Abstract
Spiders produce multiple silks with different physical properties that allow them to occupy a diverse range of ecological niches, including the underwater environment. Despite this functional diversity, past molecular analyses show a high degree of amino acid sequence similarity between C-terminal regions of silk genes that appear to be independent of the physical properties of the resulting silks; instead, this domain is crucial to the formation of silk fibers. Here, we present an analysis of the C-terminal domain of all known types of spider silk and include silk sequences from the spider Argyroneta aquatica, which spins the majority of its silk underwater. Our work indicates that spiders have retained a highly conserved mechanism of silk assembly, despite the extraordinary diversification of species, silk types and applications of silk over 350 million years. Sequence analysis of the silk C-terminal domain across the entire gene family shows the conservation of two uncommon amino acids that are implicated in the formation of a salt bridge, a functional bond essential to protein assembly. This conservation extends to the novel sequences isolated from A. aquatica. This finding is relevant to research regarding the artificial synthesis of spider silk, suggesting that synthesis of all silk types will be possible using a single process.
Similar content being viewed by others
Introduction
Spider silk proteins form multiple types of materials, including fibers and glues, each with specific mechanical properties and functions (Blamires et al. 2017; Eisoldt et al. 2011; Garb 2013; Hormiga and Griswold 2014). Gene duplication, recombination and diversification are thought to have generated this huge diversity of different silk types, each having a different ecological function (Clarke et al. 2015). These functions include safety lines, the structural frameworks of webs, external layers of protection around egg sacs, and securing prey (Hinman et al. 2000; Rising and Johansson 2015).
In the case of Argyroneta aquatica (Clerck 1757) (Araneae: Cybaeidae (Catalog 2016)), silk is used in a typical fashion to construct a web in which the spider resides and performs a number of actions, from feeding to mating and storing eggs (Schütz and Taborsky 2003, 2005, 2011; Schutz et al. 2007; Seymour and Hetz 2011). What is unusual about this spider is that it is the only known species to spin silk whilst submersed in water. The subsequent sheet web is then inflated with air drawn down from the surface and is utilised as an air reservoir. The silken “diving bell” allows for oxygen diffusion to occur, allowing the spider to avoid surfacing for extended periods of time (Seymour and Hetz 2011). Cybaeus angustiarum, a fellow cybaeid, is a terrestrial species found in dense forests (often on north facing, scree slopes) in areas under stones or in decaying wood with humidity close to 100%.
In all spiders, liquid silk dope is passed through specialised, elongated glands within the body of the spider and extruded through spinnerets (Askarieh et al. 2010; Rising and Johansson 2015; Vollrath and Knight 2001).
All silk genes have three components; a type-specific repetitive region flanked by conserved N-terminal (Motriuk-Smith et al. 2005; Rising et al. 2006) and C-terminal (Challis et al. 2006; Collin et al. 2016; Gnesa et al. 2012; Hagn et al. 2010) domains. Whilst the repetitive region determines the mechanical properties of each silk product (Hayashi and Lewis 1998; Hayashi et al. 1999), the terminal domains work together ensuring that individual proteins assemble correctly and that the fibre forms at the correct stage of the spinning process (Andersson et al. 2014; Andersson et al. 2017; Rising and Johansson 2015). The N-terminal domain restricts the formation of silk fibers to a precise point in the silk duct, preventing silk proteins stored in the silk gland from agglutinating (Askarieh et al. 2010). The C-terminal domain drives spontaneous fibre formation, likely through use of a pH-sensitive “salt bridge” (Ittah et al. 2006; Stark et al. 2007), where noncovalent interactions between one basic and one acidic residue are disrupted at low pH because the acidic residue becomes protonated and is no longer charged.
Salt bridges, either individual or paired, have been proposed to explain the dimeric bundling of 4 or 5 alpha helices in the C-termini of one particular silk, the major ampullate (Hagn et al. 2010; Sponner et al. 2004a, 2005). Whilst the miniature spidroin 4RepCT, formed of four copies of a MaSp repetitive region and one C-terminus, has been sufficient to produce self-assembling silk fibers (Stark et al. 2007), it has recently been shown in the minispidroin NT2RepCT that including the N-terminal region achieves greater efficiency in the production of synthetic fibers (Andersson et al. 2017). However, studies have shown that the level of solubility demonstrated by the terminal regions differs between spider species, leading to further questions around their function and the suitability of individual terminal regions for use in silk protein synthesis (Andersson et al. 2014, 2017; Askarieh et al. 2010).
A degree of amino acid sequence conservation has been observed from studies of small numbers of different silks (Beckwitt and Arcidiacono 1994; Challis et al. 2006; Collin et al. 2016; Gnesa et al. 2012; Hagn et al. 2010; Sponner et al. 2004b, 2005). What is not clear is the extent to which this conservation is maintained, particularly where silk proteins exhibit vastly different properties (e.g., the glue-like aggregate and piriform silk versus the superior strength of major ampullate and aciniform silks). Additionally, the diving bell spider Argyroneta aquatica spins silk whilst completely submersed in fresh water; given how silk proteins are dehydrated as part of the spinning process, does this “extreme” environment necessitate variation within the silk protein and spinning process?
Here, we analyse the genetic sequences silks of all known types from spiders of as many groups as are available in GenBank. We investigate whether the salt bridge structure is conserved across all species and silk types and ask if changes in biophysical properties or the utilisation of silk in an “extreme” environment necessitates a change in how silk proteins are formed.
Materials and methods
Transcriptome assembly
RNA was extracted from the silk glands and whole abdomen of adult females and sequenced on an Illumina NextSeq500 (DeepSeq, University of Nottingham). Transcriptomes were trimmed using Scythe (Buffalo 2014) and Sickle (Joshi and Fass 2011) and assembled using Trinity (Grabherr et al. 2011). Protein sequences were predicted using TransDecoder (Haas et al. 2013).
Identification of silk genes
Custom blast databases of silk genes downloaded from GenBank and the Nephila clavipes (Babb et al. 2017) and Stegodyphus mimosarum genomes (Sanggaard et al. 2014) were generated to screen transcriptomes for silk sequences, which were manually examined and blasted against GenBank for confirmation (e-value < 0.0005).
Phylogenetic analysis
Silk sequences from GenBank, the N. clavipes and S. mimosarum genomes and newly-identified spidroins were filtered to only include C-terminal regions, identified by the “QALLE” motif (Challis et al. 2006). Duplicate sequences were discarded, leaving the longest sequence for analysis. Two sequences were considered identical if their nucleotide sequences had only a small number of variations which did not significantly alter the amino acid composition (e.g., variation causing a polar residue to change to acidic was considered significant, whereas polar to polar was not). Selected sequences were manually trimmed leaving only the C-terminal region (maximum length: 360 bp (Hagn et al. 2010; Rising and Johansson 2015)).
Selected nucleotide sequences were aligned in Geneious v8.1.8 (Kearse et al. 2012) (Clustal W algorithm (Larkin et al. 2007)) and subsequently translated in order to allow refined alignment by eye. The refined untranslated nucleotide sequence alignment was imported into MEGA6.0 (Tamura et al. 2013) and used to construct a maximum-likelihood tree (1000 bootstrap replicates; see supplementary file 2).
Results
Diversity of silk C-termini
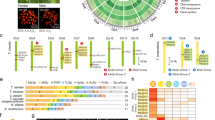
Forty-four new silk sequences isolated from transcriptomes of the spiders A. aquatica, C. angustiarum, and Pholcus phalangioides included a C-terminal domain (this study; GenBank Accession Numbers MG744694–MG744714). Once refined (see methods), 21 unique sequences remained for analysis. Three hundred and thirty-four silk sequences were retrieved from GenBank and the N. clavipes and S. mimosarum genomes, of which 150 sequences were both unique and contained a C-terminal region. Specialised silks were mostly found in the Orbiculariae and S. mimosarum genome, with the exception of one tubuliform and a small number of major ampullate sequences (Babb et al. 2017; Garb et al. 2010; Perez-Rigueiro et al. 2010; Rising et al. 2007; Sanggaard et al. 2014; Stark et al. 2007) (Fig. 1). An unrooted maximum-likelihood tree of nucleotide sequences (1000 bootstraps) shows the clustering together of sequences that are identified as belonging to the same silk type by BLAST (see supplementary figure 2).
Familial origins of silk sequences used in this study. Phylogenetic tree from Nentwig (Nentwig et al. 2013), modified to show positions of the different spider families and the silks previously described as being of a particular type. Location of Argyroneta aquatica and Cybaeus angustiarum (Cybaeidae) marked with an arrow. Each colored box represents a silk type; uncoloured boxes represent unclassified published spidroins. Numbers in each box indicate the total number of occurrences of each silk type. The total number of species sampled per family is detailed at the end of each row; * indicates where sequences have been retrieved from a genome
Conservation of a salt bridge structure across spider silk genes
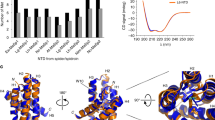
Analysis of the predicted secondary structures of sequences representing each silk type with JPred4 (Drozdetskiy et al. 2015) suggests the secondary structure of the C-terminus is conserved (data not shown), containing up to five α-helices, supporting previous studies (Hagn et al. 2010; Ittah et al. 2007). However, some predictions suggest Helix 1 may be absent, including in the N. clavipes MaSp-a sequence analysed here (Fig. 2).
Alignment of Nephila clavipes C-terminal sequences representing characterized silk types, with predicted helical secondary structure of MaSp-a below. Amino acid residues implicated in the formation of a salt bridge are marked A (acidic residue; glutamic or aspartic acid) and found within region B (basic residue; arginine or lysine). Residues are highlighted to show whether acidic (red), basic (blue), hydrophilic (green) or hydrophobic (yellow)
In all sequences analysed in this study, one acidic residue and one basic residue pair has been conserved. A conserved basic residue is consistently found in Helix 2 (position 30–31 Fig. 2, 23–37 supplementary figure 1), whilst an acidic base is always found towards the center of Helix 4 (position 85 in Fig. 2; 88 in supplementary figure 1). Additionally, in major ampullate, minor ampullate, aciniform, and piriform sequences a second acidic and basic residue appears to be partially conserved (supplementary figure 1) as identified by Sponner et al. (2004b, 2005). These pairs correspond with those already shown to form a salt bridge in the secondary protein structure in previous studies (Hagn et al. 2010; Ittah et al. 2006).
Content of the C-terminal domain
The majority of the C-terminus is composed of hydrophobic and hydrophilic amino acids (average 53.5 and 37.0% respectively) including serine, alanine, and leucine (average 21.2, 13.2, and 10.8% respectively) in an alternating arrangement. These residues promote the formation of alpha-helices in the protein structure, with the hydrophilic residues exposed, promoting solubility (Hagn et al. 2010).
Overall DNA sequence identity is highest in the acidic residue found in the “QALLE” motif (Challis et al. 2006), where the first two nucleotides (guanine, adenine) are completely conserved and the final nucleotide determines whether it is a glutamic or aspartic acid residue (89.3 and 10.7%, respectively of all sequences in this study; see supplementary figure 1). The basic residues involved in the formation of the salt bridge are usually arginine, although some are lysine or histidine (94.7, 3.3, and 2.0%, respectively).
Overall, the charged residue content of the C-terminus is typically less than 10% (5.2% acidic, 4.1% basic; average across all sequences in this study) although this varies depending on the sequence (aggregate spidroins average 13% acidic, 8% basic). However, one acidic and basic residue pair (A and region B, respectively, Fig. 2 and supplementary figure 1) are present in each sequence examined, irrespective of species or silk type.
Additionally, we find a raised percentage of the amino acids surrounding the basic residue within spider silk are hydrophilic (43.0%, region B, supplementary figure 1), reducing its pKa and hence promoting its protonation, whereas a majority of those surrounding the acidic residue are hydrophobic (68.8%, region A, supplementary figure 1), increasing its pKa and again promoting protonation.
Discussion
This analysis encompasses 19 families from within the Araneae and shows conservation of a pH-sensitive salt bridge, typically composed of an arginine-glutamic acid pairing, within all the silk types and species examined. Our finding of this degree of conservation confirms an essential role for this feature in the correct assembly of silk fibers. The extended coverage in terms of species and phylogenetic diversity suggests this feature has been conserved for the entire evolutionary history of the group—some 360 million years. Moreover, this conservation has persisted despite the diversification of other spider traits. For example, terrestrial spiders such as the mygalomorphs produce two to three silk proteins from a large, undifferentiated silk gland whereas the Orbiculariae have evolved several specialised glands from which different types of silk are extruded (Blackledge and Hayashi 2006; Blamires et al. 2017; Clarke et al. 2017; Garb 2013; Garb et al. 2010; Perez-Rigueiro et al. 2010; Rising et al. 2007; Stark et al. 2007).
Conservation of C-terminus in all A. aquatica silk genes
Argyroneta aquatica predominantly spins its silk in water, although will occasionally leave a dragline when walking in a terrestrial environment. Comparatively, individual Cybaeus did not appear to produce a dragline but given time would spin silken webs in which they resided, which appear visually similar to the dehydrated diving bells of A. aquatica. In sequences obtained from both these species, and also from the first fully-annotated spider genome of Nephila clavipes (Babb et al. 2017) in which a number of new silk sequences were identified, the residue pairing within the C-terminal domain has been conserved. This suggests that the formation of silk fibers by A. aquatica occurs using the same method as in terrestrial spiders. This leads to questions around how this spider is adapted to spinning silk underwater and the subsequent mechanical properties of A. aquatica silk in such an “extreme” environment. This is particularly relevant as studies of silk in humid conditions suggest the structure may be temporarily affected by the level of humidity; the degree to which major ampullate silk supercontracts depends on the species, whereas minor ampullate silk does not supercontract (Agnarsson et al. 2009; Blackledge et al. 2009; Boutry and Blackledge 2010).
Conservation of a physical structure
The residues responsible for the formation of the pH-sensitive salt bridge(s) are both uncommon and conserved within the restricted number of sequences studied thus far (Challis et al. 2006; Hagn et al. 2010). Analysis of the expanded range of sequences used in this study shows this trait is maintained, with >90% of the C-terminal domain typically composed of hydrophobic and hydrophilic residues. Of the charged residues present, at least one acidic and one basic residue is conserved in all sequences and as such inferred to be crucial to the formation of a salt bridge and therefore the correct folding of silk proteins into a fibre. The conservation of an arginine-glutamic acid pair of residues suggests this may be the optimum pairing for a salt bridge in silk, but the presence of lysine and aspartic acid in some sequences implies this is not always essential, although the effect of these substitutions on the final protein structure and its physical properties is currently unknown.
The higher level of hydrophilic residues around the basic residue and hydrophobic residues around the acidic residue may be necessary to ensure the correct formation of the salt bridge during the protein folding stage and allowing its regulation by smaller changes in pH due to the local environment created by the protein. Where two pairs of residues are conserved in major ampullate, minor ampullate, piriform, and some aciniform sequences it is likely that there are two salt bridges present in the C-terminus, although further structural analysis would confirm that the second is not merely sequence duplication or an evolutionary artefact suggesting the presence of a second bridge.
Conclusion
In silk research, the C-terminus is a key component of the minimal sequence used for recombinant spider silk production, as without this region fibers will not form (Stark et al. 2007). What we find is an overall model for all silk types irrespective of physical traits that illustrates the range of environments in which this single protein family may be utilised.
Our results allow us to identify conserved amino acid residues essential to the correct formation of silk proteins, thereby enabling the identification of less essential residues that may be chemically functionalised in artificially synthesised silk using techniques such as click-chemistry (Harvey et al. 2016). This study will aid researchers in selecting suitable sequences without repetitive testing in vivo whilst predicting sites which may be suitable for modification, as seen in Harvey et al. (2016), and build a repository of spidroin parts that could be combined to achieve novel and custom characteristics.
Summary
Here, we compare C-terminal sequences representing all known silk types, and from all the major clades within the Araneae. We illustrate how the salt bridge structure is conserved throughout the gene family as a whole, and show by inclusion of new data that this conservation extends to silks found in all the major divisions within the Araneae. This degree of conservation includes groups that are highly diverged in terms of their basic morphology, such as their silk spinning apparatus (Coddington 2005). We include new data on silk of the Cybaeid Argyroneta aquatica, the only extant species adapted to spin silk in an underwater environment, and a terrestrial Cybaeid for comparison, Cybaeus angustiarum. The insight gained allows us to characterize amino acid residues essential to correct protein assembly and aids the identification of residues that might be substituted to chemically functionalised alternatives for use in artificially synthesised silk proteins (Harvey et al. 2016).
Declarations
Availability of data and materials
The novel sequences generated in this study are deposited in the GenBank repository, Accession Numbers MG744694–MG744714 (https://www.ncbi.nlm.nih.gov/genbank/).
References
Agnarsson I, Boutry C, Wong SC, Baji A, Dhinojwala A, Sensenig AT et al. (2009) Supercontraction forces in spider dragline silk depend on hydration rate. Zoology 112:325–331
Andersson M, Chen G, Otikovs M, Landreh M, Nordling K, Kronqvist N et al. (2014) Carbonic anhydrase generates CO(2) and H(+) that drive Spider silk formation via opposite effects on the terminal domains. PLoS Biol 12(8):e1001921
Andersson M, Jia Q, Abella A, Lee X-Y, Landreh M, Purhonen P et al. (2017) Biomimetic spinning of artificial spider silk from a chimeric minispidroin. Nat Chem Biol. 13:262–264
Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A et al. (2010) Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465:236–238
Babb PL, Lahens NF, Correa-Garhwal SM, Nicholson DN, Kim EJ, Hogenesch JB et al. (2017) The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat Genet 49:895–903
Beckwitt R, Arcidiacono S (1994) Sequence conservation in the C-terminal region of spider silk proteins (Spidroin) from Nephila clavipes (Tetragnathidae) and Araneus bicentenarius (Araneidae). J Biol Chem 269:6661–6663
Blackledge TA, Boutry C, Wong SC, Baji A, Dhinojwala A, Sahni V (2009) How super is supercontraction? Persistent versus cyclic responses to humidity in spider dragline silk. J Exp Biol 212:1981–1989
Blackledge TA, Hayashi CY (2006) Silken toolkits: biomechanics of silk fibers spun by the orb web spider Argiope argentata (Fabricius 1775). J Exp Biol 209:2452–2461
Blamires SJ, Blackledge TA, Tso I-M (2017) Physicochemical property variation in spider silk: ecology, evolution, and synthetic production. Annu Rev Entomol 62:443–460
Boutry C, Blackledge TA (2010) Evolution of supercontraction in spider silk: structure–function relationship from tarantulas to orb-weavers. J Exp Biol 213:3505–3514
Buffalo V (2014) Scythe—A Bayesian adapter trimmer (version 0.994 BETA) [Software]
Catalog WS (2016) World Spider Catalog, version 17
Challis RJ, Goodacre SL, Hewitt GM (2006) Evolution of spider silks: conservation and diversification of the C-terminus. Insect Mol Biol 15:45–56
Clarke TH, Garb JE, Haney RA, Chaw RC, Hayashi CY, Ayoub NA (2017) Evolutionary shifts in gene expression decoupled from gene duplication across functionally distinct spider silk glands. Sci Rep 7:8393
Clarke TH, Garb JE, Hayashi CY, Arensburger P, Ayoub NA (2015) Spider transcriptomes identify ancient large-scale gene duplication event potentially important in silk gland evolution. Genome Biol Evol 7:1856–1870
Clerck CA (1757). Svenska Spindlar uti sina hufvud-slågter indelte samt under några och sextio särskildte arter beskrefne och med illuminerade figurer uplyste/Aranei Svecici, descriptionibus et figuris æneis illustrati, ad genera subalterna redacti, speciebus ultra LX determinati
Coddington JA (2005) Phylogeny and classification of spiders. In: Ubick D, Paquin P, Cushing PE, & Roth V (eds) Spiders of North America: an identification manual. American Arachnological Society, Poughkeepsie, New York, USA
Collin MA, Clarke TH, Ayoub NA, Hayashi CY (2016) Evidence from multiple species that spider silk glue component ASG2 is a spidroin. Sci Rep 6:21589
Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394
Eisoldt L, Smith A, Scheibel T (2011) Decoding the secrets of spider silk. Mater Today 14:80–86
Garb JE (2013) Spider silk: an ancient biomaterial for the 21st century. In: Penney D (ed.) Spider research in the 21st century: trends and perspectives. Siri Scientific Press, Manchester, pp 252–281
Garb JE, Ayoub NA, Hayashi CY (2010) Untangling spider silk evolution with spidroin terminal domains. BMC Evolut Biol 10:243
Gnesa E, Hsia Y, Yarger JL, Weber W, Lin-Cereghino J, Lin-Cereghino G (2012) Conserved C-terminal domain of spider tubuliform spidroin 1 contributes to extensibility in synthetic fibers. Biomacromolecules 13:304–312
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J et al. (2013) De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protocols 8(8): https://doi.org/10.1038/nprot.2013.084
Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T (2010) A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 465:239–242
Harvey D, Bardelang P, Goodacre SL, Cockayne A, Thomas NR (2017). Antibiotic spider silk: site-specific functionalization of recombinant spider silk using “Click” chemistry. Adv Mater 29:1604245-n/a.
Hayashi CY, Lewis RV (1998) Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks1. J Mol Biol 275:773–784
Hayashi CY, Shipley NH, Lewis RV (1999) Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol 24:271–275
Hinman MB, Jones JA, Lewis RV (2000) Synthetic spider silk: a modular fiber. Trends Biotechnol 18:374–379
Hormiga G, Griswold CE (2014) Systematics, phylogeny, and evolution of Orb-weaving spiders. Annu Rev Entomol 59:487–512
Ittah S, Cohen S, Garty S, Cohn D, Gat U (2006) An essential role for the C-terminal domain of A dragline spider silk protein in directing fiber formation. Biomacromolecules 7:1790–1795
Ittah S, Michaeli A, Goldblum A, Gat U (2007) A model for the structure of the C-terminal domain of dragline spider silk and the role of its conserved cysteine. Biomacromolecules 8:2768–2773
Joshi N, Fass J (2011). Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version1.33) [Software]
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H (2007) Clustal W and Clustal X version 2A0. Bioinformatics 23:2947–2948
Motriuk-Smith D, Smith A, Hayashi CY, Lewis RV (2005) Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromolecules 6:3152–3159
Nentwig W, Arnedo MA, Coddington JA, Hormiga G, Jocqué R, Kropf C et al. (2013) Appendix. Spider Phylogeny. In: Nentwig W (ed) Spider ecophysiology. Springer, Berlin
Perez-Rigueiro J, Plaza GR, Torres FG, Hijar A, Hayashi C, Perea GB (2010) Supercontraction of dragline silk spun by lynx spiders aOxyopidaea. Int J Biol Macromol 46:555–557
Rising A, Hjalm G, Engstrom W, Johansson J (2006) N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules 7:3120–3124
Rising A, Johansson J (2015) Toward spinning artificial spider silk. Nat Chem Biol 11:309–315
Rising A, Johansson J, Larson G, Bongcam-Rudloff E, Engström W, Hjälm G (2007) Major ampullate spidroins from Euprosthenops australis: multiplicity at protein, mRNA and gene levels. Insect Mol Biol 16:551–561
Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V (2014) Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun 5:3765
Schütz D, Taborsky M (2003) Adaptations to an aquatic life may be responsible for the reversed sexual size dimorphism in the water spider, Argyroneta aquatica. Evolut Ecol Res 5:105–117
Schütz D, Taborsky M (2005) Mate choice and sexual conflict in the size dimorphic water spider, Argyroneta aquatica (araneae, argyronetidae). J Arachnol 33:767–775
Schütz D, Taborsky M (2011) Sexual selection in the water spider: female choice and male-male competition. Ethology 117:1101–1110
Schutz D, Taborsky M, Drapela T (2007) Air bells of water spiders are an extended phenotype modified in response to gas composition. J Exp Zool Part A 307:549–555
Seymour RS, Hetz SK (2011) The diving bell and the spider: the physical gill of Argyroneta aquatica. J Exp Biol 214:2175–2181
Sponner A, Unger E, Grosse F, Weisshart K (2004a) Conserved C-termini of Spidroins are secreted by the major ampullate glands and retained in the silk thread. Biomacromolecules 5:840–845
Sponner A, Unger E, Grosse F, Weisshart K (2004b) Conserved C-termini of spidroins are secreted by the major ampullate glands and retained in the silk thread. Biomacromolecules 5:840–845
Sponner A, Vater W, Rommerskirch W, Vollrath F, Unger E, Grosse F et al (2005) The conserved C-termini contribute to the properties of spider silk fibroins. Biochem Biophys Res Commun 338:897–902
Stark M, Grip S, Rising A, Hedhammar M, Engström W, Hjälm G et al. (2007) Macroscopic fibers self-assembled from recombinant miniature spider silk proteins. Biomacromolecules 8:1695–1701
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Vollrath F, Knight DP (2001) Liquid crystalline spinning of spider silk. Nature 410:541–548
Acknowledgements
Spider collection and dissection by MR. RNA extracted by VT and MS, transcriptomics and analysis by MS, with genomics assistance by James Fowkes. All living authors read and approved the final manuscript. Sequencing and data quality control by Sunir Malla (DeepSeq facility, University of Nottingham). In memory of Victor Tudorica (1990–2016).
Funding
This work was supported by the Royal Society [Grant Number IE141413 to SLG and MR], Biotechnology and Biological Sciences Research Council doctoral training studentships to MS and VT [Grant Number BB/M008770/1] and the Ministry of Agriculture of the Czech Republic [Grant Number MZe RO0415 to MR].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Strickland, M., Tudorica, V., Řezáč, M. et al. Conservation of a pH-sensitive structure in the C-terminal region of spider silk extends across the entire silk gene family. Heredity 120, 574–580 (2018). https://doi.org/10.1038/s41437-018-0050-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0050-9
This article is cited by
-
Structural conversion of the spidroin C-terminal domain during assembly of spider silk fibers
Nature Communications (2024)
-
NMR assignment and dynamics of the dimeric form of soluble C-terminal domain major ampullate spidroin 2 from Latrodectus hesperus
Biomolecular NMR Assignments (2023)
-
Spidroins and Silk Fibers of Aquatic Spiders
Scientific Reports (2019)
-
Two-step self-assembly of a spider silk molecular clamp
Nature Communications (2018)