Abstract
Purpose
Genotype-guided antiplatelet therapy is increasingly being incorporated into clinical care. The purpose of this study is to determine the extent to which patients initially genotyped for CYP2C19 to guide antiplatelet therapy were prescribed additional medications affected by CYP2C19.
Methods
We assembled a cohort of patients from eight sites performingCYP2C19 genotyping to inform antiplatelet therapy. Medication orders were evaluated from time of genotyping through one year. The primary endpoint was the proportion of patients prescribed two or more CYP2C19 substrates. Secondary endpoints were the proportion of patients with a drug–genotype interaction and time to receiving a CYP2C19 substrate.
Results
Nine thousand one hundred ninety-one genotyped patients (17% nonwhite) with a mean age of 68 ± 3 years were evaluated; 4701 (51%) of patients received two or more CYP2C19 substrates and 3835 (42%) of patients had a drug–genotype interaction. The average time between genotyping and CYP2C19 substrate other than antiplatelet therapy was 25 ± 10 days.
Conclusions
More than half of patients genotyped in the setting of CYP2C19-guided antiplatelet therapy received another medication impacted by CYP2C19 in the following year. Given that genotype is stable for a patient’s lifetime, this finding has implications for cost effectiveness, patient care, and treatment outcomes beyond the indication for which it was originally performed.
Similar content being viewed by others
INTRODUCTION
Genotype-guided antiplatelet therapy in the setting of acute coronary syndromes is increasingly being incorporated into clinical care at many medical centers.1 A large body of evidence supports this practice, and many studies have found it to be cost effective.2,3,4CYP2C19 codes for the major enzyme responsible for the activation of clopidogrel. Individuals carrying one or two copies of no function variants in CYP2C19 are known as intermediate and poor metabolizers, respectively (~1/3 of the population).5 These individuals are at increased risk of major adverse cardiovascular events if they are treated with clopidogrel after percutaneous coronary intervention (PCI) compared with those with two fully functional copies of the gene.6,7,8,9 In addition to clopidogrel, CYP2C19 is responsible for metabolizing several antidepressants (e.g., certain selective serotonin reuptake inhibitors and tricyclic antidepressants), proton pump inhibitors, and the antifungal medication voriconazole. Guidelines published by the Clinical Pharmacogenetics Implementation Consortium (CPIC) exist for each of these examples to guide the choice of medication or dose based on CYP2C19 genotype.5,10,11,12CYP2C19 also has an increased function variant that results in increased gene expression. Individuals with one normal and one increased function or two copies of the increased function variant are known as rapid or ultrarapid metabolizers, respectively. In contrast to clopidogrel, for which only intermediate and poor metabolizers require a change in therapy, in the case of other CYP2C19 substrates, rapid and ultrarapid metabolizers may require a dose or medication change.
Given that genotype is stable for a patient’s lifetime, it follows that using CYP2C19 genotype to guide the dose or medication selection for other actionable CYP2C19 substrates (in addition to the one for which genotype testing was ordered), would be efficient and would provide additional “free” information that may benefit patient outcomes. Herein we evaluated the extent to which individuals genotyped for CYP2C19 for the purpose of guiding antiplatelet therapy selection after PCI were prescribed other CYP2C19 substrates addressed by CPIC guidelines and how often a drug–genotype interaction was present.
MATERIALS AND METHODS
Ethics Statement
This study was performed in accordance with the principles set out in the Declaration of Helsinki. Studies at each site were approved by their respective IRBs and only aggregate data were shared for analyses. Data from Indiana University, the University of Alabama Birmingham and the University of Pennsylvania were collected with written informed consent obtained from each participant. Data from Pittsburgh University, the University of Florida, the University of Maryland and Vanderbilt University were collected with a waiver of informed consent approved by their respective IRBs.
Eight sites participating in the Implementing GeNomics in PracTice (IGNITE) Network Pharmacogenetics Working Group that had clinically implemented genotype-guided antiplatelet therapy selection participated in this cohort analysis: Indiana University, University of Alabama at Birmingham, University of Florida (UF) Health Gainesville, UF Health Jacksonville, University of Maryland, University of Pennsylvania, University of Pittsburgh, and Vanderbilt University Medical Center.1 Genotyping for CYP2C19 occurred at the time of left heart catheterization or PCI at five sites and at the time a medication with actionable pharmacogenetic information was prescribed at one site. Medication orders were extracted from the electronic health record (EHR) for all patients who were genotyped. Medication orders were extracted starting 30 days prior to the genotype date (to account for medications started before genotype results were available in the EHR, e.g., antiplatelet therapy) through one year after the genotyping date. Time to receive a CYP2C19 substrate was assessed from the time of genotype being reported in the EHR, and negative values (i.e., the medication was ordered prior to the genotype resulting) were converted to zero. The medications that were considered were those metabolized by CYP2C19 and addressed in CPIC guidelines published or in process and included amitriptyline, citalopram, clomipramine, clopidogrel, dexlansoprazole, doxepin, escitalopram, esomeprazole, imipramine, lansoprazole, omeprazole, pantoprazole, prasugrel, rabeprazole, sertraline, ticagrelor, trimipramine, and voriconazole.5,10,11,13 If a patient received more than one drug from a class, only one was counted to avoid double counting when a medication switch may have been made due to genotype or another reason. Prasugrel and ticagrelor are not affected by CYP2C19 genotype but were included as they are the recommended alternative agents to clopidogrel for CYP2C19 intermediate and poor metabolizers. Given that all sites involved in the study performed CYP2C19 genotyping to assist with antiplatelet therapy selection, focusing specifically on clopidogrel would have omitted patients who were prescribed prasugrel or ticagrelor based on genotype results.
Genotype–drug interactions were assigned according to CPIC guideline recommendations (Table 1). Patients were considered to have a drug–genotype interaction if they (1) had a CYP2C19 phenotype for which CPIC recommends a change in dose or drug and (2) received a medication order for the relevant CYP2C19 substrate medication. For example, prescribing voriconazole in patients with the ultrarapid metabolizer phenotype would be considered a drug–genotype interaction. A person with more than one gene–drug interaction was only counted once for the calculation.
Descriptive statistics were used to calculate the means, medians, and proportions. Weighted averages were used to calculate means and standard deviations across study sites according to sample size. To determine how oftenCYP2C19 genotype could be used again to guide treatment with another medication after antiplatelet therapy, our primary endpoint was the proportion of patients who received two or more CYP2C19 substrate medication orders during the year following genotyping.
RESULTS
A total of 9191 patients with CYP2C19 genotype data were assessed for medication orders. The mean age of the genotyped patients was 68 ± 3 years and 3322 (36.1%) were female. The racial breakdown of the patients was as follows: 7398 (80.1%) white, 1202 (13.1%) black, and 534 (5.8%) other/unknown.
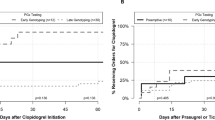
The vast majority of patients received at least one CYP2C19 substrate (including antiplatelet therapy) (92.5%). Fifty-one percent of patients received two or more CYP2C19 substrates (including clopidogrel, prasugrel, and ticagrelor) during the year following their genotyping for prediction of response to antiplatelet therapy (Fig. 1). Forty-one percent of patients received exactly one substrate, 39.3% received two substrates, 10.7% received three substrates, and 1.1% received four or more substrates in the year following genotyping. The most commonly prescribed drug class with the potential to be impacted by genotype other than antiplatelet therapy was proton pump inhibitors. The next most commonly prescribed drug class among these patients was selective serotonin reuptake inhibitors. The average time between genotyping results and receiving a CYP2C19 substrate other than antiplatelet therapy was 24.5 ± 10.6 days.
Overall, 42% of patients in the cohort had a drug–genotype interaction (i.e., received a medication order for which the dose or drug should be altered based the patient’s genotype according to CPIC guidelines including antiplatelet therapy) (Fig. 1). If we excluded interactions due to antiplatelet therapy, 30% of patients in the cohort were impacted by a genotype–drug interaction. Finally, among the sites who performed genotyping at the time of diagnostic left heart catheterization (i.e., before antiplatelet therapy was actually needed), 234 of 735 patients (31.8%) went on to receive clopidogrel, prasugrel, or ticagrelor in the year following their genotyping.
DISCUSSION
In the setting of genotyping to help guide antiplatelet therapy selection, the prescribing of additional drugs impacted by CYP2C19 was common. Over half of the cohort (51%) received two or more CYP2C19 substrate drugs in the year following genotyping, meaning they received another medication that could be impacted by CYP2C19 independent of the primary purpose for their genotyping. Overall, 42% of the cohort had an actionable drug–genotype interaction. These findings have important implications for several aspects of clinical care including downstream clinical decision support, patient education, data portability outside the institution that ordered the genotyping, and cost effectiveness.
Our findings highlight the importance of integrating CYP2C19, and other pharmacogenetic genotyping results, into the EHR given that over 50% of patients initially genotyped to assist with antiplatelet drug therapy were subsequently prescribed another CYP2C19 substrate in one year (25 days on average), and over 40% had a drug–genotype interaction. Our findings further support the need for clinical decision support to assist prescribers with selection and dosing of multiple CYP2C19 substrates based on the patient’s genotype. Such support may be most efficiently delivered through advisories or alerts that appear in the EHR when a patient with an actionable genotype, meaning a genotype that would lead to an altered prescribing decision, is prescribed a drug impacted by that genotype. An investigation by our group of implementation strategies of 12 early adopters, including those included in this paper, found that all sites integrated results into the EHR, most often as discrete genotype and phenotype fields with a full report of the genotyping testing available via a link.1 Nearly all sites participating in this cohort have activated clinical decision support for clopidogrel and most have it for additional CYP2C19 substrates. This is an important step in implementation of pharmacogenomic services, given that test results have the potential for lifelong utility, and prescribers are unlikely to seek out month- or year-old test results in the EHR or even to know that such results exist. The integration of pharmacogenetic results (genotype and/or phenotype) as discrete fields in the EHR enables more specific and actionable decision support. Simply scanning a paper pharmacogenetic test result into a patient’s record does not allow for specific, actionable recommendations based on genotype. In this situation, one could argue that alerts could inform prescribers of the existence of a paper-based scanned test result (but not its result directly). However, interruptive alerts on patients who ultimately do not have an actionable test result (e.g., CYP2C19 normal metabolizers (NMs) on clopidogrel) will contribute to alert fatigue. Some of our sites have worked around the issue of not having discrete results in the EHR by entering problem list items for the pharmacogenetic phenotypes and generating alerts based on those items.
Finally, the fact that the majority of patients received additional medications impacted by the gene they were originally tested for, CYP2C19, suggests that additional thought should be placed into how to share these results and how to shape associated clinical decision support with health-care providers outside the original institution where the results reside. PCI is often performed emergently and requires specialized facilities and personnel that are not available at all hospitals. As a result, many patients undergo PCI in a health-care institution separate from where they receive their primary care treatment. These issues also illustrate the ongoing need for data portability and patient education if they received pharmacogenetic-guided treatment. In our previous report we found a minority of early adopters actively reported the test results to downstream providers outside of their institution.1 This problem will not be unique to CYP2C19 but will hold true for many other pharmacogenetic examples as well (e.g., CYP2D6,CYP2C9). Educating patients that this testing was performed and of their test result may help to bridge this gap, but we previously reported that a minority of institutions routinely provided patients with their genotype results.1 The FDA has been concerned that this may lead to patients altering their medication regimens without health-care provider supervision.14,15 However, this type of alteration has been found to be exceptionally rare (<1%),16 and others have argued that providing patients with their results may be the best approach to ensure portability and downstream use of results.17,18
Economic analyses have focused on the single use of CYP2C19 for selecting an antiplatelet agent, and do not take into account downstream reusability of the CYP2C19 result. Because genotype does not change throughout the lifespan, these additional uses of CYP2C19 genotype can potentially improve care and may decrease health-care expenditures at minimal additional economic cost to the payer or the health-care system. Thus, economic models analyzing the cost effectiveness of genotyping for a single therapeutic decision may be underestimating the true “real world” value of genotyping. This will have to be tested in formal economic studies as there is a cost for building clinical decision support that will need to be incorporated.
Our study does have some limitations. We were only able to assess orders within the health systems where genotyping was ordered. We also only evaluated medication orders for the year following genotyping. Both of these factors could lead to an underestimate of the rate of reusability of the genotype data. It is also possible that some prescribers may have been aware of genotype results in a patient and therefore avoided substrates in patients with actionable genotypes, which again would lead to underestimation of reusability. Our study population also contained a limited number of racially and ethnically diverse patients who may have differing CYP2C19 allele frequencies (e.g., poor, intermediate, and ultrarapid metabolizers are more common in individuals of African ancestry). Finally, our study did not differentiate between incident and prevalent medication orders. For some medications, genotype would be most informative for incident prescriptions and less relevant after the patient has proven to be effective or well-tolerated.
In summary, we demonstrated that CYP2C19 genotype often has treatment implications beyond antiplatelet selection for patients undergoing PCI. The use of CYP2C19 genotype to guide treatment decisions outside of PCI adds potential clinical and economic value to CYP2C19-guided antiplatelet therapy. However, to extract this value, institutions must ensure that providers outside the catheterization laboratory know of the CYP2C19 results and know how to appropriately use it to guide treatment.
Change history
03 August 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
03 August 2020
The original version of this Article contained an error in the spelling of the author Dominick J. Angiolillo, which was incorrectly given as Dominick P. Angiolillo. This has now been corrected in both the PDF and HTML versions of the Article.
References
Empey PE, Stevenson JM, Tuteja S, et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin Pharmacol Ther. 2018;104:664–674.
Limdi NA, Cavallari LH, Lee CR, et al. Cost-effectiveness of CYP2C19-guided antiplatelet therapy in patients with acute coronary syndrome and percutaneous coronary intervention informed by real world data. Pharmacogenomics J. 2020 Feb 11; https://doi.org/10.1038/s41397-020-0162-5 [Epub ahead of print].
Reese ES, Daniel Mullins C, Beitelshees AL, Onukwugha E. Cost-effectiveness of cytochrome P450 2C19 genotype screening for selection of antiplatelet therapy with clopidogrel or prasugrel. Pharmacotherapy. 2012;32:323–332.
Sorich MJ, Horowitz JD, Sorich W, Wiese MD, Pekarsky B, Karnon JD. Cost-effectiveness of using CYP2C19 genotype to guide selection of clopidogrel or ticagrelor in Australia. Pharmacogenomics. 2013;14:2013–2021.
Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323.
Mega JL, Close SL, Wiviott SD, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–1319.
Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830.
Osnabrugge RL, Kappetein AP, Janssens AC. Carriage of reduced-function CYP2C19 allele among patients treated with clopidogrel. JAMA. 2011;305:467–468.
Claassens DMF, Vos GJA, Bergmeijer TO, et al. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. N Engl J Med. 2019;381:1621–1631.
Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98:127–134.
Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37–44.
Moriyama B, Obeng AO, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin Pharmacol Ther. 2017;102:45–51.
Cavallari LH, Lee CR, Beitelshees AL, et al. Multisite investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc Interv. 2018;11:181–191.
US Food and Drug Adminnistration. The FDA warns against the use of many genetic tests with unapproved claims to predict patient response to specific medications: FDA safety communication. 2018. https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific. Accessed 18 December 2019.
US Food and Drug Administration. FDA issues warning letter to genomics lab for illegally marketing genetic test that claims to predict patients’ responses to specific medications. 2019. https://www.fda.gov/news-events/press-announcements/fda-issues-warning-letter-genomics-lab-illegally-marketing-genetic-test-claims-predict-patients. Accessed 18 December 2019.
Carere DA, VanderWeele TJ, Vassy JL, et al. Prescription medication changes following direct-to-consumer personal genomic testing: findings from the Impact of Personal Genomics (PGen) Study. Genet Med. 2017;19:537–545.
GenomeWeb. New coalition, stakeholder groups push back against FDA ‘backdoor' attempts to regulate PGx tests. 2019. https://www.genomeweb.com/molecular-diagnostics/new-coalition-stakeholder-groups-push-back-against-fda-backdoor-attempts#.XfpZbvx7mUk. Accessed 18 December 2019.
Haidar CE, Relling MV, Hoffman JM. Preemptively precise: returning and updating pharmacogenetic test results to realize the benefits of preemptive testing. Clin Pharmacol Ther. 2019;106:942–944.
Acknowledgements
Funding for this work was provided by National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) U01HG007775 (A.L.B.); American Heart Association 17MCPRP33400175 (J.M.S.); National Heart, Lung, and Blood Institute (NHLBI) K23HL143161 and Penn Center for Precision Medicine (S.T.); RO1HL092173 and K24HL133373, Clinical and Translational Science Award UL1TR000165, University of Alabama Birmingham’s Health Service Foundations’ General Endowment Fund and Hugh Kaul Personalized Medicine Institute (N.A.L., C.D.); the American Society of Health System Pharmacists, NIH UL1TR0000005, and by an Anonymous Donor (J.M.S. and P.E.E.), NIH/NHGRI U01HG007253 and U01HG010232 (J.F.P. and E.M.F.), NIH NHGRI U01 HG007269 and NIH National Center for Advancing Translational Sciences (NCATS) UL1 TR000064 (J.A.J. and L.H.C.).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Disclosure
J.F.P. is a consultant for Color Genomics. T.C.S. is paid as a pharmacogenetics consultant for IU Health. D.J.A. declares that he has received consulting fees or honoraria from Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Boehringer Ingelheim, Bristol-Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Haemonetics, Janssen, Merck, PhaseBio, PLx Pharma, Pfizer, Sanofi, and The Medicines Company and has received payments for participation in review activities from CeloNova and St Jude Medical. D.J.A. also declares that his institution has received research grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli Lilly, Gilead, Idorsia, Janssen, Matsutani Chemical Industry Co., Merck, Novartis, Osprey Medical, Renal Guard Solutions, and Scott R. MacKenzie Foundation. F.F. declares that he has received consulting fees or honoraria from, AstraZeneca, Bayer, Sanofi. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Beitelshees, A.L., Stevenson, J.M., El Rouby, N. et al. Evaluating the extent of reusability of CYP2C19 genotype data among patients genotyped for antiplatelet therapy selection. Genet Med 22, 1898–1902 (2020). https://doi.org/10.1038/s41436-020-0894-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0894-2
Key words
This article is cited by
-
Use of the Electronic Health Record for Monitoring Adverse Drug Reactions
Current Allergy and Asthma Reports (2023)