. b PCR-based analysis of kanamycin-resistant transgenic plants. M, DNA ladder (DL 2000); OE-1 and OE-2, overexpression transgenic plants; RNA1 and RNA2, RNA interference transgenic plants. c Relative expression levels of CmWRKY15-1.
Degree of Resistance to P. horiana Infection of Chrysanthemum Transgenic Plants
We next performed a pathogen infection test on all plant genotypes (the Jinba and C029 wild types, as well as OE-1, OE-2, RNAi-1, and RNAi-2) for 14 days. We treated 30–40 leaves per line and assessed their infection phenotypes, calculated their associated disease severity index (DSI), and then determined whether the plants were resistant or susceptible to P. horiana.
Immediately after inoculation, the leaves of all three line types (WT, OE, and RNAi) were similar. We observed discontinuous teliospores in the leaves of Jinba after 14 days, as well as clear white spots and some visible spores, which is consistent with expected symptoms for a susceptible plant (S). By contrast, the OE plants showed few white spots on their leaves, and some leaves had no sporozoites, even after 14 days (Fig. 4a), making them moderately resistant (MR). In addition, we evaluated the disease resistance of the RNAi lines. As shown in Fig. 4b, compared with the control plants, the CmWRKY15-1-silenced plants exhibited a higher sensitivity to P. horiana infection after 14 days, as evidenced by the low frequency of light macula on the leaf surface and the discontinuous teliospore heaps on the abaxial side of the leaf. The degree of infection in the RNAi plants was not as severe as that in Jinba, indicating an MR-resistance type. As expected, C029 showed immunity (I) to the fungus.
The DSI of the OE lines was considerably lower than that of Jinba (Fig. 4c). By contrast, the RNAi lines had a considerably higher DSI than their corresponding C029 wild-type plants did.
At 14 days after infection, we scored the number of plants with visible symptoms and quantified the disease index for Jinba (87.8%), OE-1 (57.8%), and OE-2 (61.1%). The disease index of the RNAi-1 and RNAi-2 plants was significantly higher than that of C029, reaching 54.4 and 51.1% of the index of the blank control group, respectively. These results indicated that CmWRKY15-1 is involved in the disease resistance of chrysanthemum.
Changes in SA Levels and Expression of SA Biosynthesis Genes
To investigate whether endogenous phytohormones might participate in plant resistance to P. horiana and whether CmWRKY15-1 might be involved, we measured the levels of endogenous SA in the CmWRKY15-1 transgenic plants and corresponding wild types at 24 h after infection with P. horiana. We discovered that P. horiana infection triggered an increase in SA levels in both the OE-1 and OE-2 lines, whereas SA levels were reduced in the RNAi lines relative to those in the WT (Fig. 5a). An analysis of cis-regulatory elements within the CmWRKY15-1 promoter revealed a number of SA signal-response elements. We therefore speculated that CmWRKY15-1 might be involved in SA signaling. To test this hypothesis, we analyzed the expression level of the key SA biosynthesis genes isochorismate synthase 1 (ICS1) and phenylalanine ammonia lyase (PAL) before and after P. horiana infection. We observed no significant differences in the transcript levels of ICS1 or PAL between the noninfected transgenic plants and the WT (Fig. 5b). However, the ICS1 transcript levels increased in the OE lines 24 h after inoculation. PAL expression followed the same trend, with higher levels in both OE lines relative to those in the control group. In contrast to those in the OE lines, the transcript levels of both the ICS1 and PAL genes in the RNAi lines decreased 24 h after inoculation. Based on these results, we conclude that the changes in endogenous SA levels are closely related to CmWRKY15-1 expression.
CmWRKY15-1 Regulates Pathogenesis-Related Genes Involved in the SA Signaling Pathway
To determine the role of SA and CmWRKY15-1 in the regulation of chrysanthemum resistance to P. horiana, we tested whether CmWRKY15-1 might directly regulate the expression of defense-related genes. Here, we used RT-qPCR to quantify the relative expression levels of the pathogenesis-related (PR) genes nonexpresser of PR genes 1 (NPR1), pathogenesis-related 1 (PR1), PR2, and PR5, which are SA marker genes. PR1 expression was sharply upregulated in the OE lines after P. horiana infection compared to the expression in the infected wild-type controls (Fig. 6a). PR2 responded similarly, as did PR5, at least in the OE-1 background. The expression of NPR1 did not change significantly between the WT and OE lines. These results indicated that the resistance of transgenic chrysanthemum plants to P. horiana infection may be associated with the upregulated expression of defense-related genes. In agreement with this hypothesis, all defense-related genes showed reduced transcript levels in the RNAi lines following P. horiana infection relative to those in the wild type. Even NPR1 transcript levels were significantly lower in the RNAi lines than in the WT, although the effect was not as pronounced as that of the other PR genes (Fig. 6b).
Collectively, these results indicate that CmWRKY15-1 regulates the resistance of chrysanthemum to P. horiana infection by modulating the SA signaling pathway.
Discussion
Multiple lines of evidence suggest that WRKY transcription factors play roles in regulating pathogen infection in plants24,25,26,27. To explore the function of WRKY transcription factors in chrysanthemum, we isolated a differentially expressed gene based on transcriptomic data of chrysanthemum treated with P. horiana. This gene, CmWRKY15-1, encodes a WRKY-type transcription factor, which we speculated was related to the regulation of disease resistance, and we tested the role of this gene in the regulation of disease responses.
Plant defense systems involve a complex signal regulatory network in which plant hormones such as SA, JA, and ethylene play a crucial role28,29,30. SA enhances plant resistance to pathogen attack, regulates defense responses to various pathogens, and increases the transcript levels of pathogenesis-related genes31,32. SA is synthesized mainly through two pathways: the ICS and PAL pathways33. The exact biosynthesis pathway varies among plant species. For example, in Arabidopsis and tobacco (Nicotiana tabacum), SA is synthesized by the PAL pathway. In soybean (Glycine max), both pathways contribute to SA biosynthesis34. The ICS pathway is generally considered to be the main source of continuous SA biosynthesis, while the PAL pathway synthesizes SA rapidly only in locally necrotic cells35. In our study, the expression of the ICS and PAL SA biosynthesis genes also significantly increased in the CmWRKY15-1 OE lines after P. horiana infection (Fig. 5b), and their expression showed the opposite trend in the RNAi lines, which is consistent with the change in endogenous SA content measured across all transgenic plants. These results suggested that CmWRKY15-1 overexpression may result in stronger defense responses through increased SA accumulation.
In chrysanthemum, previous studies have reported that a number of WRKY genes are induced or repressed by pathogens. CmWRKY1, CmWRKY11, and CmWRKY15 are induced by Alternaria tenuissima inoculation36. CmWRKY1, CmWRKY6, and CmWRKY8 are also induced by Fusarium oxysporum, whereas CmWRKY4, CmWRKY8, and CmWRKY11 expression is significantly repressed by P. horiana infection. Among the 15 chrysanthemum WRKY genes (CmWRKY1 to CmWRKY15), 11 are induced by SA; 3, ABA; and 4, MeJA. These high numbers suggest that these WRKYs may be involved in disease-resistance defense pathways. Here, CmWRKY15-1 was induced by P. horiana infection in both susceptible and resistant chrysanthemum, and its expression level in resistant chrysanthemum was significantly higher than in susceptible chrysanthemum. In addition, CmWRKY15-1 was also induced by the phytohormones SA, MeJA, and ET, with SA resulting in the strongest induction. Therefore, it is reasonable to hypothesize that CmWRKY15-1 may participate in the disease resistance of chrysanthemum to P. horiana and that the disease-resistance process is mediated by phytohormone signals.
WRKY transcription factors often control, directly or indirectly, the expression of disease-resistance genes by activating signaling pathways37. To date, a variety of WRKY transcription factors have been shown to be involved in different phytohormone-dependent defense pathways. Sixteen WRKY genes are induced in response to S. sclerotiorum infection in rapeseed, some of which are involved in signaling pathways such as the SA and JA pathways38. The expression levels of PR1 and plant defensin 1.2 (PDF1.2) increased 2.5- and 3.3-fold, respectively, in plants overexpressing rapeseed WRKY33, indicating that rapeseed WRKY33 is involved in SA- and JA-mediated signaling pathways39. Mutation of Arabidopsis WRKY33 results in decreased expression of the JA signaling-related gene PDF1.2 and increased susceptibility to B. cinerea. Overexpression of Arabidopsis WRKY33 decreases the expression level of PR1 and increases plant susceptibility to P. syringae. Thus, the regulatory effects of Arabidopsis WRKY33 on defense pathways against B. cinerea and P. syringae are antagonistic40.
SA signaling is largely involved in the response to vegetative pathogens of living organisms, such as Oidium neolycopersici and Hyaloperonospora parasitica41, whereas JA/ET signals are targeted toward necrotizing pathogens42. We measured expression changes in the JA-ET pathway-related gene PDF1.2 when WT and transgenic plants were inoculated with P. horiana but observed no differences in either genotype. Simultaneously, we used RT-qPCR to analyze the expression of the PR genes NPR1, PR1, PR2, and PR5 and detected significant changes in their expression in the transgenic plants but not in the wild type. The CmWRKY15-1-overexpressing plants showed enhanced resistance to P. horiana infection, likely due to the upregulation of PR genes. Conversely, silencing CmWRKY15-1 resulted in significantly decreased NPR1, PR1, PR2, and PR5 transcript levels. However, NPR1 expression was not significantly altered in the overexpression lines. This discrepancy may be due to a signaling interaction between NPR1 and WRKY transcription factors. Several studies have shown that NPR1 activates the expression of downstream WRKY genes by interacting with a TGA-type transcription factor at their promoters, thereby promoting the expression of downstream disease-resistance genes43,44,45. The promoter region of NPR1 does contain a W-box, which indicates that NPR1 itself may be regulated by one or more WRKY transcription factors46. These results further suggest that CmWRKY15-1 acts as a positive regulator of chrysanthemum resistance to P. horiana via the SA signaling pathway. Our future research will focus on how CmWRKY15-1 transcription factors interact with PR genes. Our results provide a solid theoretical basis for breeding chrysanthemum varieties that are resistant to chrysanthemum white rust and serve as a reference point for discovering functional genes involved in the SA signaling pathway.
Materials and methods
Plant materials and growth conditions
All experiments were conducted at the Forestry College of Shenyang Agricultural University, Shenyang, China, from 2018 to 2020. The chrysanthemum resistant cultivar C029 and susceptible cultivar Jinba were provided by the flower base of the Forestry College of Shenyang Agricultural University.
Seedlings at the 6- to 8-leaf stage were grown in a potting soil mixture and placed in a greenhouse under fluorescent lights for 2 weeks at 25 ± 3 °C.
Pathogen culture
We collected teliospores of P. horiana from the abaxial side of chrysanthemum leaves infected with white rust and placed the teliospores in 1 mL of sterile water. We adjusted the concentration of the teliospores to 40–60 per field of vision under BA400 microscope (Motic, Xiamen). We removed a drop of teliospore suspension with a sterile straw and then dropped the suspension onto a glass substrate that was placed on a U-shaped rod in a culture dish covered with wet filter paper. The teliospores were allowed to germinate under a temperature of 18–21 °C for 24 h, after which we resuspended the germinated spore suspension in sterile water with 0.05% (w/v) Tween 20 pH (4–6.5). We then sprayed the abaxial side of the plant leaves evenly with the solution before covering the plants with plastic film and moving them into the dark and high humidity. After 16–24 h, we transferred the infected plants to new growth conditions of 17 °C and 50% humidity. The fungi started to release basidiospores, which germinated within 3 h Basidiospores invade leaf surfaces47,48,49,50,51.
RNA isolation and cDNA synthesis
We extracted the total RNA from the leaves of C029 using an RNA prep Pure Plant Kit (Tiangen, Beijing). We subsequently synthesized first-strand cDNAs using the Prime Script II 1st Strand cDNA Synthesis Kit following the manufacturer’s protocol (Takara, Japan).
Isolation and sequencing analysis of CmWRKY15-1
We amplified the coding sequence of CmWRKY15-1 using CmWRKY15-1 forward (F) and reverse (R) primers. The PCR program was as follows: denaturation at 94 °C for 5 min; 35 cycles of 30 s at 94 °C, 30 s at 57 °C, and 2 min at 72 °C; and a final extension at 72 °C for 10 min. We purified the PCR product and subcloned it using a p-TOPO Zero Background Kit (Aidlab, Beijing). We transformed the ligation reaction into Escherichia coli DH5α (Aidlab) and identified the clones harboring the inserts for subsequent sequencing.
Bioinformatic analysis
We analyzed the DNA and deduced the protein sequences from CmWRKY15-1 with DNAMAN software. We carried out a phylogenetic analysis of CmWRKY15-1 and related WRKY proteins via MEGA 5.0. We used the online tool ExPASy (http://expasy.org) to predict the physicochemical properties of CmWRKY15-1 and the SOPMA tool (http://npsa-pbil.ibcp.fr/) and Phyre2 database to analyze the predicted protein structure.
Analysis of CmWRKY15-1 expression under stress treatments
Four-week-old seedlings were used to determine the expression patterns of CmWRKY15-1 under different stress treatments. We sampled the leaves of cultivars C029 and Jinba at 0, 12, 24, 36, 48, and 72 h after treatment with P. horiana and at 0, 1, 6, 12, 24, and 48 h after treatment with 0.1 mM SA, 50 µM MeJA, 0.5 g L−1 ETH, and water. All the samples were stored at −80 °C, and each treatment was replicated three times. We quantified the relative expression levels of CmWRKY15-1 via quantitative real-time PCR (RT-qPCR), with CmActin used as the internal control (with the primer pair CmActin-F/R). We performed real-time qPCR according to the instructions provided with SYBR® Premix Ex Taq II. The PCR program was as follows: predenaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s; and a melt cycle of 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. All the reactions were carried out three times for three independent biological replicates. We calculated the relative transcript levels of the target genes using the 2−ΔΔCT method52. We set the expression level of CmWRKY15-1 in untreated leaves at 0–1 h for normalization across all the treatments.
Construction of the transformation vector and genetic transformation
To construct the overexpression vector, we amplified the CmWRKY15-1 full-length cDNA sequence via PCR after the addition of the enzymatic sites for XbaI and SacI using gene-specific primers (Supplemental Table 1). We purified the PCR product for ligation into a pTOPO vector to generate a p-TOPO-CmWRKY15-1 construct, which was confirmed by sequencing, and digested it with XbaI and SacI to release the PCR product for subcloning into a pBI121 vector containing the cauliflower mosaic virus (CaMV) 35S promoter53. To generate an RNAi vector, pHANNIBAL was used as an intermediate vector. By using PCR, we amplified a 200-bp sense and a 200-bp antisense fragment from CmWRKY15-1 containing the XhoI/KpnI and ClaI/HindIII restriction sites, respectively. The two fragments were digested with enzymes and then inserted into both sides of a PDK intron to yield a RNA hairpin construct. We then connected the hairpin RNA construct of pHANNIBAL to pBI121 to generate a CmWRKY15-1 gene silencing vector. We subsequently introduced the pBI121-CmWRKY15-1 overexpression construct and RNAi pRNAi-CmWRKY15-1 construct into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 and then separately transformed Jinba and C029 with pRNAi-CmWRKY15-154.
Confirmation of transgenic chrysanthemum
Putative transgenic plantlets of Jinba and C029 were rooted in Murashige and Skoog solid media supplemented with 0.2 mg L−1 NAA and 20 mg L−1 kanamycin. The positive plants were screened via PCR and RT-qPCR using the primers NptII-F/R and qRT-CmWRKY15-1-F/R (Supplemental Table 1).
Phenotypic characterization of transgenic chrysanthemum
To determine the extent of disease resistance, we inoculated plants of the OE lines and RNAi lines and WT plants with 5–10 mL of a teliospore and basidiospore suspension. After 20 days, infection was observed and recorded for 30–40 leaves per line; the resistant types were determined and classified for each leaf according to the methods of Zhu55. We counted the number of blades per disease grade and then calculated the disease severity index (DSI) according to the following formula, based on the methods of Wang56: DSI = ∑(disease grade × number of blades) × 100% / highest disease level × total number of blades.
Determination of endogenous salicylic acid levels
We sampled leaves from the WT, OE, and RNAi lines at 0 h and 24 h after pathogen infection. Plant extracts were prepared as described previously57. SA levels were then determined using a Plant Hormone Salicylic Acid ELISA Kit (ProNetsBio, Wuhan) according to the manufacturer’s instructions.
Expression analysis of genes of CmWRKY15-1 transgenic plants
We sampled the leaves of the OE lines, RNAi lines, and the wild type at 0 and 24 h after P. horiana inoculation. We quantified the relative expression of all the genes by RT-qPCR. The full list of primers used is provided in Supplementary Table S1.
Statistical analysis
Three biological replicates were evaluated, with three technical replicates per biological replicate. All the data were analyzed using ANOVA and t-tests to determine significant differences with SPSS 24.0 software.
References
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Katagiri, F. & Tsuda, K. Understanding the plant immune system. Mol. Plant Microbe Interact. 23, 1531–1536 (2010).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
Cheng, M. N. et al. The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus polyrhizus). Hortic. Res. 4, 17039 (2017).
Eulgem, T., Rushton, P. J., Robatzek, S. & Somssich, I. E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206 (2000).
Rushton, P. J., Somssich, I. E., Ringler, P. & Shen, Q. J. WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010).
Ishihama, N. & Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437 (2012).
Yu, D., Chen, C. & Chen, Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540 (2001).
Birkenbihl, R. P., Diezel, C. & Somssich, I. E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285 (2012).
Xu, X., Chen, C., Fan, B. & Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326 (2006).
Deslandes, L. et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl Acad. Sci. USA 99, 2404–2409 (2002).
Chen, L., Zhang, L., Li, D., Wang, F. & Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl Acad. Sci. USA 110, 1963–1971 (2013).
Guo, Z. F., Tao, F., Tian, W., Yang, J. R. & Hu, X. P. Functions of TaWRKY45 on the high-temperature resistance to stripe rust in Xiaoyan 6. J. Wheat Crops 37, 1318–1326 (2017).
Wang, J. et al. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 18, 649–661 (2017).
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S. & Van Wees, S. C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 (2009).
Banerjee A.Roychoudhury A., WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 1–17 (2015).
Birkenbihl, R. P., Liu, S. & Somssich, I. E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9 (2017).
Yu, Y. et al. Research progress on the role of WRKY transcription factors in plant defense. Mol. plant breed. 16, 7009–7020 (2018).
Chen, C. & Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716 (2002).
Xiao, Y. et al. Molecular characterization of PR and WRKY genes during SA- and MeJA-induced resistance against Colletotrichum musae in banana fruit. Postharvest Biol. Technol. 79, 62–68 (2013).
Luan, Q. Q. et al. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 279, 59–69 (2019).
Dong, L. et al. Transcriptome analysis of chrysanthemum in responses to white rust. Sci. Hortic. 233, 421–430 (2018).
Li, P. et al. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 34, 1365–1378 (2015).
Wang, M. et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1, 14016 (2014).
Song, Y., Chen, L. G., Zhang, L. P. & Yu, D. Q. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35, 459–471 (2010).
Rushton, D. L. et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11 (2012).
Peng, X. et al. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498 (2012).
Spoel, S. H. & Dong, X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351 (2008).
Robert-Seilaniantz, A., Navarro, L., Bari, R. & Jones, J. D. Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379 (2007).
Herrera-Vasquez, A., Salinas, P. & Holuigue, L. Corrigendum: Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 8, 964 (2017).
Yang, B., Jiang, Y., Rahman, M. H., Deyholos, M. K. & Kav, N. N. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 9, 68 (2009).
Vlot, A. C., Dempsey, D. A. & Klessig, D. F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 (2009).
Su, H. et al. Endogenous salicylic acid shows different correlation with baicalin and baicalein in the medicinal plant Scutellaria baicalensis Georgi subjected to stress and exogenous salicylic acid. PLoS One 13, e0192114 (2018).
Zhang, Y. & Li, X. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36 (2019).
Song, A. et al. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 15, 1442–1455 (2014).
Li, J., Brader, G., Kariola, T. & Palva, E. T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491 (2006).
Yang, B., Jiang, Y., Rahman, M. H., Deyholos, M. K. & Kav, N. N. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 9, 68 (2009).
Wang, D., Amornsiripanitch, N. & Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, 1042–1050 (2006).
Zheng, Z., Qamar, S. A., Chen, Z. & Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 (2006).
Mammarella, N. D. et al. Apoplastic peroxidases are required for salicylic acid-mediated defense against Pseudomonas syringae. Phytochemistry 112, 110–121 (2015).
Thomma, B. P., Penninckx, I. A., Broekaert, W. F. & Cammue, B. P. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68 (2001).
Yu, Y. C., Qiao, M., Liu, Z. H. & Xiang, F. N. Diversification function of WRKY transcription factor. Chin. Bull. Life Sci. 22, 345–351 (2010).
Lippok, B. et al. Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20, 420–429 (2007).
Bi, Y. D. et al. Isolation and sequence analysis of transcription factor GmWRKY53 from soybean (glycine max). Soybean Sci. 34, 972–976 (2015).
Wang, L. & Gao, X. Q. Advances in Research on function of WRKY transcription factor genes in plant resistance. J. Plant Genet. Resour. 12, 80–85 (2010).
Whipps, J. M. A review of white rust (Puccinia horiana Henn.) disease on chrysanthemum and the potential for its biological control with Verticillium lecanii (Zimm.) Viégas. Ann. Appl. Biol. 122, 173–187 (1993).
Ohishi, K., Okumura, Y. & K, M. Incubation of Puccinia horiana P. H. Using chrysanthemum plants cultured in vitro. J. Jpn. Soc. Hort. Sci. 69, 767–769 (2000).
Vencescontreras, C. & Vázquezgarcía, L. M. Inoculation in vitro of the white rust (Puccinia horiana Hennings) in chrysanthemum (Dendranthema grandiflora Tzvelev). Agron. Mesoam. 19, 81–85 (2008).
Takatsu, Y. et al. Use of chrysanthemum plantlets grown in vitro to test cultivar susceptibility to white rust, Puccinia horiana P. hennings. Plant Breed. 119, 528–530 (2000).
Firman, I. D. & Martin, P. H. White rust of chrysanthemums. Ann. Appl. Biol. 62, 429–442 (1968).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Po-Yen, C., Chen-Kuen, W., Shaw-Ching, S. & Kin-Ying, T. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 11, 287–293 (2003).
Hong, B. et al. Expression of the Arabidopsis DREB1A gene in transgenic chrysanthemum enhances tolerance to low temperature. J. Hortic. Sci. Biotech. 81, 1002–1008 (2006).
Zhu, P. F., Zhao, N. L., Qi, D., Liu, N. & Duan, Y. X. Optimization on identification standards and artificial inoculation methods in vitro on resistance to chrysanthemum white rust. Agric. Sci. Technol. 12, 1640–1644 (2011).
Wang, X. et al. The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 7, 57 (2020).
Li, S., Fu, Q., Chen, L., Huang, W. & Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252 (2011).
Acknowledgements
This work was supported jointly by the National Natural Science Foundation of China (31972447), the National Key R&D Program Projects (2018YFD1000400) and the Natural Science Foundation of Liaoning Province (2019-ZD-0707).
Author information
Authors and Affiliations
Contributions
H.M. and M.B. designed the project and wrote the manuscript. M.B. and X.L. performed most of the experiments. X.Y. extracted the RNA. P.Z. provided the plant materials. X.L., X.Y., G.G. and D.L. analyzed the data and discussed the article. All the authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bi, M., Li, X., Yan, X. et al. Chrysanthemum WRKY15-1 promotes resistance to Puccinia horiana Henn. via the salicylic acid signaling pathway. Hortic Res 8, 6 (2021). https://doi.org/10.1038/s41438-020-00436-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00436-4
This article is cited by
-
DkWRKY transcription factors enhance persimmon resistance to Colletotrichum horii by promoting lignin accumulation through DkCAD1 promotor interaction
Stress Biology (2024)
-
Indole-3 acetic acid negatively regulates rose black spot disease resistance through antagonizing the salicylic acid signaling pathway via jasmonic acid
Planta (2024)
-
Genome-wide analysis of the CAD gene family reveals two bona fide CAD genes in oil palm
3 Biotech (2022)