Abstract
Purpose
Newborn screening (NBS) is performed to identify neonates at risk for actionable, severe, early-onset disorders, many of which are genetic. The BabySeq Project randomized neonates to receive conventional NBS or NBS plus exome sequencing (ES) capable of detecting sequence variants that may also diagnose monogenic disease or indicate genetic disease risk. We therefore evaluated how ES and conventional NBS results differ in this population.
Methods
We compared results of NBS (including hearing screens) and ES for 159 infants in the BabySeq Project. Infants were considered “NBS positive” if any abnormal result was found indicating disease risk and “ES positive” if ES identified a monogenic disease risk or a genetic diagnosis.
Results
Most infants (132/159, 84%) were NBS and ES negative. Only one infant was positive for the same disorder by both modalities. Nine infants were NBS positive/ES negative, though seven of these were subsequently determined to be false positives. Fifteen infants were ES positive/NBS negative, all of which represented risk of genetic conditions that are not included in NBS programs. No genetic explanation was identified for eight infants referred on the hearing screen.
Conclusion
These differences highlight the complementarity of information that may be gleaned from NBS and ES in the newborn period.
Similar content being viewed by others
INTRODUCTION
Conventional state-mandated newborn screening (NBS) is routinely performed shortly after birth to diagnose severe, early-onset disorders, where prompt diagnostic evaluation and treatment can improve outcomes.1 In the United States, NBS includes a dried blood spot in addition to point-of-care tests, such as the hearing screen and screening for critical congenital heart diseases.2,3,4 As this is a screening test, false positive results commonly occur (as can false-negative results).2,5,6
Exome or genome sequencing (ES/GS) may complement traditional NBS in identifying risk for disease, ideally prior to the development of symptoms. We5,7 and others8 have previously described cases of inborn errors of metabolism that had been missed by traditional newborn screening and later detected by sequencing, raising the question of whether ES/GS should be incorporated during infancy for genetic risk assessment. Of the 35 disorders included in the Recommended Uniform Screening Panel (RUSP) that are tested using the dried blood spot, nearly all have a Mendelian genetic basis, including sickle cell disease, cystic fibrosis, and maple syrup urine disease, among others.9 Various sequencing approaches are increasingly used in NBS algorithms for second-tier confirmation, and the use of sequencing has been proposed as an alternative primary method of detection.10 Though a number of studies have demonstrated the ability of ES/GS to diagnose genetic conditions in symptomatic infants,11,12,13,14 prior comparisons of traditional NBS with ES/GS have shown that infants may be missed by either method, though these analyses generally favor traditional NBS as more sensitive than ES/GS for the RUSP-targeted disorders.15,16,17 There are relatively few studies comparing the results of ES/GS to NBS in a cohort that is not enriched for such disorders, with one prior study demonstrating a higher sensitivity of conventional NBS versus ES to detect NBS-targeted conditions.18 We therefore evaluated the relationship between NBS and ES results in the BabySeq Project19 to highlight the unique attributes of each approach in a population-based sample.
MATERIALS AND METHODS
Study design and participants
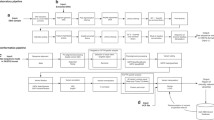
The BabySeq Project was a randomized clinical trial examining the impact of ES in the clinical care of newborns. Details of its study design have been previously described.19 In brief, healthy newborns from the Brigham and Women’s Hospital (BWH) Well Newborn Nursery and newborns admitted to Boston Children’s Hospital’s (BCH), Massachusetts General Hospital, and BWH neonatal and cardiac intensive care units (NICUs and CICUs) were randomized to receive modified standard of care consisting of a standard NBS report and a family history report (control arm) or the same plus ES (intervention arm). Families in the intervention arm received a report with results that included variants indicating monogenic disease risk, heterozygous status for variants in genes associated with autosomal recessive childhood-onset disorders, and a very limited number of actionable adult-only onset disorders, as well as variants with pediatric pharmacogenomic associations.17
ES results for this cohort have been previously reported.17 In this analysis, we analyzed NBS dried blood spot and newborn hearing screen results for the infants sequenced in the intervention arm of this study. Infants were considered “NBS positive” if any abnormal result was found on the dried blood spot conferring risk of disease and requiring follow-up diagnostic testing or a repeat sample. “NBS negative” infants either had no abnormalities found or had results indicating recent blood transfusion. “ES positive” infants had variants found conferring a diagnosis of a monogenic disease or disease risk and “ES negative” infants had either no disease-causing variants found or were heterozygous for recessive variants that were not expected to have clinical implications.
RESULTS
Characteristics of the study population
Overall, 316 infants were enrolled in the study, and 159 of them were assigned to NBS plus family history plus ES. In this intervention arm, 127 infants were enrolled from the well-baby nursery and considered healthy (80%) and 32 were enrolled from ICUs (20%).
ES and NBS results
The majority of infants were both NBS and ES negative (132/159, 84%). Of the NBS positive infants (12/159, 8%), most were enrolled from the NICU (7/12, 58%), and 4/12 (33%) were false positives commonly seen particularly in critically ill infants.
Three infants were both NBS and ES positive, but only one was positive for the same disorder by both modalities. Interestingly, this infant was initially flagged on NBS for biotinidase deficiency, but a follow-up NBS was reported normal. ES revealed two variants in BTD, one of which is associated with a mild phenotype. The baby was subsequently determined to have reduced enzyme activity indicating a mild form of the disease and was treated with biotin.7 The other 2/3 had genetic conditions found by ES that are not included on NBS and NBS results that were deemed false positives: one had G6PD deficiency identified by ES but was flagged for a thyroid abnormality on NBS while the second infant was identified to be at risk for nonclassic congenital adrenal hyperplasia by ES and this infant’s NBS was flagged for multiple abnormalities that are all false positives often seen in the NICU that later resolved (abnormal thyroid function, low T-cell receptor excision circles [T-RECs], and multiple abnormal amino acids consistent with administration of total parenteral nutrition).
Nine infants were NBS positive and ES negative, reflecting abnormalities found on dried blood spot analysis that were not corroborated by sequencing, typically because the NBS was false positive (7/9 cases). The two true positive NBS results were hemoglobin variants (FAB and FAV) that ES analytic pipelines may not be optimized to detect and were not thought to be clinically significant; both infants were enrolled from the well-baby nursery. Of the remaining seven infants who were NBS positive/ES negative, 5/7 were in the NICU and felt to illustrate common false positives seen with prematurity and critical illness and two were in the well-baby nursery and had false positive abnormal thyroid screens.
Fifteen infants were ES positive and NBS negative, reflecting risks for genetic conditions that are not included on NBS panels—such as cardiomyopathies and cancer risk syndromes that have later onset. Of the 15 infants, 10 were healthy and 5 were enrolled from a NICU. In one case enrolled from the NICU, ES results were diagnostic for an underlying genetic syndrome that had not previously been considered: this infant presented with anal atresia and was found to have a de novo likely pathogenic variant in ANKRD11 associated with KBG syndrome. These infants are described in detail in a previous study17 and summarized in Fig. 1.
Additional details on ES positive cases have been previously published.17 CAH congenital adrenal hyperplasia.
All eight infants whose initial hearing screens were abnormal (one bilateral, the rest unilateral) were ES negative. Four, including the bilateral referral, passed follow-up hearing screens, and none of the remaining four unilateral referrals (all well newborns) had any reported health issues or specialty visits noted on follow-up assessments and are therefore assumed not to have significant congenital hearing loss.
DISCUSSION
We present an analysis of infants in the BabySeq Project who underwent both ES and NBS. Our findings demonstrate that the ES and NBS results serve complementary roles in determining disease risk, with each modality having strengths and weaknesses. Regarding the utility of ES compared with NBS in diagnosing the RUSP-targeted conditions, we detected only one case in our cohort: the case of partial biotinidase deficiency as previously reported.7 Overall, the NBS results in our cohort did not reveal any baby at risk for severe, early-onset disease. The dearth of “true positive” results for NBS-targeted disorder by either modality is similar to prior population-based studies owing to the rarity of these conditions.15,18 The recent NEXUS study, which enrolled and sequenced children with known inborn errors of metabolism or hearing loss in addition to healthy newborns, found that most (15/17) inborn errors of metabolism were found on ES and that ES yielded an explanation for suspected hearing loss in only 18% (5/28). As with our study, conditions not targeted by NBS were also identified.15 Another study of 1,696 individuals undergoing trio GS, compared with NBS results, also demonstrated higher sensitivity of NBS for diagnosing inborn errors of metabolism.18 This is further supported by a study of nearly 1,500 individuals with positive NBS who underwent ES, where actionable disorders identified by NBS were missed on ES.16 These prior studies suggest that relying on ES/GS alone to detect inborn errors of metabolism, where prompt intervention is critical, would not be advised at this time due to limitations in sequencing technology and variant interpretation.
Our present cohort demonstrates that abnormal findings on NBS requiring follow-up are frequently seen in newborns, particularly those in the NICU, as evidenced by the 12/159 “NBS positive” cases. While many of these will resolve on subsequent NBS samples, particularly such findings as abnormal thyroid markers seen commonly in premature or critically ill infants, hypothyroidism warranting treatment is occasionally detected that would not be found using sequencing alone. Hemoglobin variants were also identified by NBS our cohort but not reported on the sequencing analysis, because the variants in hemoglobin genes were either difficult to detect by ES (e.g., hemoglobin Barts, FAB, due to a deletion) or because the hemoglobin variant (FAV) is likely to have uncertain or no clinical significance and therefore would not meet ES reporting standards. We similarly found a low yield of ES as a primary screen for hearing loss, with only one infant found to have a condition causing hearing loss (variant in KCNQ4) detected by sequencing. This infant passed the NBS hearing screen, which is not surprising given that hearing loss due to pathogenic KCNQ4 variant is late onset. ES was not able to be fully evaluated as a follow-up to the newborn hearing screen as none of the eight infants who screened positive were later identified to have severe hearing loss. Overall, our results demonstrate that NBS is highly sensitive for hemoglobinopathies and thyroid abnormalities while ES will allow screening for numerous Mendelian disorders for which there is no current NBS analyte or algorithm and not necessarily a therapy.
Eighteen of the 159 infants who underwent ES were found to have results indicative of monogenic disease or disease risk. This demonstrates the potential value of sequencing in the newborn period to detect conditions that are generally not targeted by NBS panels, particularly those that confer later-onset disease risk and are variably penetrant. However, this is counterbalanced by ethical challenges such as the psychosocial implications of returning such information to the parents of a newborn. The full impact of early identification of disorders detected on ES is not known, and further research is needed. As sequencing technology continues to evolve, its utility for identifying actionable conditions in the newborn must be continually assessed.
Data availability
The genomic data/analyses reported in this paper have been deposited in the NBSTRN LPDR (https://nbstrn.org/tools/lpdr) under accession identifier nbs000002.v1.p1. Data access is restricted; for information on how to request access, please contact corresponding author Monica Wojcik at monica.wojcik@childrens.harvard.edu or Pankaj Agrawal at pagrawal@enders.tch.harvard.edu.
References
Kemper, A. R. et al. Long-term follow-up after diagnosis resulting from newborn screening: statement of the US Secretary of Health and Human Services’ Advisory Committee on Heritable Disorders and Genetic Diseases in Newborns and Children. Genet. Med. 10, 259–261 (2008).
Fabie, N. A. V., Pappas, K. B. & Feldman, G. L. The current state of newborn screening in the United States. Pediatr. Clin. North Am. 66, 369–386 (2019).
Sahai, I. & Marsden, D. Newborn screening. Crit. Rev. Clin. Lab. Sci. 46, 55–82 (2009).
Wilcken, B. & Wiley, V. Newborn screening. Pathology. 40, 104–115 (2008).
Wojcik, M. H. et al. Beta-ketothiolase deficiency presenting with metabolic stroke after a normal newborn screen in two individuals. JIMD Rep. 39, 45–54 (2018).
Slaughter, J. L. et al. The effects of gestational age and birth weight on false-positive newborn-screening rates. Pediatrics. 126, 910–916 (2010).
Murry, J. B. et al. Reconciling newborn screening and a novel splice variant in BTD associated with partial biotinidase deficiency: a BabySeq Project case report. Cold Spring Harb. Mol. Case Stud. 4, a002873 (2018).
Verbeeten, K. C. et al. Carnitine uptake defect due to a 5’UTR mutation in a pedigree with false positives and false negatives on newborn screening. Mol. Genet. Metab. 129, 213–218 (2020).
American College of Medical Genetics Newborn Screening Expert Group. Newborn screening: toward a uniform screening panel and system--executive summary. Pediatrics. 11, S296–307 (2006).
Parad, R. B., Kaler, S. G., Mauceli, E., Sokolsky, T., Yi, L. & Bhattacharjee, A. Targeted next generation sequencing for newborn screening of Menkes disease. Mol. Genet. Metab. Rep. 24, 100625 (2020).
Saunders, C. J. et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med. 4, 154ra135 (2012).
Meng, L. et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 171, e173438 (2017).
Gubbels, C. S. et al. Prospective, phenotype-driven selection of critically ill neonates for rapid exome sequencing is associated with high diagnostic yield. Genet. Med. 22, 736–744 (2020).
Kingsmore, S. F. et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am. J. Hum. Genet. 105, 719–733 (2019).
Roman, T. S. et al. Genomic sequencing for newborn screening: results of the NC NEXUS Project. Am. J. Hum. Genet. 107, 596–611 (2020).
Adhikari, A. N. et al. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat. Med. 26, 1392–1397 (2020).
Ceyhan-Birsoy, O. et al. Interpretation of genomic sequencing results in healthy and ill newborns: results from the BabySeq Project. Am. J. Hum. Genet. 104, 76–93 (2019).
Bodian, D. L. et al. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet. Med. 18, 221–230 (2016).
Holm, I. A. et al. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr. 18, 225 (2018).
Acknowledgements
M.H.W. is supported by K23 HD102589–01. Research reported in this publication was supported by the National Institutes of Health under award numbers R01HG009922, R01HL143295, U01TR003201, and U19HD077671. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization: P.B.A., M.H.W. Data curation: M.H.W., T.Z., O.Z.B., C.G. Formal analysis: M.H.W., O.Z.B.,M.L. Project administration: C.G. Supervision: R.C.G., A.H.B., I.A.H., R.B.P., H.L.R. Writing—original draft: M.H.W., T.Z; Writing—review & editing: all authors.
Corresponding authors
Ethics declarations
Ethics declaration
The BCH’s Institutional Review Board (IRB) and the Partners Human Research Committee (the IRB for BWH) approved this study and informed consent was obtained from all participants as required by the IRB.
Competing interests
R.C.G. has received compensation for advising the following companies: AIA, Grail, Humanity, Kneed Media, Plumcare, UnitedHealth, Verily, VibrentHealth, Wamberg; and is co-founder of Genome Medical, Inc. P.B.A. is on the Clinical Advisory Board of Illumina Inc. and GeneDx. H.L.R. is a compensated scientific advisory board member of Genome Medical. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wojcik, M.H., Zhang, T., Ceyhan-Birsoy, O. et al. Discordant results between conventional newborn screening and genomic sequencing in the BabySeq Project. Genet Med 23, 1372–1375 (2021). https://doi.org/10.1038/s41436-021-01146-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01146-5
This article is cited by
-
Molecular genetic screening of full-term small for gestational age
BMC Pediatrics (2023)
-
Genomic newborn screening for rare diseases
Nature Reviews Genetics (2023)
-
A multicenter prospective study of next-generation sequencing-based newborn screening for monogenic genetic diseases in China
World Journal of Pediatrics (2023)
-
Genomisches Neugeborenenscreening – Forschungsansätze, Herausforderungen und Chancen
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2023)
-
Rare diseases' genetic newborn screening as the gateway to future genomic medicine: the Screen4Care EU-IMI project
Orphanet Journal of Rare Diseases (2023)