Abstract
GRAS proteins are plant-specific transcription factors that play crucial roles in plant development and stress responses. However, their involvement in the ripening of economically important fruits and their transcriptional regulatory mechanisms remain largely unclear. Here, we demonstrated that SlGRAS4, encoding a transcription factor of the GRAS family, was induced by the tomato ripening process and regulated by ethylene. Overexpression of SlGRAS4 accelerated fruit ripening, increased the total carotenoid content and increased PSY1 expression in SlGRAS4-OE fruit compared to wild-type fruit. The expression levels of key ethylene biosynthesis genes (SlACS2, SlACS4, SlACO1, and SlACO3) and crucial ripening regulators (RIN and NOR) were increased in SlGRAS4-OE fruit. The negative regulator of tomato fruit ripening, SlMADS1, was repressed in OE fruit. Exogenous ethylene and 1-MCP treatment revealed that more endogenous ethylene was derived in SlGRAS4-OE fruit. More obvious phenotypes were observed in OE seedlings after ACC treatment. Yeast one-hybrid and dual-luciferase assays confirmed that SlGRAS4 can directly bind SlACO1 and SlACO3 promoters to activate their transcription, and SlGRAS4 can also directly repress SlMADS1 expression. Our study identified that SlGRAS4 acts as a new regulator of fruit ripening by regulating ethylene biosynthesis genes in a direct manner. This provides new knowledge of GRAS transcription factors involved in regulating fruit ripening.
Similar content being viewed by others
Introduction
Fruit ripening can be classified as climacteric or nonclimacteric, depending on the presence or absence, respectively, of massive ethylene production during ripening1. Tomato (Solanum lycopersicum) is a classic model of climacteric fruit ripening. Many biological changes, including color conversion, softening, and sugar/acidity alteration, occur during the fruit ripening process, and the ethylene burst is closely related to the rise in climacteric respiration. It is important to understand the fruit ripening process that determines the nutritional quality, storage life and wastage of many fresh plant products worldwide.
Ethylene is an important phytohormone for fruit ripening, and ethylene biosynthesis is strictly regulated during fruit ripening. Ethylene biosynthesis is divided into two steps. The first step is the rate-limiting step, in which the conversion of S-adenosyl-l-Met (SAM) to 1-aminocyclopropane-1-carboxylic acid (ACC) is catalyzed by ACC synthase (ACS). Then, the conversion of ACC to ethylene is catalyzed by ACC oxidase (ACO)2,3. Fourteen ACS genes and 6 ACO genes have been identified in tomato, and the expression of ACS2, ACS4, ACO1 and ACO2 was significantly induced by fruit ripening initiation, suggesting that they may act as the main genes for ethylene biosynthesis during tomato fruit ripening4. Two ethylene biosynthesis systems, system 1 and system 2, were introduced based on the level of ethylene production during fruit development5. Ethylene production exhibited a massive increase associated with fruit ripening in climacteric fruits and was considered system 2. In tomato fruit, ethylene production shifted from system 1 to system 2 at the climacteric stage, with an accumulation of transcripts of ACS2, ACS4, ACO1, and ACO4, resulting in positive feedback regulation6,7.
In the past few years, many transcription factors involved in fruit ripening have been identified. The natural mutants rin (ripening-inhibitor), nor (nonripening) and Cnr (colorless nonripening) have been widely used for studying the regulatory mechanisms of fruit ripening. RIN is a classic MADS-box transcription factor, and previous studies have suggested that rin is a loss-of-function mutant, but a recent study indicated that rin is actually a gain-of-function mutant8. Regardless of the mechanism, a large number of RIN target genes have been identified, including SlACS2 and SlACS4, as have many other genes that participate in the regulation of ethylene signaling and fruit quality formation during ripening9,10,11,12. The nor mutant exhibited a nonripening phenotype (Patent US 6,762,347 B1)13. NOR encodes an NAC transcription factor that influences many more genes than RIN during fruit ripening14. However, unlike the natural mutant, a recent study showed that the ripening progress of knock-out mutants produced by CRISPR/Cas9 was only partly affected15,16. Formation of the Cnr mutant was caused by the increased cytosine methylation level in the promoter region of the LeSPL-CNR gene, and the epigenetic change led to a severe nonripening phenotype17. However, a recent study showed that the fruit ripening of LeSPL-CNR CRISPR/Cas9 lines was only delayed18. Moreover, a large number of other transcription factors influence fruit ripening by regulating ethylene biosynthesis genes, including SlMADS1, FUL1, FUL2, TAGL1, SlMBP8, and SlMBP15 belonging to the MADS-box family19,20,21,22,23; SlNAC1 and SlNAC4 belonging to the NAC family24,25; and SlAP2a and SlERF.B3 belonging to the ERF transcription factor family26,27.
Several studies have shown that GRAS transcription factors are widely involved in regulating plant development and resisting abiotic stress28,29,30,31. However, few studies have reported that GRAS participates in regulating fruit ripening, except for a report on SlGRAS38-silenced fruit with lower lycopene content and lower ethylene production32. The expression of SlGRAS38, also named SlFSR, increased during fruit ripening, and downregulation of SlFSR altered cell wall modification and prolonged fruit shelf life33. Our previous study identified that SlGRAS4 encodes a GRAS transcription factor and plays a positive role in enhancing chilling tolerance in tomato fruit. Fruit with SlGRAS4 overexpression could ripen normally after chilling treatment, whereas WT fruit could not turn red completely31. Here, the expression of SlGRAS4 was induced by the fruit ripening process, and overexpression of SlGRAS4 accelerated fruit ripening. We also confirmed that SlGRAS4 can directly bind to and activate the promoters of the ethylene biosynthesis genes SlACO1 and SlACO3, suggesting that SlGRAS4 influences fruit ripening mainly through SlACO1 and SlACO3. Furthermore, SlGRAS4 can also directly repress SlMADS1 expression, and our study provides new insight into the GRAS transcription factors that are involved in regulating fruit ripening.
Results
SlGRAS4 expression is induced by fruit ripening and exhibits ethylene regulation
To explore the expression pattern of SlGRAS4 (Soly01g100200) during fruit development and ripening, fruit at different stages were investigated, including 8 DPA (days post anthesis) fruit, 16 DPA fruit, mature green fruit (MG), breaker fruit (Br), 3 days post breaker (Br + 3) fruit, and 7 days post breaker (Br + 7) fruit. The expression level of SlGRAS4 gradually increased from breaker to red ripe fruit, which accompanied the fruit ripening process (Fig. 1a), suggesting that SlGRAS4 may play a role during fruit ripening in tomato. To investigate whether SlGRAS4 was under ethylene regulation, wild-type (WT) MG fruit were treated with ethylene, and SlGRAS4 was significantly induced by ethylene treatment. Simultaneously, E4, an ethylene response gene, was used as a control to validate the efficacy of the treatment (Fig. 1b). On the other hand, WT Br was treated with 1-methylcyclopropene (1-MCP), and SlGRAS4 was significantly repressed after 1-MCP treatment (Fig. 1c), suggesting that SlGRAS4 was under ethylene regulation.
a Expression patterns of SlGRAS4 in 8 DPA (days post anthesis) fruit, 16 DPA fruit, mature green fruit (MG), breaker fruit (Br), 3 days post breaker (Br + 3) fruit, and Br + 7 fruit of WT tomato. b The relative expression levels of SlGRAS4 and E4 in ethylene-treated WT mature green fruit. c The relative expression levels of SlGRAS4 and E4 in 1-methylcyclopropene (1-MCP)-treated WT breaker fruit. The data represent the mean values of three independent experiments, and error bars show the ±standard error values. In (b) and (c), ** refers to significant differences between control and treatment with P < 0.01 (two-tailed Student’s t-test)
Overexpression of SlGRAS4 accelerates fruit ripening
To investigate the function of SlGRAS4 in fruit ripening, the days at anthesis and DPA were recorded for WT and SlGRAS4 overexpression plants (the efficiency of overexpression is shown in Fig. S1). The SlGRAS4-OE fruit exhibited earlier ripening than the WT fruit, and the OE fruit showed an orange color at 37 DPA, whereas the WT was still at the mature green stage; when the WT fruit reached the orange stage at 42 DPA, the OE fruit was red ripe (Fig. 2a). The calculated days from anthesis to the breaker stage showed that the ripening period of OE fruit was ~5 days earlier than that of WT fruit (Fig. 2b). The most obvious characteristic of fruit ripening is the color changes that occur with the accumulation of carotenoids. We further measured the total carotenoid content in WT and SlGRAS4-OE fruit, and low levels of carotenoids were observed in both WT and OE fruit at 35 DPA, but the carotenoid content in OE fruit was much higher than that in WT at 40 DPA (Fig. 2c). PSY1 encodes phytoene synthase 1, which plays a critical role in the synthesis of carotenoids. The expression level of SlPSY1 in OE fruit was much higher than that in WT fruit at both 35 DPA and 40 DPA (Fig. 2d), which is consistent with the high carotenoid level in OE fruit.
a Phenotype of SlGRAS4-overexpressing (OE) fruit. The color turning of OE fruit occurred earlier than that of WT fruit. DPA, days post anthesis. b Days from anthesis to the breaker stage in WT and SlGRAS4-OE fruit. c Total carotenoid content in WT and SlGRAS4-OE fruit at 35 DPA and 40 DPA. d Expression of SlPSY1 in 35 DPA and 40 DPA fruit of WT and SlGRAS4-OE lines. In (b) to (d), the data represent the mean values of three independent experiments, and error bars show the ±standard error values. ** Refers to significant differences between WT and transgenic lines with P < 0.01 (two-tailed Student’s t-test)
Ethylene biosynthesis and ripening-related genes are induced in SlGRAS4-OE fruit
More ethylene production was found in SlGRAS4-OE fruit at the breaker stage than in WT fruit (Fig. 3a). The expression of ethylene biosynthesis genes, ethylene signaling genes and crucial transcription factors in fruit ripening was analyzed in WT and OE fruit (Fig. 3b–h, S2–4). The expression levels of SlACS4 and SlACO3 in OE fruit were significantly higher than those in WT at the breaker stage, and the expression levels of SlACO1 and SlACO3 in OE fruit were higher than those in WT at the Br + 3 stage (Fig. 3b–e). The expression levels of other ethylene biosynthesis genes, including SlACS1a, SlACS6, SlACO2, SlACO4, and SlSAM1, showed no obvious changes in OE fruit compared to WT fruit at the breaker stage, except that SlACS3 was downregulated in OE fruit (Fig. S2). There were no significant differences in most ethylene signaling genes between WT and SlGRAS4-OE fruit at the breaker stage, except that SlEBF3 expression was much higher in OE fruit (Fig. S3). As important regulators of ethylene biosynthesis and fruit ripening, the transcriptional levels of RIN and NOR were also significantly induced in OE fruit at the breaker stage (Fig. 3f, g). In contrast, the expression of SlMADS1, a negative regulator of tomato fruit ripening, was significantly downregulated in OE fruit compared to WT fruit at both the breaker and Br+3 stages (Fig. 3h). In addition, SlPG2a and SlFUL1 were expressed at higher levels in OE fruit at the breaker stage (Fig. S4). These results suggested that SlGRAS4 may accelerate fruit ripening by influencing the expression of genes involved in ethylene biosynthesis and important transcription factors.
a Ethylene production in WT and SlGRAS4-OE fruit at the breaker stage. The expression levels of SlACS2 (b), SlACS4 (c), SlACO1 (d), SlACO3 (e), RIN (f), NOR (g), and SlMADS1 (h) were analyzed by qRT-PCR. Br, breaker stage; Br + 3, 3 days post breaker stage. The data represent the mean values of three independent experiments, and error bars show the ±standard error values. * and ** refer to significant differences between WT and transgenic lines with P < 0.05 and P < 0.01, respectively (two-tailed Student’s t-test)
a Phenotypes of WT and SlGRAS4 OE #18 fruit after treatment with ethylene for 48 and 72 h, respectively. The fruit were picked at the mature green stage for ethylene treatment. b Phenotypes of WT and SlGRAS4 OE #18 fruit after treatment with 1-MCP for 72 h. The fruit were picked at the breaker stage for 1-MCP treatment. c The expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 in WT and SlGRAS4-OE fruit after treatment with ethylene for 72 h. d The expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 in WT and SlGRAS4-OE fruit after treatment with 1-MCP for 72 h. In (c) and (d), the data represent the mean values of three independent experiments, and error bars show the ±standard error values. * and ** refer to significant differences between WT and transgenic lines with P < 0.05 and P < 0.01, respectively (two-tailed Student’s t-test)
SlGRAS4-OE fruit exhibit more obvious phenotypes after exogenous ethylene and 1-MCP treatment
Ethylene is crucial for tomato fruit ripening, and 1-MCP is a potent inhibitor of ethylene perception that can inhibit fruit ripening. WT and SlGRAS4-OE fruit at the mature green stage were picked and used for exogenous ethylene treatment. Color turning in OE fruit occurred noticeably earlier than that in WT fruit after treatment for 48 h, and WT and OE fruit both turned orange after treatment for 72 h, but OE fruit looked darker (Fig. 4a). On the other hand, WT and SlGRAS4-OE fruit at the breaker stage were picked and used for exogenous 1-MCP treatment. There were obvious differences between WT and OE fruit after 1-MCP treatment. The OE fruit were light orange after treatment for 72 h, whereas the WT fruit were still at the breaker stage and exhibited no obvious color turning (Fig. 4b); in addition, more ethylene was produced in OE fruit (Fig. S5), suggesting that the accelerated color turning in OE fruit after 1-MCP treatment was caused by the higher levels of endogenous ethylene. The expression of four crucial ethylene biosynthesis genes, namely, SlACS2, SlACS4, SlACO1, and SlACO3, was higher in OE fruit than in WT fruit after ethylene treatment for 72 h (Fig. 4c). Consistent with the phenotype, the expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 in OE fruit were all significantly higher than those in WT fruit after 1-MCP treatment (Fig. 4d).
SlGRAS4-OE seedlings exhibit more intense phenotypes after exogenous ACC treatment
To ascertain whether the increased level of ethylene production persisted in nonfruit tissues in SlGRAS4-overexpressing plants, an ethylene triple-response assay was performed. The ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) was used to treat WT and OE seedlings. In the absence of ACC, there were no significant differences in hypocotyl length and root length between WT and OE seedlings (Fig. 5a–c). Hypocotyl elongation and root elongation were inhibited in both WT and OE seedlings after ACC treatment, but the inhibition in OE seedlings was more severe than that in WT seedlings, and the bending of hypocotyls was more obvious in OE seedlings (Fig. 5a–c), suggesting that more ethylene may be produced in SlGRAS4-OE seedlings. Furthermore, the expression of SlACS2, SlACS4, SlACO1 and SlACO3 was detected in WT and SlGRAS4-OE seedlings with or without ACC treatment. There were no changes in SlACO1 and SlACO3 in OE seedlings in the absence of ACC, whereas SlACS2 and SlACS4 were downregulated (Fig. 5d). However, after ACC treatment, all four genes were significantly upregulated in OE seedlings compared to WT seedlings (Fig. 5e). These results provide molecular evidence for the likely increase in ethylene production in SlGRAS4-OE seedlings.
a Phenotypes of WT and SlGRAS4-OE seedlings in the absence or presence of 2.0μM 1-aminocyclopropane-1-carboxylate (ACC). Hypocotyl length (b) and root length (c) of WT and SlGRAS4-OE seedlings in the absence or presence of 2.0 μM ACC. d The expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 in WT and SlGRAS4-OE seedlings without ACC treatment. e The expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 in WT and SlGRAS4-OE seedlings with ACC treatment. In (b) to (e), the data represent the mean values of three independent experiments, and error bars show the ±standard error values. * and ** refer to significant differences between WT and transgenic lines with P < 0.05 and P < 0.01, respectively (two-tailed Student’s t-test)
SlGRAS4 directly binds to and activates the SlACO1 and SlACO3 promoters
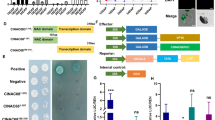
The results described above suggest that SlGRAS4 accelerates fruit ripening by influencing ethylene biosynthesis. The expression of SlACS2, SlACS4, SlACO1, and SlACO3 was enhanced in OE fruit regardless of whether they were on the vine or treated with ethylene and 1-MCP. On the other hand, several SlGRAS4-binding motifs were identified on the promoters of SlACS2, SlACS4, SlACO1, and SlACO3 by an in silico search (Table S1), suggesting that SlGRAS4 may regulate the expression of these genes in a direct manner. A yeast one-hybrid assay was performed to verify the interaction of SlGRAS4 with these promoter fragments, and the results showed that SlGRAS4 can directly bind to the SlACO1 and SlACO3 promoter fragments (Fig. 6a), whereas there was no interaction between SlGRAS4 and the SlACS2 and SlACS4 promoters. The dual-luciferase assay further confirmed that SlGRAS4 directly activates the SlACO1 and SlACO3 promoters (Fig. 6b, c). These results confirmed that SlGRAS4 influences ethylene biosynthesis by regulating SlACO1 and SlACO3 expression in a direct manner.
a SlGRAS4 binding with SlACO1 and SlACO3 promoter fragments assessed by a yeast one-hybrid assay. b Effector and reporter constructs used for the dual-luciferase assay. c SlGRAS4 activates SlACO1 and SlACO3 promoter activity as determined by a dual-luciferase assay. The empty effector was used as a control (set as 1). In (c), the data represent the mean values of five independent experiments, and error bars show the ±standard error values. ** Refers to significant differences between the SlGRAS4-effector group and empty group with P < 0.01 (two-tailed Student’s t-test)
SlGRAS4 directly binds to the SlMADS1 promoter and represses its expression
SlMADS1 negatively regulates fruit ripening in tomato, and silencing of SlMADS1 accelerates fruit ripening. SlACS2, SlACO1, and SlACO3 were upregulated in SlMADS1-silenced fruit21. SlMADS1 was significantly downregulated in SlGRAS4-OE fruit during fruit ripening (Fig. 3h). In our previous study, SlMADS1 was identified as one of the SlGRAS4 target genes through a ChIP-seq approach31. Several SlGRAS4-binding motifs on the SlMADS1 promoter were identified by an in silico search (Table S1). A yeast one-hybrid assay showed that SlGRAS4 can directly bind to the SlMADS1 promoter fragment (Fig. 7a, S6), and a dual-luciferase assay revealed that SlGRAS4 can directly repress the promoter activity of the SlMADS1 gene (Fig. 7b, c). These results indicated that the accelerated fruit ripening of SlGRAS4-OE fruit is also caused by the repression of SlMADS1 in a direct manner.
a SlGRAS4 binding with the SlMADS1 promoter fragment assessed by a yeast one-hybrid assay. b Effector and reporter constructs used for the dual-luciferase assay. c SlGRAS4 represses SlMADS1 promoter activity as determined by a dual-luciferase assay. The empty effector was used as a control (set as 1). In (c), the data represent the mean values of five independent experiments, and error bars show the ±standard error values. ** Refers to significant differences between the SlGRAS4-effector group and empty group with P < 0.01 (two-tailed Student’s t-test)
Discussion
Previous studies have revealed that tomato GRAS transcription factors are involved in regulating plant growth and development and participate in regulating abiotic stress resistance29,30,31,34,35,36. Silencing of SlGRAS38 significantly lowered the lycopene content and ethylene production in tomato fruit, and several ripening-related genes were influenced by SlGRAS3832. In the SlFSR-RNAi fruit, the expression of several cell wall modification-related genes was decreased, and the related enzyme activities were decreased, which prolonged the fruit shelf life, and overexpression of SlFSR in rin resulted in upregulation of multiple cell wall modification-related genes and shortened fruit shelf life33. Moreover, there is no other study on GRAS regulating fruit ripening. Here, we identified that SlGRAS4 was induced by the fruit ripening process (Fig. 1), and overexpression of SlGRAS4 accelerated fruit ripening (Fig. 2a). The transgenic fruit ripened ~5 days earlier than the wild type (Fig. 2b), suggesting that SlGRAS4 acts as a novel regulator of tomato fruit ripening. In this study, a much higher carotenoid content was observed in SlGRAS4-OE fruit than in WT fruit at 40 DPA (Fig. 2c), which is consistent with the earlier ripening phenotype. Accordingly, we found that the expression level of SlPSY1 in SlGRAS4-OE fruit was much higher than that in WT (Fig. 2d), which was also consistent with the increased carotenoid content phenotype in OE fruit.
Early studies have demonstrated the importance of ethylene biosynthesis genes during fruit ripening; the antisense activity of tomato ACS2 or downregulation of ACO1 in tomato plants causes reduced ethylene biosynthesis, and fruit ripening is thus retarded37,38,39. Similar to the result for SlMADS1, suppression of SlMBP8 accelerated fruit ripening, and more ethylene production and enhanced SlACS2, SlACO1, and SlACO3 expression were found in SlMBP8-silenced fruit22. On the other hand, delayed ripening was also observed in SlCMB1-downregulated tomato fruit, SlACS2, SlACS4, SlACO1, and SlACO3 were repressed, and ethylene production and carotenoid content were all reduced40. Downregulation of SlNAC48 and SlNAC19 inhibits fruit ripening, and SlACS2, SlACS4, and SlACO1 are seriously repressed41. These studies indicated that the effect of ripening regulators from different transcription factor families on fruit ripening may occur through the regulation of ethylene biosynthesis genes. Similarly, in our study, the expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 were all enhanced in SlGRAS4-OE fruit, and no changes in other ethylene biosynthesis genes were observed in OE fruit (Fig. 3, S2), suggesting that the earlier ripening of SlGRAS4-OE fruit may be caused by the higher level of these four crucial ethylene biosynthesis genes. In addition, the color turning of SlGRAS4-OE fruit after ethylene treatment occurred earlier than that of WT fruit, which was consistent with the increased expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 (Fig. 4). Furthermore, the ripening of WT fruit was severely inhibited by 1-MCP, whereas SlGRAS4-OE fruit ripened naturally after 1-MCP treatment, more endogenous ethylene was detected in SlGRAS4-OE fruit after 1-MCP treatment (Fig. S5), and the expression levels of SlACS2, SlACS4, SlACO1, and SlACO3 were significantly higher than those in WT fruit after 1-MCP treatment (Fig. 4). These results indicated that overexpression of SlGRAS4 accelerates fruit ripening by enhancing the expression of SlACS2, SlACS4, SlACO1, and SlACO3. In addition, SlGRAS4-OE seedlings exhibited more intense phenotypes after exogenous ACC treatment, and the expression of SlACS2, SlACS4, SlACO1, and SlACO3 in OE seedlings was much higher than that in WT seedlings after ACC treatment (Fig. 5). These results will help us determine whether there is an interaction between SlGRAS4 and these ethylene biosynthesis genes.
Recently, many transcription factors involved in the regulation of fruit ripening through the regulation of ethylene biosynthesis genes in a direct manner have been reported. For example, MaERF11 can suppress the expression of MaACS1 and MaACO1 by directly binding to their promoters42. The banana C2H2 zinc-finger protein 1/2 can bind to the MaACO1 promoter and repress its expression43. In kiwifruit, AdEIL2 and AdEIL3 can activate AdACO1 expression to affect ripening44. In tomato, RIN can interact with the promoters of SlACS2 and SlACS49,10,11,12. FUL1 and FUL2 can bind to the SlACS2, SlACS4, and RIN promoters20. TAGL1 can directly bind to the promoter region of SlACS2, thus regulating ethylene biosynthesis19,45. LeHB-1 can directly regulate SlACO1, thus influencing fruit ripening46. Moreover, NOR-like1 influences ethylene biosynthesis in tomato fruit by regulating SlACS2 and SlACS4 in a direct manner47. However, no new direct regulator of the ACS and ACO genes has been identified in tomato. Our study showed that SlGRAS4 directly binds to and activates SlACO1 and SlACO3 promoters by yeast one-hybrid and dual-luciferase assays (Fig. 6), identifying a novel regulator of fruit ripening that directly modulates the expression of ethylene biosynthesis genes.
On the other hand, MADS-box transcription factors are key regulators of tomato fruit ripening and are usually formed as complexes by protein-protein interactions. Several MADS-box transcription factors, including SlMADS1, SlCMB1, and SlMBP8, can interact with RIN21,22,40. SlNAC4 can also interact with RIN24. However, there was no protein-protein interaction between SlGRAS4 and RIN, and considering that SlGRAS4 cannot interact with the SlACS2 and SlACS4 promoters, the increased expression of SlACS2 and SlACS4 in SlGRAS4-OE fruit may be caused by the enhanced expression level of RIN (Fig. 3f). Furthermore, there are three RIN-binding motifs in the SlGRAS4 promoter (Table S2), but a yeast one-hybrid assay confirmed that there was no interaction between RIN and the SlGRAS4 promoter region, suggesting that there was no direct regulatory relationship between RIN and SlGRAS4. In addition, the downregulated expression of SlMADS1 that was directly repressed by SlGRAS4 (Figs. 3h, 7) also contributed to the enhanced ACS and ACO gene expression in OE fruit.
Notably, there were no obvious changes in the fruit ripening process of SlGRAS4-RNAi fruit compared to that of WT fruit. We obtained three effective SlGRAS4-repressing lines (Fig. S7), and four SlGRAS4 homologous genes had no influence on SlGRAS4-RNAi fruit (Fig. S8). There were no differences in the days from anthesis to breaker stage between WT and SlGRAS4-RNAi plants (Fig. S9a). The SlGRAS4 target genes SlACO1 and SlACO3 were both repressed in RNAi fruit at the Br+3 stage, and the expression of SlACS2 and SlACO3 in RNAi fruit was significantly lower than that in WT fruit at the breaker stage. RIN was also downregulated in RNAi fruit at the Br + 3 stage, and the expression of SlMADS1 in RNAi fruit showed an increasing trend (Fig. S9). The expression level of SlACS4 was significantly enhanced in SlGRAS4-RNAi fruit at the Br + 3 stage (Fig. S9), and the expression of other ethylene biosynthesis genes, including SlACS1a, SlACS3, SlACS6, SlACO2, and SlACO4, was significantly increased in RNAi fruit at the breaker stage (Fig. S10). Similar to OE fruit, the expression levels of ethylene signaling genes also showed no changes in RNAi fruit compared to WT fruit (Fig. S11), except that SlEBF3 was upregulated, but this gene was also upregulated in OE fruit. The expression levels of ripening-related transcription factors, including SlPG2a, SlFUL1, SlFUL2, SlTAGL1, SlHB1, and CNR, also showed no changes in SlGRAS4-RNAi fruit compared to WT fruit (Fig. S12). These results for SlGRAS4-RNAi fruit indicated that although downregulation of SlGRAS4 resulted in the repression of SlACO1 and SlACO3, the expression levels of other ethylene biosynthesis genes were significantly upregulated, leading to normal ripening of SlGRAS4-RNAi fruit, which may be caused by other fruit ripening regulators, as discussed above, that function in a complementary manner.
In our previous study, SlGRAS4 was identified as a positive regulator of fruit chilling tolerance. After chilling treatment, WT fruit could not turn red completely, whereas SlGRAS4-OE fruit ripened normally31. In addition to the metabolic pathways related to chilling tolerance regulated by SlGRAS4, as presented in our previous study, we hold the opinion that ethylene biosynthesis regulated by SlGRAS4 also contributes to the normal ripening of SlGRAS4-OE fruit after chilling treatment. Whether SlGRAS4 participates in the convergence of the fruit ripening process and cold response needs to be further studied, as they are both very complex regulatory networks. Overall, our work revealed a novel regulator (SlGRAS4) of fruit ripening and provided new insight into the complex network associated with fruit ripening.
Materials and methods
Plant materials and growth conditions
Tomato plants (Solanum lycopersicum cv. Micro-Tom) and transgenic lines in this background (three OE lines OE #12, OE #18, and OE #27 and three RNAi lines RNAi #10, RNAi #15 and RNAi #16 were used as in our previous study31) were grown in a greenhouse under controlled conditions (18-h light/6-h dark cycles, 25 °C day/18 °C night, and 60% relative humidity). For tissue expression analysis, 8 DPA fruit, 16 DPA fruit, MG, Br, 3 days post breaker (Br + 3) fruit, and 7 days post breaker (Br + 7) fruit were collected from six plants. Each tissue was sampled three independent times.
Fruit ripening time and carotenoid determination
The ripening period was indicated as DPA, twelve plants of each line were included, and three independent observations were performed. The total carotenoid content was detected according to the method described previously with minor modification48. In brief, tomato fruit powder was extracted with acetone/hexane solution (2:3 by volume), and the supernatant was used for absorbance measurements at 663, 645, and 470 nm. The total carotenoid content was calculated by the following equation: total carotenoid (μg/g) = [1000 × A470 − 3.27 × (12.72 × A663 − 2.59 × A645) − 104 × (22.88 × A645 − 4.67 × A663)] × 10/229.
Ethylene production measurement
The measurement of fruit ethylene production was performed according to a previous study49. In brief, fruit was placed in 50-mL airtight containers for 16 h, and then 1 mL of gas was injected into a gas chromatograph (Varian CP-3800 GC gas chromatograph, USA). Ethylene production was normalized to fruit weight, and ethylene standard gas was used as a control. The measurement was carried out with at least 10 fruit for each line, and three independent biological replicates were performed.
Ethylene and 1-MCP treatment of tomato fruit
For SlGRAS4 ethylene response testing, WT MG fruit was dipped in 10 ppm ethephon solution for 6 h, and WT Br was harvested and treated with 1-MCP (1 mg/L) for 1 h. For phenotypic observation, WT and transgenic MG fruit were dipped in 10 ppm ethephon solution for 3 h once a day for three days. Fruit at the breaker stage were harvested and treated with 1-MCP (1 mg/L) for three days. The fruit was placed in an incubator under a 18-h light (25 °C)/6-h dark (18 °C) cycle. The differences in color were observed, and the fruit were frozen in liquid nitrogen and stored at −80 °C after 72 h of treatment.
Triple response assay
WT and transgenic seeds were sterilized and sown on ½ × MS alone and ½ × MS containing 2.0 μM ACC and then incubated in the dark for 7 days. The hypocotyl length and root length were assessed for the seedling triple response. For each line, at least 20 seedlings were measured, and three independent treatments were performed.
Expression analyses by qRT-PCR
Total RNA extraction, first-strand cDNA synthesis and quantitative real-time PCR were performed using commercial kits following the manufacturer’s instructions (TAKARA, Japan). The PCR procedure was performed using a Bio-Rad CFX system (Bio-Rad, USA). The 2−ΔΔCt method was used for calculation of the relative fold change by Bio-Rad CFX Manager 3.0, and SlActin was used as an internal reference gene. Three biological replicates were performed for each sample. All the primers used for qRT-PCR are listed in Table S3.
Yeast one-hybrid assay
The yeast one-hybrid assay was performed according to our previous study31. In brief, SlACO1, SlACO3, and SlMADS1 promoter regions containing the SlGRAS4-binding motif were cloned into the pAbAi vector and transformed into the Y1HGold yeast strain. After screening the inhibitory aureobasidin A concentration, the SlGRAS4-pGADT7 plasmid and empty pGADT7 plasmid were transformed into the recombinant yeast strain. The interaction between SlGRAS4 and promoter regions was determined according to the growth of the colony on SD/-Leu/AbA culture medium.
Dual-luciferase assay
The full-length SlGRAS4 ORF was cloned into the pGreenII 62-SK vector, which was transformed into Agrobacterium tumefaciens strain GV3101 as an effector. The promoter of the target genes was cloned into the pGreenII 0800-LUC vector, which was transformed into GV3101 as a reporter. The effector and reporter strains were cotransfected into tobacco (Nicotiana benthamiana) leaves, and the commercial Dual-Luciferase® Reporter Assay System (Promega, USA) was used for LUC assays.
Data availability
All relevant data are provided within the paper and its supplementary files.
References
Giovannoni, J., Nguyen, C., Ampofo, B., Zhong, S. & Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu Rev. Plant Biol. 68, 61–84 (2016).
Adams, D. O. & Yang, S. F. Ethylene biosynthesis-identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl Acad. Sci. USA 76, 170–174 (1979).
Boller, T., Herner, R. C. & Kende, H. Assay for and enzymatic formation of an ethylene precursor, 1-aminocyclopropane-1-carboxylic acid. Planta 145, 293–303 (1979).
Liu, M., Pirrello, J., Chervin, C., Roustan, J. P. & Bouzayen, M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 169, 2380–2390 (2015).
Mcmurchie, E. J., Mcglasson, W. B. & Eaks, I. L. Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237, 235–236 (1972).
Nakatsuka, A. et al. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118, 1295–1305 (1998).
Barry, C. S., Llop-Tous, M. I. & Grierson, D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986 (2000).
Ito, Y. et al. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866–874 (2017).
Fujisawa, M., Nakano, T. & Ito, Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 11, 26 (2011).
Martel, C., Vrebalov, J., Tafelmeyer, P. & Giovannoni, J. J. The tomato MADS-Box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579 (2011).
Masaki, F., Toshitsugu, N., Yoko, S. & Yasuhiro, I. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386 (2013).
Zhong, S. L. et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159 (2013).
Giovannoni, J. J. Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180 (2004).
Sonia, O. et al. Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (Nor, Rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425 (2011).
Wang, R. et al. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9, 1696 (2019).
Gao, Y. et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot. 71, 3560–3574 (2020).
Manning, K. et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952 (2006).
Gao, Y. et al. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 6, 39 (2019).
Itkin, M. et al. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095 (2009).
Marian, B. et al. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24, 4437–4451 (2012).
Dong, T. T. et al. A tomato MADS-Box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 163, 1026–1036 (2013).
Yin, W. et al. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato (Solanum lycopersicum). Plant Physiol. Biochem. 118, 235 (2017).
Yin, W. et al. Suppression of SlMBP15 inhibits plant vegetative growth and delays fruit ripening in tomato. Front Plant Sci. 9, 938 (2018).
Zhu, M. K. et al. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55, 119–135 (2014).
Meng, C. et al. Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 193, 88–96 (2016).
Rumyana, K. et al. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23, 923–941 (2011).
Liu, M. C. et al. The chimeric repressor version of an ethylene response factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. N. Phytol. 203, 206–218 (2014).
Li, X. Y. et al. Control of tillering in rice. Nature 422, 618–621 (2003).
Huang, W. et al. Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol. J. 15, 472–488 (2017).
Liu, Y. et al. Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Sci. 8, 1659 (2017).
Liu, Y. et al. SlGRAS4 mediates a novel regulatory pathway promoting chilling tolerance in tomato. Plant Biotechnol. J. 18, 1620–1633 (2020).
Shinozaki, Y. et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 9, 364 (2018).
Zhang, L. et al. The SlFSR gene controls fruit shelf-life in tomato. J. Exp. Bot. 69, 2897–2909 (2018).
Carrera, E. et al. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 160, 1581–1596 (2012).
Zhou, S. E. et al. Manipulation of plant architecture and flowering time by down-regulation of the GRAS transcription factor SlGRAS26 in Solanum lycopersicum. Plant Sci. 271, 81–93 (2018).
Li, M. et al. Silencing GRAS2 reduces fruit weight in tomato. J. Integr. Plant Biol. 60, 498–513 (2018).
Hamilton, A. J., Lycett, G. W. & Grierson, D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346, 284–287 (1990).
Oeller, P. W. et al. Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439 (1991).
Picton, S. et al. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 3, 469–481 (1993).
Zhang, J. L. et al. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 8, 3413 (2018).
Kou, X. et al. SNAC4 and SNAC9 transcription factors show contrasting effects on tomato carotenoids biosynthesis and softening. Postharvest Biol. Tec. 144, 9–19 (2018).
Xiao, Y. Y. et al. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 64, 2499–2510 (2013).
Han, Y. C., Fu, C. C., Kuang, J. F., Chen, J. Y. & Lu, W. J. Two banana fruit ripening-related C2H2 zinc finger proteins are transcriptional repressors of ethylene biosynthetic genes. Postharvest Biol. Tec. 116, 8–15 (2016).
Yin, X. R., Allan, A. C., Chen, K. S. & Ferguson, I. B. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 153, 1280–1292 (2010).
Vrebalov, J. et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene. TAGL1. Plant Cell. 21, 3041–3062 (2009).
Lin, Z. et al. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55, 301–310 (2010).
Gao, Y. et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 5, 75 (2018).
Deng, H. et al. A novel tomato F-box protein, SlEBF3, is involved in tuning ethylene signaling during plant development and climacteric fruit ripening. Plant J. 95, 648–658 (2018).
Yang, L. et al. Functional characterization of SlSAHH2 in tomato fruit ripening. Front. Plant Sci. 8, 1312 (2017).
Acknowledgements
This work was sponsored by the Natural Science Foundation of Chongqing, China (cstc2019jcyj-bshX0008 to Y.L.). This work was also supported by funding provided by the National Key Research and Development Program (2016YFD0400101) and the National Natural Science Foundation of China (No. 31772370, 31972470) to Z.L.
Author information
Authors and Affiliations
Contributions
Y.L. and Z.L. designed the research; Y.L., Y.S., D.S., and W.L. performed the experiments; Y.L. analyzed the data; Y.L. wrote the paper; Z.L. revised the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Shi, Y., Su, D. et al. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic Res 8, 3 (2021). https://doi.org/10.1038/s41438-020-00431-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00431-9
This article is cited by
-
Transcriptional regulation of tomato fruit ripening
Physiology and Molecular Biology of Plants (2024)
-
A comprehensive review on speed breeding methods and applications
Euphytica (2024)
-
AgMYB5, an MYB transcription factor from celery, enhanced β-carotene synthesis and promoted drought tolerance in transgenic Arabidopsis
BMC Plant Biology (2023)
-
Different regulatory mechanisms of plant hormones in the ripening of climacteric and non-climacteric fruits: a review
Plant Molecular Biology (2021)