Abstract
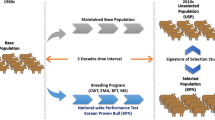
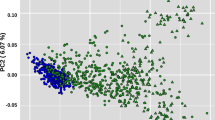
The Lori-Bakhtiari fat-tailed sheep is one of the most important heavyweight native breeds of Iran. The breed is robust and well-adapted to semi-arid regions and an important resource for smallholder farms. An established nucleus-based breeding scheme is used to improve their production traits but there is an indication of inbreeding depression and loss of genetic diversity due to selection. Here, we estimated the inbreeding levels and the distribution of runs of homozygosity (ROH) islands in 122 multi-generational female Lori-Bakhtiari from different half-sib families selected from a breeding station that were genotyped on the 50k array. A total of 2404 ROH islands were identified. On average, there were 19.70 ± 1.4 ROH per individual ranging between 6 and 41. The mean length of the ROH was 4.1 ± 0.14 Mb. There were 1999 short ROH of length 1–6 Mb and another 300 in the 6–12 Mb range. Additionally long ROH indicative of inbreeding were found in the ranges of 12–24 Mb (95) and 24–48 Mb (10). The average inbreeding coefficient (FROH) was 0.031 ± 0.003 with estimates varying from 0.006 to 0.083. Across generations, FROH increased from 0.019 ± 0.012 to 0.036 ± 0.007. Signatures of selection were identified on chromosomes 2, 6, and 10, encompassing 55 genes and 23 QTL associated with production traits. Inbreeding coefficients are currently within acceptable levels but across generations, inbreeding is increasing due to selection. The breeding program needs to actively monitor future inbreeding rates and ensure that the breed maintains or improves on its current levels of environmental adaptation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Alberto FJ, Boyer F, Orozco-terWengel P, Streeter I, Servin B, De Villemereuil P et al. (2018) Convergent genomic signatures of domestication in sheep and goats. Nat Commun 9:813
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Bijma P (2000) Long-term Genetic Contributions: Predictions of Rates of Inbreeding and Genetic Gain in Selected Populations. Wageningen University, Veenendaal, the Netherlands
Biscarini F, Cozzi P, Gaspa G, Marras G (2018) detectRUNS: Detect runs of homozygosity and runs of heterozygosity in diploid genomes. CRAN. The Comprehensive R Archive Network
Burren A, Neuditschko M, Signer-Hasler H, Frischknecht M, Reber I, Menzi F et al. (2016) Genetic diversity analyses reveal first insights into breed-specific selection signatures within Swiss goat breeds. Anim Genet 47:727–39.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 25; 4:7.
Ciani E, Crepaldi P, Nicoloso L, Lasagna E, Sarti FM, Moioli B et al. (2014) Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim Genet 45:256–266
Collier RJ, Baumgard LH, Zimbelman RB, Xiao Y (2019) Heat stress: physiology of acclimation and adaptation. Anim Front 9(1):12–19
Dixit SP, Singh S, Ganguly I, Bhatia AK, Sharma A, Kumar NA et al. (2020) Genome-wide runs of homozygosity revealed selection signatures in Bos indicus. Front Genet 11:92
Dzomba EF, Chimonyo M, Pierneef R, Muchadeyi FC (2021) Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genomics 22:7
Eydivandi S, Sahana G, Momen M, Moradi M, Schönherz A (2020) Genetic diversity in Iranian indigenous sheep vis-à-vis selected exogenous sheep breeds and wild mouflon. Anim Genet 51:772–787
FAO (1998) Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans. United Nations, Rome, Italy.
Forutan M, Ansari Mahyari S, Baes C, Melzer N, Schenkel FS, Sargolzaei M (2018) Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genomics 19:98
Ghernouti N, Bodinier M, Ranebi D, Maftah A, Petit D, Gaouar SBS (2017) Control Region of mtDNA identifies three migration events of sheep breeds in Algeria. Small Rumin Res 155:66–77.
Gibson J, Morton NE, Collins A (2006) Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 15:789–795
Huang DW, Sherman BT, Lempicki RA (2009a) Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4(1):44–57
Huang DW, Sherman BT, Lempicki RA (2009b) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13
Keshavarzpour M, Bahreini Behzadi MR, Muhaghegh, Dolatabadi M (2018) Pedigree analysis and inbreeding investigation in Lori-Bakhtiari Sheep. Iran J Anim Sci Res 9:376–386
Lawson Handly LJ, Byrne K, Santucci F, Townsend S, Taylor M, Bruford MW et al. (2007) Genetic structure of European sheep breeds. Heredity 99:620–631
Leroy G (2014) Inbreeding depression in livestock species: review and meta-analysis. Anim Genet 45:618–628
Marras G, Gaspa G, Sorbolini S, Dimauro C, Ajmone‐Marsan P, Valentini A et al. (2015) Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim Genet 46:110–121
Mastrangelo S, Tolone M, Sardina MT, Sottile G, Sutera AM, Di Gerlando R et al. (2017) Genome‑wide scan for runs of homozygosity identifies potential candidate genes associated with local adaptation in Valle del Belice sheep. Genet Sel Evol 49:84
Nascimento AV, Cardoso DF, Santos DJA, Romero ARS, Scalez DCB, Borquis RRA et al. (2021) Inbreeding coefficients and runs of homozygosity islands in Brazilian water buffalo. J Dairy Sci 104(2):1917–1927
Nosrati M, Asadollahpour Nanaei H, Javanmard A, Esmailizadeh A (2021) The pattern of runs of homozygosity and genomic inbreeding in world-wide sheep populations. Genomics 113:1407–1415
Pasqui M, Di Giuseppe E (2019) Climate change, future warming, and adaptation in Europe. Anim Front 9(1):6–11
Pedrosa S, Arranz JJ, Brito N, Molina A, San Primitivo F, Bayón Y (2007) Mitochondrial diversity and the origin of Iberian sheep. Genet Sel Evol 39:91
Pereira F, Davis SJM, Pereira L, McEvoy B, Bradley DG, Amorim A (2006) Genetic signatures of a Mediterranean Influence in Iberian Peninsula Sheep Husbandry. Mol Biol Evol 23(7):1420–1426
Peter C, Bruford M, Perez T, Dalamitra S, Hewitt G, Erhardt G, the ECONOGENE Consortium (2007) Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim Genet 38:37–44
Purcell SN, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J et al. (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis [Online]. Am J Hum Gen 81:559–575
Purfield DC, Berry DP, McParland S, Bradley DG (2012) Runs of homozygosity and population history in cattle. BMC Genet 13:70
Purfield DC, McParland S, Wall E, Berry DP (2017) The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS ONE 12:e0176780
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Rashedi Dehsahraei A, Fayazi J, Vatankhah M (2013) Investigating inbreeding trend and its impact on growth traits of Lori-Nakhtiari sheep. J Rumin Res 1:65–78
Sheikhlou M, Abbasi MA (2016) Genetic diversity of Iranian Lori-Bakhtiari sheep assessed by pedigree analysis. Small Rumin Res 141:99–105
Signer-Hasler H, Burren A, Ammann P, Drögemüller C, Flury C (2019) Runs of homozygosity and signatures of selection: a comparison among eight local Swiss sheep breeds. Anim Genet 50(5):512–525
Sobieraj-Kmiecik A, Drobik-Czwarno W, Nowak-Życzyńska Z, Wojciechowska M, Demars J, Bodin L et al. (2020) Genome analysis in local breeds: A case study on Olkuska sheep. Livest Sci 231:103880
Szpiech ZA, Xu J, Pemberton T, Peng W, Zöllner S, Rosenberg NA et al. (2013) Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet 93(1):90–102
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Vatankhah M, Sigdel A, Abdollahi-Arpanahi R (2019) Population structure of Lori-Bakhtiari sheep in Iran by pedigree analysis. Small Rumin Res 174:148–155
Vatankhah M, Talebi MA, Blair H (2016) Genetic analysis of Lori-Bakhtiari lamb survival rate up to yearling age for autosomal and sex-linked. Small Rumin Res 136:121–126
Vatankhah M, Talebi MA, Edriss MA (2008) Estimation of genetic parameters for reproductive traits in Lori–Bakhtiari sheep. Small Rumin Res 74:216–220
Vazquez AI, Bates DM, Rosa GJ, Gianola D, Weigel KA (2010) Technical note: an R package for fitting generalized linear mixed models in animal breeding. J Anim Sci 88(2):497–504
Wright S (1922) Coefficients of inbreeding and relationship. Am Nat 56(645):330–338
Yeomans L, Martin L, Richter T (2017) Expansion of the known distribution of Asiatic mouflon (Ovis orientalis) in the Late Pleistocene of the Southern Levant. R Soc Open Sci 4:170409
Zhang Q, Calus MPL, Guldbrandtsen B, Lund MS, Sahana G (2015) Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet 16:88
Acknowledgements
The authors are grateful to Guilan and Bu-Ali Sina Universities for providing financial support for this study.
Author information
Authors and Affiliations
Contributions
Conceptualization: RA; analyses: RA, SZM, CG, MS & MHF; draft preparation: RA, SZM, NGHZ, PZ & MHM; review and editing: CG, MS, SZM & NGHZ. All authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor Christine Baes.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abdoli, R., Mirhoseini, S.Z., Ghavi Hossein-Zadeh, N. et al. Runs of homozygosity and cross-generational inbreeding of Iranian fat-tailed sheep. Heredity 130, 358–367 (2023). https://doi.org/10.1038/s41437-023-00611-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-023-00611-y