Abstract
In plants, low molecular weight terpenes produced by terpene synthases (TPS) contribute to multiple ecologically and economically important traits. The present study investigates a carrot terpene synthase gene cluster on chromosome 4 associated with volatile monoterpene production. Two carrot mutants, yellow and cola, which are contrasting in the content of low molecular weight terpenes, were crossed to develop an F2 mapping population. The mapping analysis revealed overlapping QTLs on chromosome 4 for sabinene, α-thujene, α-terpinene, γ-terpinene, terpinen-4-ol and 4-carene. The genomic region of this locus includes a cluster of five terpene synthase genes (DcTPS04, DcTPS26, DcTPS27, DcTPS54 and DcTPS55). DcTPS04 and DcTPS54 displayed genotype- and tissue-specific variation in gene expression. Based on the QTL mapping results and the gene expression patterns, DcTPS04 and DcTPS54 were selected for functional characterization. In vitro enzyme assays showed that DcTPS54 is a single-product enzyme catalysing the formation of sabinene, whereas DcTPS04 is a multiple-product terpene synthase producing α-terpineol as a major product and four additional products including sabinene, β-limonene, β-pinene and myrcene. Furthermore, we developed a functional molecular marker that could discriminate carrot genotypes with different sabinene content in a set of 85 accessions.
Similar content being viewed by others
Introduction
The cultivated carrot (Daucus carota L.) is an important horticultural crop due to characteristic flavour, nutritional value and health benefits of the root1. In addition, the above-ground tissues of carrot plants are gaining popularity in modern nutrition2. With a current annual world production of more than 40 million tons and a total growing area of about 1.2 million hectares, it ranks among the top ten vegetable crops3. In the past decades, breeding efforts mainly focused on increasing yield and enhancing disease and pest resistance. Nowadays, to address the consumer’s demands, the improvement of nutritional and flavour quality traits were added to the list of breeding aims3.
Low molecular weight terpenes are involved in plant-to-plant communication and plant protection against abiotic and biotic stresses4,5. The biological role of low molecular weight terpenes in plant protection against insect, fungal and bacterial pathogens has been demonstrated in several studies6,7. For example, overexpression of the rice terpene synthase gene OsTPS19 functioning as a (S)-limonene synthase in planta lead to enhanced resistance to the blast fungus Magnaporthe oryzae8. In another report, it was shown that (E,E)-α-farnesene production is an important factor involved in pathogenesis of three post-harvest fungal pathogens (Colletotrichum acutatum, Penicillium expansum and Neofabraea alba) in apple fruit9. Furthermore, γ-terpinene displayed antibacterial activity against Xanthomonas oryzae in rice10.
Moreover, relevant to this study, the low molecular weight terpenes contribute to carrot flavour11. The flavour combines taste and aroma and is a complex trait that depends on the relative amount of sugars, non-volatiles (amino acids, phosphates, polyacetylenes, nitrogenous substances) and volatiles (terpenes, phenylalanine- and fatty acid-derived compounds)12. The typical flavour of carrots has been attributed mainly to volatile terpenes, with mono- and sesquiterpenes representing approximately 98% of the volatile compounds11,13. Few studies have focused on the correlation between terpenoid volatiles and carrot sensory attributes14,15,16. Because carrots contain a complex blend of many different terpene compounds, it is challenging to relate single terpenes to specific aroma characteristics13. Using an GC-olfactometry (GC-O) approach, a link between the carrot aroma and flavour and certain isolated terpenes could be established13. Odour sensation notes such as “carrot top”, “terpene-like”, “green”, “fruity”, “spicy” or “woody” were defined as odour descriptors. The monoterpenes sabinene, β-myrcene and p-cymene were reported to be important contributors to the “carrot top” aroma, whereas limonene, γ-terpinene and terpinolene are related to fruity notes. Sesquiterpenes like β-caryophyllene, α-humulene and γ-bisabolene contributed to “spicy” and “woody” notes13. Among some non-volatile compounds like polyacetylenes or laserine derivatives terpenes are often involved in harsh or bitter taste15. A combination of a chemical and a sensorial approach predicted sabinene, α-terpinolene and β-pinene as candidates for bitterness5.
The huge variety of different terpenes is formed by members of terpene synthases family (TPS, EC 4.2.3) from few substrates by similar carbocation-based reaction mechanisms17,18. Thus, monoterpene synthases synthesize monoterpenes from the substrates geranyl diphosphate (GPP) and its cis-isomer neryl diphosphate (NPP); sesquiterpene synthases from sesquiterpenes from both isomer forms farnesyl diphosphate (trans-FPP and cis-FPP). In this reaction, the initial ionization of the substrates results in a cationic intermediate, which undergoes a series of rearrangements, until the reaction is terminated by a proton loss or addition of a nucleophile19,20. The family of terpene synthases has been divided into eight clades (TPS-a to TPS-h) based on sequence properties21. In the carrot genome, 65 putative terpene synthase genes have been identified. Most of them belong to the clades TPS-a and TPS-b22,23.
Several studies are focusing on the genomic regions and genes potentially involved in synthesis of carrot terpenes including flavour-associated terpenes. Based on a genome-wide association study (GWAS), 21 QTLs have been described for 11 terpenes in roots23. Clusters of terpene synthase genes have been found on chromosomes 3, 4, 5, 7 and 9. A recent study, which focused on bitterness trait variation in a carrot F2:3 population, identified 71 QTLs for 25 terpenes5. These QTLs were distributed on chromosomes 3, 4 and 9. Both studies described major QTLs for low molecular weight terpenes on chromosome 4. However, the map-based cloning and further characterization of candidate genes has not been performed yet. Two studies have reported on functional characterization of terpene synthases from carrot. The first study presented data on an in vitro test for DcTPS1 and DcTPS2 belonging to the TPS-a and TPS-b subfamilies, respectively. Recombinant DcTPS1 produced the sesquiterpenes (E)-β-caryophyllene and α-humulene, while DcTPS2 synthesized the monoterpenes geraniol and β-myrcene24. Another work showed that a monoterpene synthase (WtDcTPS1) from a wild carrot produced geraniol and β-myrcene in an in vitro assay25.
The utilization of molecular markers linked to QTLs can support modern carrot breeding. The application of molecular markers can help to select putative crossing parents by analysis of functional allelic diversity of Daucus germplasm and to screen the progenies for desired gene combinations. Marker-assisted selection in carrot has been performed for resistance against the root-knot nematode Meloidogyne javanica26 and root quality traits, particularly carotenoid and sugar composition27. However, no functional molecular markers that might be used for breeding of different terpene profiles in carrot genotypes have been reported yet.
In this study, we investigated a terpene synthase gene cluster on carrot chromosome 4. To define the genomic regions associated with terpene production, a QTL analysis followed by expression analysis of DcTPS04, DcTPS26, DcTPS27, DcTPS54 and DcTPS55 genes was performed. Finally, the DcTPS04 and DcTPS54 were functionally characterized in an in vitro assay.
Results
Phenotyping and QTL mapping in a F2 population
A prerequisite for QTL analysis is two parent genotypes that are different in the trait of interest. We observed that the yellow and the cola carrot genotypes display significant phenotypic differences in the content of low molecular weight terpenes. In comparison to the yellow genotype, the cola genotype showed reduced amounts of terpinen-4-ol, α-terpinene, sabinene in leaf and root tissue and γ-terpinene in leaves. The monoterpene 4-carene was detectable in roots of the yellow mutant but not in the leaves of this genotype (Fig. 1).
Leaf (a) and root (b) tissue of seven plants was analysed by headspace GC-MS. The results are presented in box and whisker plots (min to max, show all points) with a fold change of the abundance on a log2 scale. The data are statistically analysed with Mann–Whitney test (*P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns) and not detectable (nd))
These two contrasting parental genotypes were crossed to develop a QTL mapping population. The F2 generation of this population comprised 320 individual plants. They were analysed for volatile terpene compounds using headspace SPME-gas chromatography. This analysis allowed us to quantify 20 monoterpenes and 15 sesquiterpenes in root tissue, and 18 monoterpenes and 18 sesquiterpenes in leaf tissue, respectively (Tab. S1). In leaves, the most abundant monoterpenes were β-myrcene, sabinene and limonene, and the major sesquiterpenes were β-caryophyllene and β-bisabolene. The terpenes with the highest abundance in roots were the monoterpenes terpinolene, γ-terpinene and sabinene and the sesquiterpenes β-caryophyllene, guaia-3,9-diene and α-bisabolene (Tab. S1). Phenotypic variation in the contents of sabinene, α-terpinene and terpinen-4-ol (leaf and root tissue), 4-carene, α-thujene (root tissue) and γ-terpinene (leaf tissue) was observed in the F2 population. No significant differences could be measured for the non-circular compound β-myrcene and the circular limonene, functioning as unaffected controls (Fig. 2a).
a Abundance of terpene in the VOM14 F2 biparental mapping population sorted after marker genotyping. Three hundred and twenty plants were analysed by headspace GC-MS and the terpene content was sorted by the results of the marker analysis (KASP 4-31,228,189 CAPS 4-31,218,832). Association consistencies between the abundance of β-myrcene, limonene, sabinene, α-terpinene, γ-terpinene, terpinen-4-ol, α-thujene and 4-carene, and genotype in leaf and root tissue of carrot are presented in box and whiskers (min to max, show all points) with a fold change of the abundance in log10 scale. The data were statistically analysed with Tukey’s multiple comparison test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, not significant (ns)). b LOD score of terpenes associated with chromosome 4 based on markers flanking the terpene synthase gene cluster. Significance threshold (TH) was set using 1.000 permutations. c Validation of the KASP 4-31,228,189 marker. KASP-based genotyping of 85 carrot accessions. Association consistency between the abundance of β-myrcene, limonene, sabinene, α-terpinene, γ-terpinene, terpinen-4-ol, α-thujene and 4-carene and genotype in leaf and root tissue of carrot. All measured points were statistically analysed by Tukey’s multiple comparison test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, not significant (ns)) and presented in box and whisker plots. The abundance is shown with a fold change on a log10 scale. The horizontal x-axis (a, c) indicates genotypic groups a, b and h. The y-axis (a, c) indicates an abundance of terpenes in investigated plants and is on a log10 scale. The plants in group a carry the homozygous maternal allele (a), whereas the plants in group b harbour the homozygous paternal allele (b). Individuals in group h are the heterozygous plants
Genotypic analysis was performed using 89 molecular markers that were polymorphic in the F2 mapping population. This set of molecular markers was distributed over all carrot chromosomes and included 33 SSR-, 13 CAPS-, 9 KASP-, 24 InDel- and 10 RAPD markers. Twenty-eight molecular markers close to TPS genes are presented in Tab. S2. Markers with segregation distortion were excluded from map construction. The resulting partial genetic map had a length of 536.6 cM encompassing all nine Daucus carota chromosomes (data not shown).
QTL analysis was conducted using phenotypic and genotypic data from the F2 mapping population. In total, 14 QTLs with LOD scores >10 were identified for terpenes in roots and leaves (Table 1).
In carrot roots, QTLs associated with bornyl acetate and β-pinene were detected on chromosomes 1 and 5, respectively. QTLs located on chromosome 3 were calculated for β-elemene, δ-elemene and β-farnesene in leaves and for guaia-3,9-diene and δ-elemene in roots. A genomic region spanning 105 kb on chromosome 4 includes QTLs for sabinene, α-thujene, α-terpinene, γ-terpinene, terpinen-4-ol and 4-caren. Five molecular markers, namely KASP 4-31,228,189, CAPS 4-31,218,832, KASP 4-31,147,906, KASP 4-31,258,391 and KASP 4-31,239,271 were associated within this region (Fig. 2b). These molecular markers segregated in the F2 mapping population according to the Mendel’s law of segregation (Fig. S1, Tab. S3). Highly significant QTLs were detected for sabinene (LOD: 27.49), α-terpinene (LOD: 20.37), γ-terpinene (LOD: 20.48) in leaves and for 4-carene (LOD: 28.68) in roots. Although the LOD scores for other analysed low molecular weight terpenes showed the same tendency, they did not exceed the significance threshold calculated individually for each terpene.
Molecular markers associated with sabinene content
Genotyping of the F2 mapping population with the molecular markers KASP 4-31,228,189 and CAPS 4-31,218,832 allowed the identification of three groups ‘a’, ‘h’ and ‘b’ with different content of specific monoterpenes (Fig. 2a). The plants in these groups differed regarding the homozygous maternal allele (a), the homozygous paternal allele (b) and the heterozygous genotype (h). The highest average content of specific monoterpenes was always detected in group ‘a’ and the lowest in group ‘b’ (Fig. 2a).
To test the applicability of the molecular marker KASP 4-31,228,189, we investigated a panel of 85 carrot genotypes described previously23. The group ‘a’ harbouring genotypes, homozygous for the allele (a), consists of nine genotypes, including the yellow mutant. Fifty-one accessions and the cola mutant are arranged in group ‘b’, which is homozygous for allele (b). Twenty-five accessions are members of group ‘h’ displaying the heterozygous genotype (Fig. S3). As expected, marker-trait association analysis for the control substances β-myrcene and limonene showed no significant differences. For the sabinene content in leaf tissue a strong correlation was found. The homozygous alleles (a) and (b) were associated with high and low sabinene content, respectively. The same tendency was observed for sabinene in root tissue (Fig. 2c). For α-terpinene, terpinen-4-ol, and 4-carene no significant differences were observed, due to their low detectability. For γ-terpinene, the marker-trait association in leaves showed the same tendency as for sabinene (Fig. 2c). Our data show that the molecular marker KASP 4-31,228,189 can discriminate carrot genotypes by a specific allelic configuration for sabinene production. To validate the major QTL for sabinene, we investigated candidate genes with tentative terpene synthase activity within the terpene synthase gene cluster on chromosome 4.
Terpene synthase cluster on chromosome 4 includes putative monoterpene synthases
For in silico analysis of the genomic region on chromosome 4 associated with sabinene production, we performed a reannotation of the carrot genome using 12 reference genomes in GeMoMa 1.6.2beta22,28. This gene cluster with the genomic coordinates 31,120,000–31,280,000 bp includes genes encoding proteins of different functional groups. Two zinc finger transcription factors, two methyltransferases, one phosphatase, one galacturonosyl transferase (GAUT), seven glycosyl hydrolases (GH) and five terpene synthases were predicted (Fig. 3a).
a Schematic representation of the TPS-gene cluster. Structural analysis of this region was performed based on reannotation of carrot genome assembly by GeMoMa (version 1.6.3.beta) using 12 reference genomes22,28. Receptor-like kinase (RLK) and galacturonosyltransferases (GAUT). b Amino acid sequence alignment of candidate terpene synthases from the yellow and cola mutant. Background in grey shows different residues in all five candidate terpene synthases. Differences between DcTPS04 and DcTPS54 sequences are marked in red. N-terminus of the catalytic domain (RR) and the DDIYD-conserved motif are marked in bold. The N-terminal plastid transit peptide predicted by Target P2.0 is marked with a horizontal line. The phylogram in the lower-left corner shows similarity of the catalytic domain of the 5 terpene synthases based on the translation of the nucleotide sequences from the yellow and the cola mutant. TPS-b marks the as TPS-b clade. A phylogenetic tree of all carrots TPS was previously published in ref. 23
The putative glycosyl hydrolase genes are homologous to plant xyloglucan endotransglucosylases (XET). Out of 36 XET genes that were recognized in the carrot genome, seven XETs surround the DcTPS26 gene.
We identified five terpene synthase genes, DcTPS04 (DCAR_013298), DcTPS26 (DCAR_013310), DcTPS27 (DCAR_013293), DcTPS54 (DCAR_013297) and DcTPS55 (DCAR_013294) confirming the terpene synthase gene prediction reported previously22,23. The terpene synthase genes spread over a region of 105 kb. The genes DcTPS04, DcTPS27, DcTPS54 and DcTPS55 are located physically within a genomic region of 35 kb and genetically within 0.11 cM. The gene DcTPS26 is located 70 kb upstream of DcTPS04 (Fig. 3a). In addition, a 609 bp TPS-fragment at position 31,157,047 –31,157,656 bp was found. Its amino acid sequence has 86% identity to the first 150 residues of the N-terminus of DcTPS26. However, a large part of the catalytic domain is missing. Thus, it can be assumed that this terpene synthase gene fragment is non-functional. The amino acid sequence alignment of the five predicted terpene synthase genes revealed their high sequence similarity, varying from 78.5 to 91% (Fig. 3b). Based on high sequence similarity and chromosomal orientation of DcTPS04, DcTPS26, DcTPS27, DcTPS54 and DcTPS55, the same evolutionary origin can be suggested. Although DcTPS26 has a high sequence similarity to other monoterpene synthases, the lack of a proper transit peptide indicates a sesquiterpene producing activity (Fig. 3b, Fig. S3). Proper transit peptides were predicted by Target P2.0 algorithm for DcTPS04 (the cleavage site position CS pos: 54–55, between the residues VSR-GD, with a prediction Pr. of 0.2131), for DcTPS54 (CS pos: 44–45, IRC-TS. Pr: 0.7755), for DcTPS55 (CS pos: 34–35, TCC-TS, Pr: 0.2387) and for DcTPS27 (CS pos: 45–46, IRC-TG, Pr: 0.7556), indicating their monoterpene synthase activity.
Expression pattern of terpene synthase candidates
We analysed the expression patterns of DcTPS04, DcTPS26, DcTPS27, DcTPS54 and DcTPS55 clustered on chromosome 4 to assign a priority to candidate genes, which could be involved in sabinene production. For this purpose, total RNA was isolated from leaf and root tissue of the two 16 weeks old carrot mutants (cola and yellow) and was analysed with gene-specific primers by qRT-PCR. Transcript accumulation was obtained for DcTPS04, DcTPS26 and DcTPS54, whereas the transcripts of DcTPS27 and DcTPS55 genes could hardly be identified (Fig. 4).
The levels of RNA transcripts for DcTPS04, DcTPS26, DcTPS27, DcTPS54 and DcTPS55 genes were analysed in roots (mix of cortex, xylem and phloem tissues) and leaves (mix of young leaf and petiole tissues). Each sample was pooled from three plants. Each experiment was performed in biological and technical triplicates. Relative expression for the five TPS gene in yellow and cola mutants was calculated using the Pfaffl method with the reference genes DcTub, DcHSP, DcPP2A and presented with a fold change in a log10 scale. Relative to DcTub expression levels are presented in plots with standard deviation. Data were statistically analysed by unpaired t-Test (*P < 0.05, **P < 0.01, ****P < 0.0001). Samples with non-detectable CTs for three biological replicates are marked as nd
Our results show that DcTPS04, DcTPS26 and DcTPS54 transcript levels are higher in root tissue compared to leaf tissue (Fig. 4). For DcTPS26 we detected lower expression levels in leaf tissue of yellow compared to cola. In root tissue, the expression of DcTPS26 was higher as in cola. Due to these contrasting expression patterns, DcTPS26 was not selected for further analysis. We also observed differences in the expression of DcTPS04 and DcTPS54 in the yellow and cola mutant. The transcript of DcTPS04 was detected in root and leaf tissue of cola and it was not detectable in yellow. In contrast, higher expression levels of DcTPS54 were found in yellow compared to cola (Fig. 4). The expression profile of DcTPS54 is in good agreement to the abundance of sabinene in yellow and cola mutants (Fig. 1). The expression pattern of DcTPS04 and DcTPS54 in yellow and cola gives an indication for their involvement in sabinene synthesis in these genotypes. Based on the gene expression and QTL analysis, we selected DcTPS04 and DcTPS54 for further functional analysis.
Functional characterization of sabinene synthase candidates
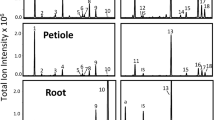
For functional characterization of DcTPS04 and DcTPS54, we cloned the ORF of the catalytic domains excluding the N-terminal region with the transit peptide. The amplified sequences of 1632 bp and 1692 bp for DcTPS04 and DcTPS54, respectively, were integrated upstream of the 6-His tag and following stop codon in pET28c plasmid and expressed in E. coli RosettaTM 2 strain. The synthesised terpene synthases were assayed for monoterpene (GPP and NPP substrates) and sesquiterpene ((E,E)-FPP and (Z,Z)-FPP substrates) synthase activity. The products were analysed by mass spectrometry. Enzymatic formation of terpenes was observed only with GPP substrate identifying DcTPS04 and DcTPS54 as functional monoterpene synthases (Fig. 5, S4).
DcTPS54 yielded the single monoterpene product sabinene. The major product of DcTPS04 is α-terpineol along with the minor products sabinene, β-limonene, β-pinene and myrcene. Extracts prepared from E. coli served as controls for terpene formation in vitro. These extracts did not produce any detectable amounts of terpenes.
Our data clearly show that both proteins catalyse sabinene formation in vitro. Moreover, the functional test revealed DcTPS54 as a single-product terpene synthase and DcTPS04 as a multiple-product terpene synthase. Therefore, we were able to confirm the sabinene-associated QTL on chromosome 4.
Discussion
In the present study, we investigated the genomic region on carrot chromosome 4 associated with monoterpene production by terpene metabolite profiling, QTL mapping and functional analysis of terpene synthase candidate genes. This combinatorial approach allowed us to identify terpene synthase genes responsible for sabinene synthesis.
To dissect the genetic basis of a quantitative trait, such as the content of a natural product, two main methods are available, conventional QTL mapping and GWAS. QTL mapping depends on genetic diversity of two parents and is time-consuming due to the necessity to develop a mapping population. Moreover, QTL regions can be quite large and may include many potential candidate genes. Nevertheless, this method has been used in carrot research and breeding to elucidate genetic control for morphological, disease resistance and root quality traits3,29. GWAS can overcome the limitations of QTL mapping. This technique has great potential for detection of QTLs with high resolution in diverse sequenced genotypes. However, GWAS also can have high false-positive rates due to the population structure of most germplasm sets30,31. GWAS addressing volatile compounds has already been conducted in studies applied to tomato, apple and blueberry32,33,34.
There are only few reports on the use of GWAS for QTL identification in carrot. This technique was used, to discover the Or gene on chromosome 335 and to link volatile terpenes and their potential synthases23. A combination of QTL mapping and GWAS is a promising strategy to compensate the limitations of each method. However, a combined approach has not been performed in carrot, yet.
Our analysis of a F2 mapping population revealed 14 QTLs associated with volatile terpenes. To some extent, these results are in agreement with those obtained by GWAS for 85 carrot genotypes23. However, we were not able to confirm a QTL for the monoterpene ester bornyl acetate associated with the DcTPS03 gene on chromosome 2. Instead, we identified a QTL for bornyl acetate on chromosome 1 that might be associated with the DcTPS10 gene. We identified a large QTL cluster on chromosome 4 for sabinene, α-terpinene, γ-terpinene and terpinen-4-ol in leaves and 4-caren, sabinene and α-thujene in roots. QTLs associated with sabinene and terpinen-4-ol in roots have also been detected by a previous GWAS in the same genomic region23. A consistency of QTLs detected by QTL mapping with those identified by GWAS was observed in several crops such as soybean36, maize37, tea38 and rice39.
In silico analysis of the terpene synthase gene cluster on chromosome 4 revealed that it includes two zinc finger transcription factors, two methyltransferases, one phosphatase, one galacturonosyl transferase (GAUT), seven glycosyl hydrolases (GH) and five terpene synthases. A close association of terpene synthases with genes encoding enzymes that could modify terpenes or provide substrate for terpene synthases has already been described previously. For example, in Arabidopsis thaliana, gene groups including two to five terpene synthases clustered together with prenyltransferase-, cytochrome P450- and glycosyltransferase genes40. Similar results were obtained for tomato terpene synthase gene clusters on chromosomes 6, 7 and 8, which also contain several putative cytochromes P450, methyltransferases, acyltransferases and glycosyltransferases41. The location of these genes within or close to terpene synthase gene clusters might be beneficial for the inheritance of alleles from a single metabolic pathway and for the regulation of genes in the same biosynthetic pathway42. We found seven glycosyl hydrolases located within the terpene synthase gene cluster on chromosome 4. These enzymes are known to be involved in plant defence against pathogens and diverse physiological functions such as glycan biosynthesis, mobilization of energy, symbiosis, signalling and metabolism of glycolipids43. The role of glycosyl hydrolases in terpene metabolism remains to be investigated. A number of publications provide data suggesting a common evolution and functional specialization of terpene synthases and glycosyl hydrolases. A structural similarity of glycosyl hydrolases and monoterpene synthase has been shown for a limonene-type synthase of tobacco (5EAS). Two glycosyl hydrolase-like domains were found at its N-terminal region44,45. An early work showed that the GH activity resulted in the release of nerol, linalool, geraniol and terpineol in apricots46.
All five terpene synthase genes of the investigated gene cluster share a high amino acid sequence identity and belong to the TPS-b clade. They have the terpene synthase specific motives NSE/DTE and DDxxD that are essential for catalytic activity. These terpene synthases have the RR(x8)W motif at the N-terminus, which is characteristic for clade TPS-b. We predicted an N-terminal transit peptide for DcTPS04, DcTPS54, DcTPS55 and DcTPS27 suggesting their plastid localization. In contrast, DcTPS26 that displays a high sequence similarity to other terpene synthases of the cluster lacks a proper transit peptide. This indicates a function as cytosolic terpene synthase21. Such a subcellular differentiation of closely related terpene synthases in the cytosol and plastids has been documented for (E,E)-β-farnesene synthase (AtTPS03) and β-ocimene synthase (AtTPS02) from A. thaliana47.
DcTPS04 and DcTPS54 were expressed differently in yellow and cola genotypes, which was consistent with the phenotypic data for volatile monoterpenes. Therefore, their enzymatic activity was investigated in an in vitro assay. The monoterpene synthase DcTPS54 is a single-product sabinene synthase. DcTPS04 catalysed the production of α-terpineol as a major product and four by-products including sabinene, β-limonene, β-pinene and myrcene. DcTPS04 and DcTPS54 share a sequence identity of 89.5%. They differ in several residues at the C-terminus that are reported to be critical for product profile48. DcTPS04 has Gly and Asp at positions 589 and 594, respectively. DcTPS54 has Ser at position 589 and Gln at position 594. Previous work on monoterpene synthases from Sitka spruce (Picea sitchensis) has shown that differences in individual amino acids may determine deviating product profile. Reciprocal mutagenesis analysis of a sabinene synthase and 3-carene synthase revealed that Phe at position 596 led to synthesis of sabinene, whereas its substitution to Leu directed product profile to 3-carene48. Sequence similarity and production outcome of DcTPS04 and DcTPS54 make them an attractive target for site-directed mutagenesis to identify, which particular structural feature of these enzymes affects their function.
Although we have provided several evidences (QTL and expression analysis, results of in vitro assay) suggesting the sabinene producing activity of DcTPS04 and DcTPS54, the in vivo function of these TPSs needs to be proven conclusively.
Due to the high sequence identity, it could be suggested that the five terpene synthase genes have the same evolutionary origin through gene duplication. Terpene synthases associated with different products evolve by differential neofunctionalization and subfunctionalization. These evolutionary mechanisms are widely accepted for terpene synthases21. DcTPS26 from the gene cluster on chromosome 4 lacks a transit peptide indicating its possible localization in cytosol and sesquiterpene synthase activity. Comparative functional characterization of DcTPS26, DcTPS04 and DcTPS054 should be performed to confirm this hypothesis. Good examples of such functional divergence are the (E)-α-bergamotene synthase of the TPS-b clade from Lavendula angustifolia and the sesquiterpenes synthases SauSesquiTPS of the TPS-a clade from Santalum austrocaledonicum49,50.
Sabinene appears to be among the major terpenes related to the preferred carrot flavour characteristics. However, it may also have negative impact due to its putative involvement in harsh and bitter notes. The information in the relevant literature is somewhat contradictory. Previous investigations suggested that sabinene is responsible for both “harsh” and “carrot top” flavour13,15. Another study showed that it might be also a major contributor to “fruity” flavour16. It may be assumed that the amount of sabinene is one of the critical factors for human gustatory sensation in terms of acceptance or rejection. Sensorial investigations based on large consumer panels and carrot cultivars selected by their variability for amounts of key volatile monoterpenes might be used to reveal the impact of single substances like sabinene.
Sabinene is not only one of the volatiles associated with carrot flavour and taste11,12,13 but also has ecological and economic relevance. For instance, it has been reported that sabinene can be involved in the plant defence against insects in Sitka spruce7. Besides its ecological benefits, sabinene is a major component of carrot seed oil51. Essential oil from wild carrot seeds possesses strong biological activity, probably due to the high content of sabinene and α-pinene. For instance, these monoterpenes were reported to be components of pharmaceuticals to treat the protozoan Trypanosoma brucei causing the African sleeping sickness disease52.
The results observed in this study are in good agreement with findings reported in a very recent paper describing functional characterization of carrot TPSs53 in the fully sequenced DH1 genotype22. This study also shows that the genomic region on chromosome 4 encodes TPSs catalysing the production of sabinene53. In complement to the findings of Muchlinski et al.53 we examined a sabinene-deficient carrot genotype. Taken together the complementary results of both studies are applicable in breeding programmes focused on modification of sabinene content in carrot. For this approach, we developed and validated a functional molecular marker that might serve as a useful breeding tool to select carrot accessions with high or low amounts of sabinene.
Materials and methods
Plant material and growth conditions
Two carrot mutants, cola and yellow, were involved in the experiments. The cola mutant possesses dwarfish morphology, whereas reduced amount of chlorophyll resulted in yellowish leaves of yellow mutant54. The F2 mapping population (VOM14) of 320 individuals was developed from an initial cross of a homozygous recessive yellow mutant as a female parent and a homozygous recessive cola mutant as pollen parent54,55. Plants were grown in 19 cm/30 cm w/h plastic pots in a sand/humus mixture (3/1) (v/v) under optimized greenhouse conditions at 25 °C/20 °C (day/night) and 18 h light photoperiod. They were drop irrigated and fertilized each week with 200 ml of a 0.3% Wuxal Super solution (8% N/8% P/6% K, Wilhelm Haug GmbH and Co.KG Düsseldorf, Germany).
Volatile compound analysis of parent genotypes and F2 mapping population
The volatile compounds in root and leaf tissue of the parent genotypes and of the F2 mapping population were analysed by headspace SPME-gas chromatography as described previously23. Seven yellow and seven cola plants were involved in the initial parental phenotyping. Three hundred and twenty individual plants were investigated from the F2 mapping population.
Marker-based QTL analysis
Genomic DNA was extracted from 0.5 g of young leaf tissue. The DNA concentration of samples was estimated by a Nanodrop 8000 (Thermo Scientific, Waltham, USA) and normalized to 10 ng μL−1. A universal fluorescent labelling strategy56 with M13 tailed forward primers was used to analyse SSRs and small InDels. The DNA fragments were separated on 6.5% polyacrylamide gels and detected by a LI-COR 4300 automatic sequencer (LI-COR Biosciences, Lincoln, NE, USA). The fragment sizes were calculated using fluorescent labelled 50–350 bp or 50–1500 bp size ladders, respectively. InDels and SNPs within or near to predicted terpene synthase (TPS) genes23 were detected by sequencing homologous PCR products of 800 bp to 1200 bp length of the yellow and cola parental lines (Tab. S3). SNPs within recognition sites of restriction endonucleases were converted to CAPS markers and analysed on 1.5% agarose gels. For other SNPs, KASP assays were developed (LGC Genomics, London, UK) and the marker analysis was performed using a CFX96 real-time cycler (Bio-Rad Laboratories, Hercules, USA). Amplification conditions were as follows: initial denaturation at 94 °C for 15 min; 10 cycles of 94 °C for 20 s, 61 °C for 1 min with a decrement of 0.6 °C/cycle followed by 29 cycles of 94 °C for 20 s, 55 °C for 1 min and final incubation for 10 min at 37 °C before plate reading. If necessary, additional three cycles with 55 °C annealing temperature (recycling) were performed. Data collection and allelic discrimination has been done with the MaestroTM software (Bio-Rad Laboratories, Hercules, USA).
The obtained molecular marker data were analysed with JoinMap vers. 5.0 software (Kyazma B.V., Wageningen, The Netherlands)57. Loci were grouped using LOD thresholds from 2.0 to 6.0 in steps of 1.0 and recombination frequency lower than 0.4. The jump threshold was set to 5.0 and a third mapping round was not considered. The recombination frequencies were converted to mapping distances (in cM) using the Kosambi function. The linkage groups were assigned to the reference map using SSRs, SCARs and InDel markers with known chromosomal location58. QTL analysis was performed using the interval mapping (IM) mode of MapQTL vers. 5.0 software (Kyazma B.V., Wageningen, The Netherlands). Permutation tests (1000 permutations) were run for each root and leaf VOC to determine genome-wide significance thresholds (TH) (P = 0.05).
Cloning of candidate genes
The DcTPS04 (DCAR_013310) and DcTPS54 (DCAR_013297) genes lacking the N-terminal transit peptide and the termination codons were amplified from carrot cDNA using gene-specific primers (Tab. S4), cloned into the pCR2.1-TOPO vector (Invitrogen/Thermo Scientific, Waltham, USA) and verified by sequencing. The DcTPS04 and DcTPS54 sequences were released from the pCR2.1-TOPO plasmid by NcoI and NotI restriction sites and ligated to identically digested pET28c expression vector (Novagen/Merck, Darmstadt, Germany) resulting in pET28c-DcTPS04-His and pET28c-DcTPS54-His constructs.
In vitro enzyme assay and product identification by GC-MS
Following transformation of pET28c-DcTPS04-His and pET28c-DcTPS54-His plasmids in E. coli strain RosettaTM 2 (Novagen/Merck, Darmstadt, Germany), the individual colonies were inoculated in 100 mL LB medium supplemented with 50 mg l−1 kanamycin, cultivated at 37 °C until OD600 of 0.8, induced with 0.1 mM isopropylthio-β-galactoside (IPTG) and then grown for additional 16 h at 20 °C. Protein expression was verified by SDS-PAGE gel and Coomassie staining. Harvested bacterial pellets were resuspended in 5 ml assay buffer (250 mM HEPES-KOH buffer [pH 8], 100 mM MgCl2, 2.5 mM MnCl2, 50% glycerol, 1 mg µl−1 lysozyme), sonificated and collected by centrifugation. To perform the enzyme assay the crude bacteria extracts were mixed with 5 µl 1 M DTT, either 4 µl geranyl diphosphate (GPP, Sigma-Aldrich, St. Louis, USA) neryl diphosphate (NPP), trans-farnesyl diphosphate (E,E)-FPP and cis-farnesyl diphosphate (Z,Z)-FPP (Echelon Biosciences, Salt Lake City, USA) and filled with water to a final volume of 200 µl. After incubation at 41.5 °C for 0, 0.5, 1.5 and 4.5 h the terpene product was extracted with 200 µl hexane. For GC-MS analysis, an MPS2 autosampler was used (Gerstel, Mühlheim, Germany) and 1 µL of hexane extract was injected in splitless mode at 250 °C. Separation and detection of the products were performed by an Agilent Technologies 6890 GC equipped with a HP-INNOWax column (0.25 mm i.d., 30 m length, 0.5 μm film thickness) and an Agilent Technologies 5973N MS quadrupole analyser by electron ionization (source temperature: 230 °C, ionisation voltage: 70 eV). Carrier gas was helium with a flow rate of 1.0 mL min−1. Temperature programme: 40 °C (5 min), from 40 °C to 250 °C at 7 K min−1 and held 3 min at 250 °C, was used. Terpene synthase assay products were identified using authentic standards: myrcene, linalool, linalylisovalerat, 3-carene, α-terpineol, γ-terpinene, (+)-terpinen-4-ol, β-citronellol, β-limonene, sabinene, β-pinene, α-bisabolene, γ-bisabolene, (−)-α-bisabolol and α-farnesene (Sigma-Aldrich, St. Louis, USA).
Expression analysis of terpene synthase genes in leaf and root tissue
For the expression analysis of candidate genes, we harvested leaf and root tissue from 16 week old carrot plants (cola and yellow mutants). The root samples are heterogeneous samples including cortex, xylem and phloem tissues. The leaf samples comprise of young leaf and petiole tissues. To reduce plant-specific differences, each sample was pooled from three individual plants, frozen in liquid nitrogen and ground to a fine powder. Total RNA was extracted from 100 mg tissue powder using the innuPREP Plant RNA Kit (Analytik Jena, Jena, Germany) according to the manufacturer’s instructions. RNA integrity was confirmed by agarose gel electrophoresis. Random hexamers were used as primers for first-strand cDNA synthesis with 2 µg total RNA as a template and Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, USA). Carrot tubulin 3α (DcTUB), heat shock 70 (DcHSP) and protein phosphatase 2 (DcPP2A) were used as internal reference genes. The expression levels of DcTPS04, DcTPS26, DcTPS27, DcTPS54, DcTPS55, DcTUB, DcHSP and DcPP2A genes were analysed with the qTOWER3 G touch (Analytic Jena, Jena, Germany) equipment and gene-specific primers (Tab. S4). The results were calculated using the Pfaffl method59.
Reannotation of the carrot genome using GeMoMa 1.6.2beta
The published carrot genome was annotated using GeMoMa 1.6.2beta22,28. The 12 genomes (Arabidopsis thaliana (TAIR10), Brachypodium distachyon (v3.1), Glycine max (Wm82.a2.v1), Lactuca sativa (v5), Mimulus guttatus (v2.0), Oryza savita (v7.0), Prunus persica (v2.1), Populus trichocarpa (v3.1), Sorghum bicolor (v3.1.1), Setaria italica (v2.2), Solanum lycopersicum (v2.5) and Theobroma cacao (v1.1)) were used as reference with the software parameters: GeMoMa 1.6.2beta; SIMPLE PARAMETERS: reads: 1; splice: true; coverage: UNSTRANDED; gap opening: 11; gap extension: 1; maximum intron length: 15000; intron-loss-gain-penalty: 25; e-value: 100.0; contig threshold: 0.4; region threshold: 0.9; hit threshold: 0.9; predictions: 10; avoid stop: true; approx: true; prefix:; tag: prediction; verbose: false; timeout: 3600; sort: false; Score: ReAlign and GAF 1.6.2beta; SIMPLE PARAMETERS: tag: prediction; sorting: evidence, score; common border filter: 0.75; maximal number of transcripts per gene: 2147483647; prefix: AT; weight: 1.0; prefix: BD; weight: 1.0; prefix: GM; weight: 1.0; prefix: LS; weight: 1.0; prefix: MG; weight: 1.0; prefix: OS; weight: 1.0; prefix: PP; weight: 1.0; prefix: PT; weight: 1.0; prefix: SB; weight: 1.0; prefix: SI; weight: 1.0; prefix: SL; weight: 1.0; prefix: TC; weight: 1.0; filter: start==‘M’ and stop ==‘*’ and score/AA>=0.75; alternative transcript filter: tie==1 or evidence>1.
References
Sharma, K. D., Karki, S., Thakur, N. S. & Attri, S. Chemical composition, functional properties and processing of carrot-a review. J. Food Sci. Technol. 49, 22–32 (2012).
Boroski, M. et al. Enhancement of pasta antioxidant activity with oregano and carrot leaf. Food Chem. 125, 696–700 (2011).
Simon, P., Iorizzo, M., Grzebelus, D. & Baranski, R. The Carrot Genome (Springer, Cham, 2019).
Huang, A. C. & Osbourn, A. Plant terpenes that mediate below-ground interactions: prospects for bioengineering terpenoids for plant protection. Pest Manag. Sci. 75, 2368–2377 (2019).
Le Clerc, V. et al. Breeding for carrot resistance to Alternaria dauci without compromising taste. Mol. Breed 39, 1–15 (2019).
Pichersky, E. & Gershenzon, J. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5, 237–243 (2002).
Hall, D. E. et al. An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J. 65, 936–948 (2011).
Chen, X. et al. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 16, 1778–1787 (2018).
Souleyre, E. J. F. et al. Genetic control of α-farnesene production in apple fruit and its role in fungal pathogenesis. Plant J. 100, 1148–1162 (2019).
Yoshitomi, K. et al. Rice terpene synthase 24 (OsTPS24) encodes a jasmonate-responsive monoterpene synthase that produces an antibacterial γ-terpinene against rice pathogen. J. Plant Physiol. 191, 120–126 (2016).
Kjeldsen, F., Christensen, L. P. & Edelenbos, M. Quantitative analysis of aroma compounds in carrot (Daucus carota L.) cultivars by capillary gas chromatography using large-volume injection technique. J. Agric. Food Chem. 49, 4342–4348 (2001).
Simon, P. W., Peterson, C. E. & Lindsay, R. C. Correlations between sensory and objective parameters of carrot flavor. J. Agric. Food Chem. 28, 559–562 (1980).
Kjeldsen, F., Christensen, L. P. & Edelenbos, M. Changes in volatile compounds of carrots (Daucus carota L.) during refrigerated and frozen storage. J. Agric. Food Chem. 51, 5400–5407 (2003).
Alasalvar, C., Grigor, J. M., Zhang, D., Quantick, P. C. & Shahidi, F. Comparison of volatiles, phenolics, sugars, antioxidant vitamins, and sensory quality of different colored carrot varieties. J. Agric. Food Chem. 49, 1410–1416 (2001).
Kreutzmann, S., Thybo, A. K., Edelenbos, M. & Christensen, L. P. The role of volatile compounds on aroma and flavour perception in coloured raw carrot genotypes. Int. J. Food Sci. Technol. 43, 1619–1627 (2008).
Fukuda, T., Okazaki, K. & Shinano, T. Aroma characteristic and volatile profiling of carrot varieties and quantitative role of terpenoid compounds for carrot sensory attributes. J. Food Sci. 78, S1800–S1806 (2013).
Bohlmann, J., Meyer-Gauen, G. & Croteau, R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl Acad. Sci. USA 95, 4126–4133 (1998).
Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 9, 297–304 (2006).
Degenhardt, J., Köllner, T. G. & Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70, 1621–1637 (2009).
Karunanithi, P. S. & Zerbe, P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 10, 1–23 (2019).
Chen, F., Tholl, D., Bohlmann, J. & Pichersky, E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229 (2011).
Iorizzo, M. et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48, 657–666 (2016).
Keilwagen, J. et al. The terpene synthase gene family of carrot (Daucus carota L.): identification of QTLs and candidate genes associated with terpenoid volatile compounds. Front. Plant Sci. 8, 1–18 (2017).
Yahyaa, M. et al. Identification and characterization of terpene synthases potentially involved in the formation of volatile terpenes in carrot (Daucus carota L.) roots. J. Agric. Food Chem. 63, 4870–4878 (2015).
Yahyaa, M., Ibdah, M., Marzouk, S. & Ibdah, M. Profiling of the terpene metabolome in carrot fruits of wild (Daucus carota L. ssp. carota) accessions and characterization of a geraniol synthase. J. Agric. Food Chem. 66, 2378–2386 (2016).
Boiteux, L. S. et al. Employment of flanking codominant STS markers to estimate allelic substitution effects of a nematode resistance locus in carrot. Euphytica 136, 37–44 (2004).
Ellison, S., Senalik, D., Bostan, H., Iorizzo, M. & Simon, P. Fine mapping, transcriptome analysis, and marker development for Y2, the gene that conditions β-carotene accumulation in carrot (Daucus carota L.). G3 7, 2665–2675 (2017).
Keilwagen, J. et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44, 1–11 (2016).
Que, F. et al. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic. Res. 6, 69 (2019).
Zhu, C., Gore, M., Buckler, E. S. & Yu, J. Status and prospects of association mapping in plants. Plant Genome 1, 5–20 (2008).
Flint-Garcia, S. A., Thornsberry, J. M. & Buckler, E. S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374 (2003).
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394 (2017).
Larsen, B. et al. Genome-wide association studies in apple reveal loci for aroma volatiles, sugar composition, and harvest date. Plant Genome 12, 1–15 (2019).
Ferrão, L. F. V. et al. Genome-wide association of volatiles reveals candidate loci for blueberry flavor. N. Phytol. 226, 1725–1737 (2020).
Ellison, S. L. et al. Carotenoid presence is associated with the or gene in domesticated carrot. Genetics 210, 1497–1508 (2018).
Cao, Y. et al. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front. Plant Sci. 8, 1–10 (2017).
Zhang, X. et al. Combined GWAS and QTL analysis for dissecting the genetic architecture of kernel test weight in maize. Mol. Genet. Genomics 295, 409–420 (2020).
Koech, R. K., Mose, R., Kamunya, S. M. & Apostolides, Z. Combined linkage and association mapping of putative QTLs controlling black tea quality and drought tolerance traits. Euphytica 215, 1–19 (2019).
Lou, Q. et al. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66, 4749–4757 (2015).
Aubourg, S., Lecharny, A. & Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745 (2002).
Zhou, F. & Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. N. Phytol. 226, 1341–1360 (2020).
Chu, H. Y., Wegel, E. & Osbourn, A. From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J. 66, 66–79 (2011).
Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 227, 723–740 (2008).
Starks, C. M., Back, K., Chappell, J. & Noel, J. P. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820 (1997).
Cseke, L., Dudareva, N. & Pichersky, E. Structure and evolution of linalool synthase. Mol. Biol. Evol. 15, 1491–1498 (1998).
Krammer, G., Winterhalter, P., Schwab, M. & Schreier, P. Glycosidically bound aroma compounds in the fruits of Prunus species: apricot (P. armeniaca, L.), peach (P. persica, L.), yellow plum (P. domestica, L. ssp. Syriaca). J. Agr. Food Chem. 39, 778–781 (1991).
Huang, M. et al. Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol. 153, 1293–1310 (2010).
Roach, C. R., Hall, D. E., Zerbe, P. & Bohlmann, J. Plasticity and evolution of (+)-3-carene synthase and (−)-sabinene synthase functions of a sitka spruce monoterpene synthase gene family associated with weevil resistance. J. Biol. Chem. 289, 23859–23869 (2014).
Landmann, C. et al. Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Arch. Biochem. Biophys. 465, 417–429 (2007).
Jones, C. G. et al. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J. Biol. Chem. 286, 17445–17454 (2011).
Mockute, D. & Nivinskiene, O. The sabinene chemotype of essential oil of seeds of Daucus carota L. ssp. carota growing wild in lithuania. J. Essent. Oil Res. 16, 277–281 (2004).
Ngahang Kamte, S. L. et al. Identification of highly effective antitrypanosomal compounds in essential oils from the Apiaceae family. Ecotoxicol. Environ. Saf. 156, 154–165 (2018).
Muchlinski, A. et al. Diversity and function of terpene synthases in the production of carrot aroma and flavor compounds. Sci. Rep. 10, 1–14 (2020).
Budahn, H. et al. Mapping genes governing flower architecture and pollen development in a double mutant population of carrot. Front. Plant Sci. 5, 1–10 (2014).
Nothnagel, T., Ahne, R. & Straka, P. Morphology, inheritance and mapping of a compressed lamina mutant of carrot. Plant Breed. 124, 481–486 (2005).
Oetting, W. S. et al. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30, 450–458 (1995).
van Ooijen, J. W. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93, 343–349 (2011).
Cavagnaro, P. F. et al. A gene-derived SNP-based high resolution linkage map of carrot including the location of QTL conditioning root and leaf anthocyanin pigmentation. BMC Genomics 15, 1–17 (2014).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research 29, 2002–2007 (2001).
Acknowledgements
The financial support of the German Federal Ministry of Food and Agriculture (BMEL) is greatly acknowledged. The authors thank Dr. T. Nothnagel (JKI, Germany) for providing the carrot plant material and Dr. J. Keilwagen (JKI, Germany) for his help with bioinformatic analysis. We are also grateful to Karla Müller, Vicky Bartels and Tina Stein for excellent technical assistance.
Author information
Authors and Affiliations
Contributions
S.R., H.B., F.D. and L.K. designed the study. S.R., H.B., D.L. and D.U. performed the experiments. S.R., H.B., D.R. and D.L. analysed the data and SR prepared the figures. S.R., H.B., F.D. and L.K. wrote the manuscript. All authors read and approved the final manuscript. Open Access funding enabled and organized by Projekt DEAL.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Reichardt, S., Budahn, H., Lamprecht, D. et al. The carrot monoterpene synthase gene cluster on chromosome 4 harbours genes encoding flavour-associated sabinene synthases. Hortic Res 7, 190 (2020). https://doi.org/10.1038/s41438-020-00412-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00412-y