Abstract
Coconut (Cocos nucifera L.) is an important economic crop in tropical countries. However, the lack of a complete reference genome and the limitations of usable DNA markers hinder genomic studies and the molecular breeding of coconut. Here, we present the results of simple sequence repeat (SSR) mining from a high-throughput genotyping-by-sequencing (GBS) study of a collection of 38 coconut accessions. A total of 22,748 SSRs with di-, tri-, tetra-, penta- and hexanucleotide repeats of five or more were identified, 2451 of which were defined as polymorphic loci based on locus clustering in 38 coconut accessions, and 315 loci were suitable for the development of SSR markers. One hundred loci were selected, and primer pairs for each SSR locus were designed and validated in 40 coconut accessions. The analysis of 74 polymorphic markers identified between 2 and 9 alleles per locus, with an average of 3.01 alleles. The assessment of the genetic diversity and genetic relationships among the 40 coconut varieties based on the analysis of population structure, principal coordinate analysis (PCoA), and phylogenetic tree analysis using the 74 polymorphic SSR markers revealed three main groups of coconuts in Thailand. The identified SSR loci and SSR markers developed in this study will be useful for the study of coconut diversity and molecular breeding. The SSR mining approach used in this study could be applied to other plant species with a complex genome regardless of the availability of reference genome.
Similar content being viewed by others
Introduction
Coconut (Cocos nucifera L.) is one of the most important economic crops in many tropical countries1. It is regarded as the “tree of life” and a symbol of the tropics and presents economic value because of its myriad edible and inedible products. Coconut is the only species of the genus Cocos in the family Aracaceae (Palmaceae). It is a dioecious plant (2n = 2x = 32) with an ~2.4 Gb haploid genome2. Coconut is native to coastal areas of Melanesia and southeast Asia, probably Malaysia, Indonesia (Moluccas Islands), Philippines, and Papua New Guinea3. It is widely distributed in tropical and subtropical regions of the world in over 80 countries across Asia, Africa, America, and Oceania3. Based on plant morphology and breeding habits, coconut is classified into two ecotypes: “tall” (typica) and “dwarf” (nana)4. Tall coconuts are commonly grown for commercial purposes and can be divided into two major groups: Pacific and Indo-Atlantic5. Dwarf coconuts are native to the Pacific region and cultivated worldwide, typically near human dwellings1. Tall coconut palms are predominately outcrossing and exhibit varying degrees of heterozygosity, while dwarf palms are normally self-pollinating and present higher levels of homozygosity, showing common morphological characteristics such as dwarf stature due to short internodes and slow growth of height, a slender trunk, a smaller crown, and large numbers of relatively small nuts with a low copra content6. Traditionally, genetic diversity assessment in coconut is based on morphological trait characterization and coconut breeding is performed through conventional methods, which are laborious, time consuming, and inefficient due to environmental factors and the limited number of phenotypic markers available7,8. DNA markers could help to overcome these limitations, as they are abundant and highly polymorphic and are not influenced by the environment.
The development of DNA markers in coconut presents potential for application to molecular breeding through marker-assisted selection (MAS). A number of DNA markers have been used to characterize genetic diversity in coconut, such as restriction fragment length polymorphisms (RFLPs) that defined two genetically distinct groups of tall coconut palms9, randomly amplified polymorphic DNA (RAPD) markers that revealed a moderate level of genetic diversity of 17 distinct South Pacific populations10, amplified fragment length polymorphisms (AFLPs) that revealed more variation in tall varieties (typica), rather than intermediate (aurantiaca) and dwarf (nana) varieties11, and simple sequence repeats (SSRs) that also defined two subgroups within tall coconuts12,13. SSRs or microsatellites are tandem DNA repeats of 1–6 nucleotides per unit located mostly in noncoding regions of eukaryotic genomes14. SSRs are useful for developing DNA markers because they are abundant, highly polymorphic, multiallelic, and codominantly inherited15. Noncoding SSRs are widely used to analyze genetic diversity and population structure12,13,16,17, construct linkage maps18,19,20, and detect quantitative trait loci21,22,23, while SSRs located in coding and untranslated regions may be effective functional markers24. Although SSR markers have been proven to be useful in coconut research for decades, the number of validated SSR markers is currently limited.
Despite the many advantages of SSR markers, genomic SSR identification and subsequent marker conversion were once expensive and time-consuming techniques25. Due to the advent of next-generation sequencing technologies, genomic SSR mining and marker development are currently fast and inexpensive. Because of its adaptability, high-throughput genotyping-by-sequencing (GBS) has had a significant impact on plant breeding and genetic research26,27. This approach can provide accurate results regardless of the target species, and does not require previous genomic information. In this study, we identified SSRs and developed new SSR markers using coconut genomic sequencing data from Illumina GBS. A total of 2451 SSR loci were identified, and PCR primers for these loci were obtained. Three hundred and fifteen of these markers were proposed as a potential set of SSR markers based on marker diversity. One hundred pairs of PCR primers were synthesized for the selected loci and tested in 40 diverse coconut accessions; 74 markers were shown to be polymorphic. These markers were proven to be useful for evaluating the genetic diversity of coconut accessions in Thailand and could be applied to other coconut germplasms.
Results
GBS sequencing
For the 38 coconut accessions, ApeKI-GBS libraries were constructed, and 100-bp long GBS reads were subsequently generated. In these samples, the total GBS raw reads ranged from 8.71 million to 28.14 million, with total nucleotides ranging from 0.84 Gb to 2.72 Gb. GBS sequence analysis and microsatellite mining were carried out using a modified GBS analysis workflow without using a reference genome (Fig. 1). After filtering low-quality reads and reads that lacked enzyme cutting sites by the process_radtags component of Stacks v1.39 software28, the total number of clean reads retained in the 38 libraries ranged from 8.62 to 27.89 million, with an average of 16.41 million. The total clean-read nucleotides ranged from 0.83 Gb to 2.69 Gb, with an average of 1.59 Gb, equivalent to 0.66x coconut genome coverage (2.42 Gb) (Table 1). The total number of filtered reads of all 38 samples was ~624 million reads, and the number of unique reads after reduction by ustacks was ~21.6 million reads. All the unique sequences were incorporated into sequence catalogs using cstacks, resulting in 3,644,337 consensus locus sets.
Coconut SSR locus identification and the frequency and distribution of SSRs
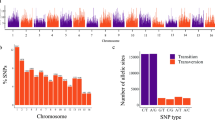
We used the MIcroSAtellite identification tool (MISA)29 with the default setting to identify SSR-containing regions among the 21,599,434 unique sequences. The search for perfect SSR-containing regions was restricted to motifs of di-, tri-, teta-, penta-, and hexanucleotides. As a result, a total of 204,068 sequences containing 22,748 loci of SSR motifs were identified (Fig. 1). This collection of SSR loci consisted of 15,165 dinucleotide repeats (66.67%), 6570 trinucleotide repeats (28.88%), 659 tetranucleotide repeats (2.90%), 116 pentanucleotide repeats (0.51%) and 238 hexanucleotide repeats (1.05%) (Table 2). Dinucleotide repeats were identified as the most abundant microsatellite class (15,165 regions), followed by trinucleotide (6570) and tetranucleotide repeats (659). The assessment of the nucleotide composition of the repeat motifs of the two most abundant classes (dinucleotide repeats and trinucleotide repeats) revealed that the most frequent type of dinucleotide repeat was AG/CT (10,408 motifs), representing 68.63% of the total dinucleotides, while the most common type of trinucleotide repeat was CCG/CGG (1663 motifs), representing 25.31% of the trinucleotides (Fig. 2).
Coconut SSR primer design and marker validation
For each individual coconut accession, we used VSEARCH30 to build SSR locus clusters and used a custom Python script to generate an allele frequency table and identify polymorphic SSR loci. As a result, a total of 2451 putative polymorphic SSR loci were identified, and flanking primers for each locus were subsequently designed by using BatchPrimer3 (version 2.3.7)31 (Supplementary Information; Table S1). Among these sequences, 315 were selected as suitable for SSR primer design based on the following criteria: (1) the expected PCR product size was 80–100 bp, and (2) the selected loci were supported by GBS sequences from at least 20 coconut accessions. (Supplementary Information; Table S1). We randomly selected 100 of those 315 primer pairs to be validated by PCR. These 100 SSR loci included 74 dinucleotide repeats, 21 trinucleotide repeats, two tetranucleotide repeats, two pentanucleotide repeats, and one hexanucleotide repeat (Supplementary Information; Table S2).
To validate the efficacy of the newly developed SSR markers, 40 diverse coconut accessions, including both tall and dwarf coconut palms, were used in the analysis with the 100 SSR markers (Supplementary Information; Table S3). Based on the polyacrylamide gel electrophoresis (PAGE) results, we found that 74 out of 100 SSR markers clearly showed polymorphic patterns, presenting consistent, interpretable amplified products (Supplementary Information; Fig. S1). Among these markers, the SSR markers CnSSR5, CnSSR9, CnSSR16, and CnSSR28 were the five most polymorphic, exhibiting 5, 9, 6, and 7 alleles, respectively, in the 40 coconut genotypes (Fig. 3). An allele of the markers CnSSR16 (94 bp) was probably shared among Thai dwarf coconut accessions. Considering tall and dwarf accessions separately, all 74 SSR markers were polymorphic among all tall coconut accessions, but only 52 of them were polymorphic among the 18 dwarf coconut accessions (Table 3).
Estimation of the genetic diversity and population structure of coconuts in Thailand using the newly developed SSR markers
To evaluate the utility of the newly developed SSR markers, we used these markers to characterize the genetic diversity of a collection of 40 coconut accessions consisting of 35 accessions of coconuts grown in Thailand collected from different locations across the country, and the other five accessions were foreign varieties present in the country (Supplementary Information; Table S3). The results of the genotyping analysis of the 40 coconut accessions with the 74 polymorphic SSR markers revealed 223 alleles. The number of observed alleles for all polymorphic SSR markers among all 40 coconut accessions ranged from 2 to 9, with an average of 3.01 alleles per locus; that among tall accessions ranged from 2 to 9, with an average of 2.95; and that among dwarf accessions ranged from 1 to 4, with an average of 1.99 alleles per locus. The estimated polymorphism information content (PIC) of the 74 markers for all 40 coconut accessions ranged from 0.05 to 0.68, with an average of 0.31; that for tall coconut accessions ranged from 0.04 to 0.74, with an average of 0.36; and that for dwarf coconut accessions ranged from 0 to 0.53, with an average of 0.17 (Table 3). The gene diversity (expected heterozygosity: He) for all coconut accessions ranged from 0.05 to 0.73, with an average of 0.35; that for tall coconut accessions ranged from 0.04 to 0.78, with an average of 0.42; and that for dwarf coconut accessions ranged from 0 to 0.60, with an average of 0.20. The observed heterozygosity (Ho) for all coconut accessions ranged from 0 to 0.58, with an average of 0.19; that for tall coconut accessions ranged from 0 to 0.62, with an average of 0.26; and that for dwarf coconut accessions ranged from 0 to 0.56, with an average of 0.10. We found that the CnSSR9 marker yielded the highest number of alleles (9 alleles), with a PIC of 0.57 (Table 3). Considering the heterozygous genotypes of each individual among the 40 accessions, the heterozygosity of dwarf accessions ranged from 1.35 to 31.08%, and that of tall accessions ranged from 15.05 to 47.95% (Supplementary Information; Table S4). The accessions exhibiting the lowest and highest heterozygosity among the dwarf accessions were ACC.34 (Ratchaburi 3) and ACC.44 (Khom), respectively. The accessions exhibiting the lowest and highest heterozygosity among the tall accessions were ACC.21 (Thailand tall Sawi #2) and ACC.24 (MaWa), respectively.
The genetic distance based on a dissimilarity matrix calculated from the 223 SSR alleles between each pair of the 40 coconut accessions ranged from 0.04 to 0.96, with an average of 0.35 (Fig. 4). The genetic distance within the group of dwarf coconut accessions was markedly lower than that within the group of tall coconut accessions, as the genetic distance of each pair among the dwarf accessions ranged from 0.04 to 0.30, with an average of 0.18, and that among the tall accessions ranged from 0.16 to 0.96, with an average of 0.42. The most distant accessions based on the dissimilarity matrix were ACC.08 (West African tall), ACC.10 (Thalai Roi) and ACC.12 (Pak Chok #2), followed by ACC.52 (Phuang Roi Si Thong), ACC.19 (Sri Lanka tall), ACC.4 (Pak Chok #1), and ACC.24 (MaWa: Malayan Yellow dwarf x West African tall).
Based on the independent STRUCTURE analysis of the 40 accessions, the maximum delta K was detected at K = 3, indicating three subgroups (Fig. 5 and Supplementary Information; Fig. S2). For each K-value, genotypes with membership probability >60% were assigned to the same group, while those with < 60% probability in any group were assigned as “admixed”32. The three clusters of subpopulations among the 40 coconut accessions identified by STRUCTURE analysis were similar to those revealed by phylogenetic tree analysis (Fig. 5). Cluster I contained seven exclusively tall accessions, including the Thailand tall accessions ACC.04 (Pak Chok #1), ACC.10 (Thalai Roi), ACC.12 (Pak Chok #2) and ACC.52 (Phuang Roi Si Thong); two foreign tall accessions, ACC.08 (West African tall) and ACC.19 (Sri Lanka tall); and a hybrid accession, ACC.24 (MaWa). Among these, four accessions (ACC.08, ACC.10, ACC.12, and ACC.19) had pure genotypes, the other two (ACC.04 and ACC.52) exhibited mixed genotypes from Cluster I and Cluster III, and another accession (ACC.24) exhibited mixed genotypes from Cluster I and Cluster II.
Cluster II contained 18 coconut accessions, including 14 Thailand tall accessions, ACC.26 (Thailand tall Ko Samui), ACC.37 (Thailand tall Ko Chang), ACC.46 (Tha Nan), ACC.50 (Mu Si Mo), ACC.45 (Maprao So #2), ACC.51 (Maphrao Fai), ACC.18 (Thailand tall Thap Sakae), ACC.20 (Thailand tall Sawi #1), ACC.22 (Thailand tall Sawi #3), ACC.17 (Thailand tall Nakhon Si Thammarat), ACC.48 (Maphrao Fai Kathi), ACC.09 (Kalok), ACC.21 (Thailand tall Sawi #2), ACC.27 (Thailand tall Sawi #3), one foreign tall accession, ACC.11 (Tahiti tall), and three dwarf accessions, including two Thailand dwarf coconut accessions, ACC.44 (Khom) and ACC.15 (Nok Khum), and one foreign dwarf coconut, ACC.06 (Cameroon yellow dwarf). Among these, six accessions (ACC.17, ACC.18, ACC.22, ACC.27, ACC.48, and ACC.51) had pure genotypes. The rest of the accessions in this cluster mostly exhibited mixed genotypes from Cluster II and Cluster III, except for two accessions (ACC.11 and ACC.21), which exhibited mixed genotypes from all three clusters.
Cluster III was a homogenous genetic group of dwarf accessions containing 15 dwarf accessions, including 14 Thailand dwarf accessions, ACC.02 (Nam Wan #1), ACC.03 (Thung Kled), ACC.13 (Mu Si Som), ACC.16 (Nali-ke), ACC.35 (Ratchaburi 1), ACC.33 (Ratchaburi 2), ACC.34 (Ratchaburi 3), ACC.36 (Nam Wan #2), ACC.40 (Nam Hom #2), ACC.41 (Pathiu), ACC.42 (Nam Wan #3), ACC.43 (Nam Hom Kathi), ACC.47 (Nam Hom #3) and ACC.49 (Nam Wan #4), and one foreign dwarf coconut accession, ACC.05 (Papua New Guinea brown dwarf). Among these, ten accessions (ACC.03, ACC.05, ACC.13, ACC.16, ACC.35, ACC.36, ACC.40, ACC.41, ACC.42, and ACC.47) had pure genotypes. The rest of the accessions in this cluster (ACC.02, ACC.33, ACC.34, and ACC.43) exhibited mixed genotypes from Cluster II and Cluster III. The accession ACC.49 was considered admixed as the value of membership probability revealed by STRUCTURE was less than the threshold of 0.60.
The three clusters were also supported by principal coordinate analysis (PCoA) as analyzed by DARwin 6.033software with 74 SSR markers. The total proportions of the variation explained by the first and second principal components were 38.54% and 9.30%, respectively (Fig. 6). Cluster I, exclusively containing tall coconuts, was clearly separated from the other two clusters. The number of SSR markers could be reduced to 49 to evaluate PCoA and achieve the same results (Supplementary Information; Fig. S3, Table S5). Discriminant Analysis of Principal Component (DAPC) was also performed using the adegenet package34 to cluster 40 coconut accessions. The results obtained from DAPC analysis supported the PCoA analysis (Supplementary Information; Fig. S4). We also performed a Mantel test using GenAlEx (6.51)35 to check the occurrence of a positive correlation (r > 0) between the Nie’s genetic distance and geographic distances among the 40 genotypes. As a result, the genetic and geographic distance were not significantly correlated (r = 0.12, p = 0.13; Supplementary Information; Fig. S5).
The x- and y-axes are indicated by the first and second coordinates, respectively, and the values show the percentages of variation explained. Circles and triangles indicate dwarf and tall accessions, respectively. Each cluster is indicated by an oval. Scree plot of PCoA is provided at the bottom of the figure.
Discussion
Coconut (Cocos nucifera) is a plant species with a large genome size (>2 Gb) in which no complete reference genome has been released. Therefore, studies of coconut genomics using a genome resequencing-based approach that requires a complete reference genome are limited. Microsatellites (SSRs) are currently markers of choice for several types of genetic diversity studies in coconut36,37,38,39,40. SSR loci have been conventionally identified through the laborious technique of genomic hybridization. The SSR markers developed based on this strategy include 296 validated SSR markers available at Tropgenedb (tropgenedb.cirad.fr). The SSR markers from this source have been frequently used in coconut genetic diversity studies6,13,41. Alternatively, with the advancement of next-generation sequencing, NGS-based approaches for identifying a large number of SSR loci have been proposed; for example, RNA sequencing (RNA-seq) analysis identified 6608 gene-based SSR loci from 57,304 unigenes8. From these loci, 309 primer pairs were developed, 191 of which were polymorphic across a set of ten coconut accessions. Recently, a total of 7139 genome-wide SSR markers were designed on the basis of a whole-genome assembly of coconut42. However, the validation of these markers was not reported. In this study, we applied a genotyping-by-sequencing (GBS) approach to generate DNA sequences from 38 different coconut accessions and subsequently identified coconut SSR loci from these GBS sequences. The restriction enzyme ApeKI, used in this study, has been widely used in the preparation of GBS libraries in several crops because it can produce an appropriate number of fragments for sequencing43,44. In addition, because of the methylation sensitivity of this enzyme, it can eliminate fragments generated from repetitive methylated genomic regions in the coconut genome, as coconut has been estimated to contain ~73% of repetitive sequences2. In fact, the ApeKI enzyme has been documented to show bias towards the coding areas. As a result, we identified 22,748 SSR loci, and flanking primers were designed for 2451 of them. A final set of 100 SSR loci were employed to develop SSR markers, and the markers were validated using 40 diverse coconut accessions collected from different parts of Thailand. Seventy-four of these markers were polymorphic among the 40 coconut accessions. The most abundant class of SSRs identified from GBS sequences in this study was dinucleotide repeats. Similar results regarding abundant classes of coconut SSRs have been reported previously42. The most frequently identified type of dinucleotide repeat in the present study was AG/CT, similar to that identified for gene-based SRRs in a previous report8. However, this dinucleotide category was different from that identified from a whole-genome assembly, which was AT/TA42. Our newly developed SSR markers were tested for their effectiveness by using them to assess the genetic diversity of a collection of 40 coconut accessions. PIC values of these SSR markers ranged from 0.05 to 0.68. The majority of the SSR markers (64 markers) displayed low to moderate PIC values (PIC values < 0.50; Table 3). Low PIC values in these SSR markers due to low levels of polymorphism among the genotypes evaluated (Supplementary Information; Fig. S1). Gene diversity (He) based on the 74 SSR markers was observed to be higher in tall coconut accessions than in dwarf accessions, as the average gene diversity values in tall coconut and dwarf coconut accessions were 0.42 and 0.20, respectively. These values were lower than the overall mean gene diversity values previously reported for world coconut populations, which were 0.70 and 0.37 for tall and dwarf coconuts, respectively12. Similar to what was reflected by the observed gene diversity, the overall allelic richness of the dwarf accessions was lower than that of the tall accessions. The dwarf accessions included in the present study were less heterozygous; ten out of the 18 accessions were heterozygous at one to ten loci. However, there were three dwarf accessions (ACC.06 (Cameroon yellow dwarf), ACC.15 (Nok Khum), and ACC.44 (Khom)) that were substantially heterozygous. According to the PCoA analysis, these three dwarf accessions likely clustered together with the other five tall accessions, i.e., ACC.09 (Kalok), ACC.11 (Tahiti tall), ACC.26 (Thailand tall Ko Samui), ACC.24 (MaWa) and ACC.37 (Thailand tall Ko Chang). Moreover, according to the STRUCTURE results, the three dwarf accessions exhibited mixed genotypes from Cluster II (mainly tall accessions) and Cluster III (dwarf accessions). Therefore, it is possible that these three dwarf accessions with high heterozygosity were derived from outcrosses between dwarf and tall coconuts. It might also be the case that some other tall and dwarf coconut accessions of Thailand were cross-pollinated or crossbred between the two groups. It is interesting that a foreign dwarf accession, ACC.05 (Papua New Guinea brown dwarf), clustered together with other Thailand dwarf accessions in Cluster III. The SSR markers clearly divided the tall accessions into two groups as in Cluster I and Cluster II. This indicates that two classes of Thai tall coconut varieties derived from two different origins. In comparison, Cluster II included all Thai tall varieties in the Pacific region and was closer to Cluster III (dwarf accessions) than Cluster I, which included Thai tall varieties from Thailand’s Indian Ocean coast. This result confirms the previous findings12. Based on the analysis of 94 coconut varieties/populations, also comprising four tall and seven dwarf coconut varieties from Thailand, Perera et al. identified two main groups of coconut genotypes, one group comprising all the Talls from Southeast Asia, the Pacific, the west cost of Panama and all Dwarf, and another group comprising all Talls from south Asia, Africa, and the Indian Ocean coast. With more Thai coconut accessions and more SSR markers, our findings expanded previous knowledge on genetic diversity and coconut population structure in Thailand.
The method of SSR mining using GBS sequences applied in this study has proven to be efficient in coconut and could be applied for other tree plants with a large genome size or those without a complete genome reference. The newly developed and validated SSR markers could be useful for genetic diversity studies, which are an essential component of the characterization and utilization of coconut germplasms and are useful for the molecular breeding of coconuts.
Materials and methods
Plant materials
A total of 51 coconut accessions collected from plantations in different locations in Thailand were used in this study. Among these coconut accessions, 38, comprising 20 tall accessions and 18 dwarf accessions, were used to generate GBS sequence data. Forty coconut accessions were used to validate SSR markers. Among these accessions, 27 overlapped with those used in GBS sequencing. (Supplementary Information; Table S1).
DNA extraction, GBS library preparation, and sequencing
Genomic DNA was isolated from 100 mg of young leaf tissue using the DNeasy Plant Mini Kit (Qiagen, USA) following the manufacturer’s instructions. The DNA of each sample was quantified using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, USA) and adjusted to 50 ng/μl. GBS libraries for each genotype were generated from 50 μl of genomic DNA using the restriction enzyme ApeKI following the GBS protocol as previously described43. GBS sequencing was performed using the Illumina HiSeq 2000 platform at the Beijing Genomics Institute (BGI, Shenzen, China).
GBS data processing and microsatellite mining
The Illumina raw reads were preprocessed by simultaneously demultiplexing and removing low-quality reads using the process_radtags component of Stacks v1.39 software28. Reads that did not contain the restriction site of ApeKI were also discarded in the preprocessing step. The preprocessed clean reads were then analyzed using the core Stack pipeline containing the ustacks and cstacks components of Stacks v.1.39 software with default parameters and without using a reference genome. Each consensus sequence resulting from the Stack pipeline was then screened for simple sequence repeats (SSRs) using MISA with default parameters29. The acquired SSRs were considered to only represent those containing perfect repeats of SSRs whose basic motifs ranged from 2 to 6 bp with defined minimum repeat units of six iterations for dinucleotide repeats and five iterations for tri-, tetra-, penta-, and hexanucleotide repeats.
Coconut SSR primer design
The BatchPrimer3 (version 2.3.7) program31 was employed to design oligonucleotide primers using the SSR flanking sequences. Putative SSR markers were selected based on the following parameters: primer length between 18–23 bp, PCR product length between 50 and 100 bp, primer melting temperature (Tm) between 52–60 °C with an optimum of 55 °C, and GC content of 30–67%. All putative markers were computationally validated across 38 genotypes to verify their polymorphism. Polymorphic SSR loci were defined as SSR polymorphisms present in the consensus sequences of the supporting accessions (at least 20 accessions).
PCR validation of SSR markers
A total of 100 SSR primer pairs were selected for PCR screening in 40 diverse coconut accessions. PCR amplification was conducted in reaction mixtures with a total volume of 10 µl containing 2 µl of 20 ng/µl genomic DNA, 5X KAPA2G Buffer B with MgCl2, 5 U/μl of KAPA2G Robust HotStart DNA Polymerase (KAPA2G Robust HotStart PCR Kits, Kapa Biosystems, USA), 2 mM MgCl2, each SSR primer at 0.5 µM and 0.2 µM dNTP mix. Briefly, the PCR cycles consisted of initial denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, primer annealing at a temperature depending on the primers for 30 s, and extension at 72 °C for 90 s, with a final extension at 72 °C for 10 min. The PCR products were electrophoretically separated on 4.5% denaturing polyacrylamide gels and visualized by silver staining. The genotypes characterized according to each marker were determined by allelic size differences in comparison to a 100 bp DNA ladder.
SSR data analysis
We examined the genetic diversity of the 40 coconut accessions using the newly developed SSR markers. The individual bands amplified by the SSR primers in the SSR banding profile were scored as present (1) or absent (0). PowerMarker version 3.2545 was used to calculate polymorphic information content (PIC), the numbers of alleles, gene diversity, and the major allele frequency (MAF) and to construct the neighbor-joining phylogram using Nei’s distance dissimilarity matrix46. The STRUCTURE algorithm47 was run using a model with admixture and correlated allele frequencies, with 3 independent replicates run for each genetic cluster (K) value, with K ranging from 1 to 8, using a burn-in of 100,000 steps and a run length of 100,000 Markov Chain Monte Carlo (MCMC) iterations. Ln(PD) values were derived for each K and plotted to identify the plateau of ΔK48. The final population structure was calculated using the web-based software STRUCTURE HARVESTER version 0.6.9249. Individuals were placed into the respective subpopulation based on the highest percentage of membership (q). Principal coordinate analysis (PCoA) was performed using DARwin 6.0 software33 based on dissimilarity distances estimated between pairs of individuals. The exploratory Discriminant Analysis of Principal Components (DAPC) was applied using the adegenet package34 (function dapc). The analysis was performed without prior information on individual populations. The optimal number of clusters for assessing the best supported model was selected based on the Bayesian Information Criterion (BIC), as suggested by Jombart et al.50. The correlation between pairwise genetic distance and geographic distance among each Thai coconut accession was performed using a Mantel test51 as implemented in the GenAlEx 6.435.
Data availability
The GBS sequencing data used in this study can be freely and openly accessed at the NCBI Sequence Read Archive via the identifier PRJNA645608.
References
Gunn, B. F., Baudouin, L. & Olsen, K. M. Independent origins of cultivated coconut (Cocos nucifera L.) in the old world tropics. PLoS ONE 6, e21143 (2011).
Xiao, Y. et al. The genome draft of coconut (Cocos nucifera). Gigascience 6, 1–11 (2017).
Chan, E. & Elevitch, C. R. Cocos nucifera (coconut). Species profiles Pac. Isl. Agrofor. 2, 1–27 (2006).
Menon K. P. V., Pandalai K. M. The Coconut. A Monograph. (Indian Central Coconut Committee, 1958).
Lebrun, P. Berger, A. Hodgkin, T. Baudouin, L. Biochemical and molecular methods for characterizing coconut diversity. In: Batugal, P. Ramanatha Rao, V. Oliver, J. (eds) Coconut genetic resources. International Plant Genetic Resources Institute, Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, Selangor DE, Malaysia, pp 225–245 (2005).
Kamaral, L. C. J., Perera, S. A. C. N., Perera, K. L. N. S. & Dassanayaka, P. N. Genetic diversity of the Sri Lanka yellow dwarf coconut form as revealed by microsatellite markers. Trop. Agric. Res. 26, 131 (2015).
Kamaral, L. C. J., Perera, S., Perera, K. & Dassanayaka, P. N. Characterisation of Sri Lanka Yellow Dwarf Coconut (Cocos nucifera L.) by DNA fingerprinting with SSR markers. J. Natl Sci. Found. 45, 405 (2017).
Xia, W. et al. Development of gene-based simple sequence repeat markers for association analysis in Cocos nucifera. Mol. Breed. 34, 525–535 (2014).
Lebrun, P., N’cho, Y. P., Seguin, M., Grivet, L. & Baudouin, L. Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 101, 103–108 (1998).
Ashburner, G. R., Thompson, W. K. & Halloran, G. M. RAPD analysis of south pacific coconut palm populations. Crop Sci. 37, 992–997 (1997).
Perera, L., Russell, J. R., Provan, J., McNicol, J. W. & Powell, W. Evaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling. Theor. Appl. Genet. 96, 545–550 (1998).
Perera, L., Russell, J. R., Provan, J. & Powell, W. Studying genetic relationships among coconut varieties/populations using microsatellite markers. Euphytica 132, 121–128 (2003).
Geethanjali, S., Anitha Rukmani, J., Rajakumar, D., Kadirvel, P. & Viswanathan, P. L. Genetic diversity, population structure and association analysis in coconut (Cocos nucifera L.) germplasm using SSR markers. Plant Genet. Resour.: Charact. Utilization 16, 156–168 (2018).
Grover, A., Aishwarya, V. & Sharma, P. C. Searching microsatellites in DNA sequences: approaches used and tools developed. Physiol. Mol. Biol. Plants 18, 11–19 (2012).
Powell, W., Machray, G. C. & Provan, J. Polymorphism revealed by simple sequence repeats. Trends plant Sci. 1, 215–222 (1996).
Guan, R. et al. Genetic diversity comparison between Chinese and Japanese soybeans (Glycine max (L.) Merr.) revealed by nuclear SSRs. Genet. Resour. Crop Evol. 57, 229–242 (2010).
Ali, M. L. et al. Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Mol. Breed. 21, 497–509 (2008).
Shoemaker, R. C. et al. Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome 51, 294–302 (2008).
Stàgel, A., Portis, E., Toppino, L., Rotino, G. L. & Lanteri, S. Gene-based microsatellite development for mapping and phylogeny studies in eggplant. BMC Genomics 9, 357 (2008).
Lebrun, P. et al. Construction of a linkage map of the Rennell Island Tall coconut type (Cocos nucifera L.) and QTL analysis for yield characters. Genome 44, 962–970 (2001).
Baudouin, L. et al. QTL analysis of fruit components in the progeny of a Rennell Island Tall coconut (Cocos nucifera L.) individual. Theor. Appl. Genet. 112, 258–268 (2006).
Neeraja, C. N. et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 115, 767–776 (2007).
Danson, J., Lagat, M., Kimani, M. & Kuria, A. Quantitative trait loci (QTLs) for resistance to gray leaf spot and common rust diseases of maize. Afr J Biotechnol. 7, 3247–3254 (2008).
Vieira, M. L. C., Santini, L., Diniz, A. L., Munhoz, C. & de, F. Microsatellite markers: what they mean and why they are so useful. Genet Mol. Biol. 39, 312–328 (2016).
Squirrell, J. et al. How much effort is required to isolate nuclear microsatellites from plants? Mol. Ecol. 12, 1339–1348 (2003).
Poland, J. A. & Rife, T. W. Genotyping-by-sequencing for plant breeding and genetics. The plant genome. Journal 5, 92 (2012).
Chung, Y. S., Choi, S. C., Jun, T.-H. & Kim, C. Genotyping-by-sequencing: a promising tool for plant genetics research and breeding. Hortic. Environ. Biotechnol. 58, 425–431 (2017).
Catchen, J. M., Amores, A., Hohenlohe, P., Cresko, W. & Postlethwait, J. H. Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 1, 171–182 (2011).
Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585 (2017).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
You, F. M. et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinforma. 9, 253 (2008).
Yang, X. et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 28, 511–526 (2011).
Perrier X., Jacquemoud-Collet J. P. DARwin software. 2006. 2015.
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Peakall, R. O. D. & Smouse, P. E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 (2006).
Liu, X., Tang, H., Li, D. & Hou, L. Genetic diversity of coconut cultivars in China by microsatellite (SSR) markers. Mol. Plant Breeding. 2, 83–91 (2011).
Loiola, C. M. et al. Genetic relationships among tall coconut palm (Cocos nucifera L.) accessions of the international coconut genebank for Latin America and the Caribbean (ICG-LAC), Evaluated using microsatellite markers (SSRs). PLoS ONE 11, e0151309 (2016).
Martinez, R. T., Baudouin, L., Berger, A. & Dollet, M. Characterization of the genetic diversity of the Tall coconut (Cocos nucifera L.) in the Dominican Republic using microsatellite (SSR) markers. Tree Genet. Genomes 6, 73–81 (2010).
Meerow, A. W. et al. Analysis of genetic diversity and population structure within Florida coconut (Cocos nucifera L.) germplasm using microsatellite DNA, with special emphasis on the Fiji Dwarf cultivar. Theor. Appl. Genet. 106, 715–726 (2003).
Perera, L., Russell, J. R., Provan, J. & Powell, W. Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 43, 15–21 (2000).
Teulat, B. et al. An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. TAG Theor. Appl. Genet. 100, 764–771 (2000).
Lantican, D. V. et al. De novo genome sequence assembly of dwarf coconut (Cocos nucifera L.‘Catigan Green Dwarf’) provides insights into genomic variation between coconut types and related palm species. G3: Genes, Genomes. Genetics 9, 2377–2393 (2019).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379 (2011).
Lu, F. et al. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9, e1003215 (2013).
Liu, K. & Muse, S. V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129 (2005).
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292 (1972).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Earl, D. A. & vonHoldt, B. M. Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012).
Jombart, T., Pavoine, S., Devillard, S. & Pontier, D. Putting phylogeny into the analysis of biological traits: a methodological approach. J. Theor. Biol. 264, 693–701 (2010).
Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 (1967).
Acknowledgements
This work was supported by Biodiversity-based Economy Development Office (Public Organization) and National Research Council of Thailand, grant number BEDO-NRCT 48/2562.
Author information
Authors and Affiliations
Contributions
S.W., S.A., T.T., and A.V. conceived and designed the experiment. K.R., W.A., P.N., T.K., and C.S. conducted the experiments. K.R., S.W., W.A., V.R., and C.S. analyzed the data. K.R., S.W., and S.A. wrote the manuscript. K.R., S.W., and S.A. revised the final version of the paper. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Riangwong, K., Wanchana, S., Aesomnuk, W. et al. Mining and validation of novel genotyping-by-sequencing (GBS)-based simple sequence repeats (SSRs) and their application for the estimation of the genetic diversity and population structure of coconuts (Cocos nucifera L.) in Thailand. Hortic Res 7, 156 (2020). https://doi.org/10.1038/s41438-020-00374-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-00374-1
This article is cited by
-
Genetic diversity of kale (Brassica oleracea L. var acephala) using agro-morphological and simple sequence repeat (SSR) markers
Genetic Resources and Crop Evolution (2024)
-
Genome-wide diversity analysis to infer population structure and linkage disequilibrium among Colombian coconut germplasm
Scientific Reports (2022)
-
Development and characterization of non-coding RNA-derived simple sequence repeat markers in coconut (Cocos nucifera L.)
Functional & Integrative Genomics (2022)
-
Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions
Scientific Reports (2021)