Abstract
Purpose
Polymerase proofreading-associated polyposis is a dominantly inherited colorectal cancer syndrome caused by exonuclease domain missense variants in the DNA polymerases POLE and POLD1. Manifestations may also include malignancies at extracolonic sites. Cancer risks in this syndrome are not yet accurately quantified.
Methods
We sequenced POLE and POLD1 exonuclease domains in 354 individuals with early/familial colorectal cancer (CRC) or adenomatous polyposis. We assessed the pathogenicity of POLE variants with yeast fluctuation assays and structural modeling. We estimated the penetrance function for each cancer site in variant carriers with a previously published nonparametric method based on survival analysis approach, able to manage unknown genotypes.
Results
Pathogenic POLE exonuclease domain variants P286L, M294R, P324L, N363K, D368N, L424V, K425R, and P436S were found in ten families. The estimated cumulative risk of CRC at 30, 50, and 70 years was 11.1% (95% confidence interval [CI]: 4.2–17.5), 48.5% (33.2–60.3), and 74% (51.6–86.1). Cumulative risk of glioblastoma was 18.7% (3.2–25.8) at 70 years. Variants interfering with DNA binding (P286L and N363K) had a significantly higher mutagenic effect than variants disrupting ion metal coordination at the exonuclease site.
Conclusion
The risk estimates derived from this study provide a rational basis on which to provide genetic counseling to POLE variant carriers.
Similar content being viewed by others
INTRODUCTION
Polymerase proofreading-associated polyposis (PPAP; OMIM 61259 and 615083), is an autosomal dominant cancer syndrome caused by the inability of the main replicative DNA polymerases, POLE and POLD1, to proofread newly synthesized DNA strands1. Individuals with PPAP carry germline variants of the 3´–5´ proofreading exonuclease domain of POLE or POLD1 and are at increased risk of colorectal cancer and adenoma. Several other tumor types, including endometrial, ovarian, duodenal, and brain cancers, have also been described in these families2. These tumors often show an “ultramutator” phenotype characterized by mutational loads exceeding 100 variants per megabase2,3,4,5. Knowledge of the age-specific cumulative cancer risks associated with PPAP could lead to improved cancer screening strategies and risk-reducing treatments. So far, only one publication has attempted to give colorectal cancer penetrance estimates in PPAP6. In the present study, we analyze the structural and functional consequences of eight different POLE exonuclease variants found in ten new PPAP families and give new estimates of the cumulative cancer risks associated with PPAP.
MATERIALS AND METHODS
Patients
We recruited 354 consecutive patients exhibiting features of early-onset/familial colorectal cancer (CRC) (218 individuals) and/or attenuated adenomatous polyposis (136 individuals) between 2014 and 2017. Early-onset/familial CRC patients met the revised Bethesda criteria for microsatellite instability (MSI) testing7. Multiple colonic adenomas patients met the French National Cancer Institute criteria for MUTYH screening8. We also included in this group patients for whom endoscopy and pathology reports gave descriptions like “polyposis” or “multiple polyps” rather than the exact number of adenomas. A group of 253 patients were analyzed for the first time, whereas 101 patients had been previously tested negative for Lynch syndrome or adenomatous polyposis at other institutions. Written informed consent was obtained from all subjects and the study received the approval of the Ethical Review Committee of the Cochin University Hospital.
Variant detection
A custom-made Ion Torrent Ion AmpliSeq panel (Thermo Fisher Scientific, Waltham, MA) and a Ion PGM system (Thermo Fisher) were used to sequence the entire coding regions of 11 genes associated with hereditary digestive cancers (APC, MUTYH, AXIN2, MLH1, MSH2, MSH6, CDH1, STK11, PTEN, SMAD4, BMPR1A) and the exons coding the exonuclease domains of POLE and POLD1 (codons 268–471 of POLE and 304–517 of POLD1). A search for large rearrangements in the genes APC, MLH1, MSH2, MSH6, and EPCAM was done by multiplex ligation-dependent probe amplification (MLPA) (MRC Holland).
Cases with loss of MLH1 protein expression in the tumor by immunohistochemistry and without germline MLH1 deleterious variants were further studied by methylation-specific MLPA (MRC Holland) searching for constitutional epimutation of MLH1.
Estimation of penetrance
The objective was to estimate the penetrance function in the context where most genotypes are unobserved. For this purpose, we use a nonparametric method based on a survival analysis approach. Since many genotypes are unobserved, we use a classical expectation–maximization (EM) algorithm9 to estimate iteratively both the penetrance function and the posterior carrier probability. All genetic computations (integration over all possible genotypes) are done using classical sum-product algorithms derived from the graphical model theory10, which are similar to the most recent Elston–Stewart-like algorithms11. The approach we used allows to obtain previously published parametric estimates (e.g., Weibull survival12) as particular cases as well as more general Kaplan–Meier nonparametric estimates. However, the nonparametric approach allows an easier adjustment to the data and therefore provides more accurate estimates of the penetrance. Covariates can also be taken into account using a proportional hazard model. Finally, ascertainment correction is taken into account as described12. The method has been described in detail elsewhere13. Analysis was performed by using R software. In the present study, the covariate tested was gender and a difference between groups was tested using a likelihood ratio test.
Strains and plasmid construction
The Saccharomyces cerevisiae (MATa ade5-1 lys2-InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52) yeast strain used in the study is isogenic to E13414. Nucleotides 372–6669 of POL2 (the yeast homolog of human POLE) were amplified, cloned in pGEMT (Promega) and assembled in the pFA6a-KanMX6 integration plasmid by restriction enzyme cloning. Exonuclease domain variants Pol2-P301L, Pol2-M309R, Pol2-P339L, Pol2-N378K, Pol2-D383N, Pol2-L439V, Pol2-K440R, Pol2-A445T, and Pol2-P451S (encoding variants equivalent to human POLE P286L, M294R, P324L, N363K, D368N, L424V, K425R, A430T, and P436S) were created with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). Plasmids were integrated into the POL2 locus after linearization with Bsu36I and selection for G418 resistance, following the protocol described by Gietz and Schiestl15. Constructs were verified by sequencing. Two control strains were also constructed using the same procedure, a negative control Pol2-wt (wild-type) strain and a positive control Pol2-AIA strain defective in 3’–5’ proofreading exonuclease by variants of the highly conserved Exo I motif DIE into an AIA motif (residues 290–292).
Fluctuation assays
The spontaneous variant rate was measured by fluctuation analysis according to Lang and Murray16. For mutant selection, yeast cultures were plated onto two types of selective media: CAN10X (synthetic complete medium without arginine, 0.5 g/liter of L-canavanine) and HIS- (synthetic complete medium without histidine). Canavanine is a toxic arginine analog, whose uptake requires the arginine transporter encoded by the CAN1 gene. The Canr forward variant assay selects for loss-of-function mutants of this transporter, which occur through a variety of base substitutions, frameshifts, and larger rearrangements. The his7-2 reporter is a -1 frameshift variant creating a run of 7 adenines. Gain-of-function mutants selected by the His+ reverse variant assay occur primarily by +1 or -2 indel changes. All experiments were done in duplicate with two independent clones. The colonies were counted manually for each clone. Fluctuation data were analyzed by the Ma–Sandri–Sarkar maximum-likelihood method17. Calculations were performed with the FALCOR web tool18.
Model building and in silico mutagenesis
The X-ray crystal structure of Saccharomyces cerevisiae DNA polymerase POL219 (pdb code: 4PTF) was used as template for protein in silico mutagenesis. Double-strand DNA in editing mode was positioned into the exonuclease domain using the DNA model contained in the X-ray crystal structure of Pyrococcus abyssi B family DNA polymerase20 (pdb code: 4FLU). The two magnesium ions required for exonuclease activity were then manually placed in A and B sites after comparison with magnesium-bound exonuclease structures. Variants effects on protein stability were estimated by calculating the change in Gibbs free energy with the FoldX suite21 using the apo- and the DNA-bound models.
RESULTS
Contribution of POLE variants to early/familial colorectal cancer and adenomatous polyposis
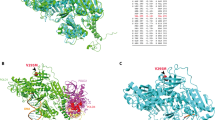
We identified eight rare (ExAC minor allele frequency <0.0001) POLE exonuclease domain missense variants (c.857C>T p.(Pro286Leu), c.881T>G p.(Met294Arg), c.971C>T p.(Pro324Leu), c.1089C>A p.(Asn363Lys), c.1102G>A p.(Asp368Asn), c.1270C>G p.(Leu424Val), c.1274A>G p.(Lys425Arg), and c.1306C>T p.(Pro436Ser); NM_006231.4) in ten independently ascertained individuals (pedigrees shown in Supplementary Figure S1) in our study population of 354 patients (2.8%). Variants L424V and K425R were found in two families each. Five of these variants were novel (P286L, M294R, P324L, D368N, P436S). All variants, with the exception of P324L, were located in or around the DNA binding pocket (Fig. 1a, b). All variants were located on residues invariant in POLE orthologs (Fig. 1c). Five variants (N363K, D368N, L424V, K425R, and P436S) were located in the conserved Exo domains II, IV, and V of the exonuclease22, whereas P286L flanked Exo I and two variants (M294R and P324L) were outside the Exo domains (Fig. 1c). We found five of these variants in the adenomatous polyposis group (of 136 individuals, frequency 3.7%) and five in the early-onset/familial CRC group (of 218 individuals, frequency 2.3%) (Supplementary Figure S2 and Supplementary Table S1).
(a) Surface representation of the exonuclease domain (residues 268–471). (Inset) Mutated residues are highlighted to demonstrate clustering of the variants in and around the DNA binding pocket. (b) Schematic representation of human POLE gene showing the exonuclease (Exo) and polymerase domains. Exons are shown as multicolored boxes. (c) Sequence alignment demonstrating that the eight residues (P286, M294, P324, N363, D368, L424, K425, P436) studied in this work are invariant in POLE orthologs. The missense variants are indicated by closed arrows. Conserved Exo motifs I–V are highlighted in pink and the four conserved acidic metal-binding residues are indicated by an asterisk.
We also estimated the contribution of the Lynch syndrome genes and other adenomatous polyposis genes in the group without previous germline testing (n = 253; left part of Supplementary Figure S2A and B). We identified a Lynch syndrome gene (MLH1, MSH2, MSH6) deleterious germline variant or constitutional promoter epimutation in 20.7% (36/174) of patients with early-onset/familial CRC, whereas APC or MUTYH biallelic deleterious variants were found in 15.2% (12/79) of patients with adenomatous polyposis.
One other individual had co-occurence of a POLE K425R variant and a pathogenic MSH2 c.942+3A>T variant. A rare variant of unknown significance c.1040C>T p.(Pro347Leu) of the exonuclease domain of POLD1 (NM_001256849.1) was also identified in a woman who had an endometrial cancer at 34 years and two colorectal cancers at 48 and 52 years. These two cases were excluded from analyses of disease expression and cancer penetrance.
Phenotypic expression of the disease
To identify the types of cancer associated with PPAP, we recorded all cancer diagnoses that had been made in POLE variant carriers (Table 1 and Supplementary Table S2). In our cohort of ten families, we identified 51 individuals (27 women and 24 men) carrying one of the 8 POLE variants studied in this paper. Ten were the probands and the 41 others were relatives who had tested positive for the presence of the variant identified in the proband or were considered obligate carriers of that variant based on Mendel’s laws of inheritance.
We found that 34/51 (67%) carriers were affected with at least one cancer (15 women and 19 men). Of 34 affected individuals,14 (41%) had at least two synchronous or metachronous cancers. The most frequently observed cancer both in men and women was colorectal cancer, occurring in 25/34 (74%) of the affected carriers (mean age ± SD = 48.9 ± 13.7 years). The only other digestive cancers observed in affected carriers were duodenal or jejunal cancers (4/34 [12%]). Brain tumors were observed in 5/34 (15%) affected carriers (we previously reported a detailed pathological description of four of these tumors23) and four other brain tumors were diagnosed in first-degree relatives of carriers. Of 15 affected women carriers, 11 had at least one CRC, 5 had an endometrial cancer, 5 had a breast cancer, and 3 had an ovarian carcinoma.
Penetrance of cancers associated with POLE variants
To better assess cancer risks in PPAP, we extended our analysis beyond the known variant carriers and used a newly described statistical framework that incorporated both genotyped and nongenotyped relatives to obtain penetrance estimates (see “Materials and Methods”). Penetrance at 30, 50, and 70 years was 11.1% (95% confidence interval [CI]: 4.2–17.5), 48.5% (33.2–60.3), and 74% (51.6–86.1) for colorectal cancers. Cumulative risk of colorectal cancer was not significantly different between men and women (p = 0.0572): penetrance at 30, 50, and 70 years was 14.9% (5.2–23.6), 61.2% (40.1–74.9), and 86.4% (60.6–95.3) for men; and 8.3% (1.9–14.2), 39.8% (19.1–55.2), and 65.6% (33.5–82.2) for women (Fig. 2a). The estimated cumulative risk of brain tumors, one of the most striking features of PPAP, was 18.7% (3.2–25.8) at 70 years. Taking into account all cancers, the estimated penetrance for cancers at any site at 30, 50, and 70 years was 21.3% (12.1–29.5), 61% (47.3–71.2), and 89% (72.6–95.6) and was similar for both sexes (p = 0.245) (Fig. 2b).
Functional significance of the variants determined by yeast fluctuation assays
All tested POL2 variants had significantly increased mutagenesis levels compared with the wild-type control on Canr and His+ variant assays (Fig. 3 and Supplementary Table S3). Differences between variants were more readily seen with the more sensitive His+ assay that tests for instability of a short mononucleotide repeat. Whereas some variants (pol2-D383N, pol2-K440R, and pol2-P451S) had mutagenesis levels similar to those of the exonuclease-deficient pol2-AIA, other variants had mutagenesis levels that significantly exceeded those of the exonuclease-null pol2-AIA, sometimes by as much as one order of magnitude (pol2-P301L and pol2-N378K).
Structural significance of the variants determined by in silico mutagenesis
We calculated the change in Gibbs free energy between mutant and wild-type models using both apo- (ΔΔGAPO) and DNA-bound (ΔΔGDNA) yeast DNA polymerase POL2 models (Supplementary Table S4). Five variants (P286L, M294R, P324L, L424V, and P436S) were predicted to significantly increase the ΔΔGAPO, corresponding to a protein structure destabilization. We also estimated the influence of variants on DNA binding by calculating the difference between ΔΔGDNA and ΔΔGAPO. Two variant (P286L and N363K) were predicted to destabilize DNA binding. Indeed, modeling of these two variants suggested that they affect DNA positioning in the exonuclease active site (Fig. 4a, b, and Supplementary Movie S1). A third mechanism of exonuclease deficiency was alteration of ion metal coordination as seen with D368N, a variant that eliminates a carboxylate from the DEDDy motif required for ion metal coordination at the catalytic site24, and M294R, predicted by in silico modeling to alter ion coordination by interacting with E277 (also a DEDDy residue, Fig. 4c).
In our models, P301L (hP286L) leads to the (294-307) loop destabilization right next to the exonuclease site (green background) and modify the DNA binding pocket structure (a). N378K (hN363K) does not appear to have a destabilizing effect on the apo-model, but considerably affects the DNA binding capacity by causing direct steric clashes (red springs) with DNA (see also Video S1) (b). M309R (hM294R) modifies exonuclease site structure by establishing new hydrogen bonds (green dotted line) with catalytic E292 (hE277) (c).
These different mechanisms of exonuclease domain disruption correlated nicely with the results of our yeast mutagenesis assays (Fig. 3): variants that directly destabilized DNA binding (P286L and N363K) were the strongest mutators, with mutagenesis rates one order of magnitude above the exonuclease-null allele, whereas variants selectively affecting ion metal coordination (D368N, M294R) had variant rates similar to those of the null allele. The other variants, with various degrees of protein destabilization, ranked between these two groups (with the exception of P436S, a variant located further away from the DNA binding interface and producing milder effects in yeast assays).
Pathogenic classification of POLE missense variants
We gathered the results of our functional assays with all other available evidence pertaining to the American College of Medical Genetics and Genomics (ACMG) categories25. Five variants (P286L, N363K, D368N, L424V, P436S) were classified as pathogenic and the three others (M294R, P324L, K425R) as likely pathogenic (details in Supplementary Table S5).
DISCUSSION
DNA polymerase proofreading-associated polyposis is a recently described hereditary cancer syndrome1 for which penetrance of colorectal cancer has not yet been accurately quantified, which is an impediment to the establishment of cancer screening guidelines. Cancer penetrance data have been difficult to obtain because the disease is rare in the population, with only a few dozen cases described since the discovery of the syndrome five years ago2, and because all putative causal variants known to date are exonuclease domain missense variants, which are notoriously difficult to classify2. Establishing the pathogenicity of missense variants requires gathering extensive phenotypic, functional, and population data25, something that has not been done in most published studies reporting germline POLE or POLD1 variants.
In this study we carefully classified all rare exonuclease domain variants discovered in early/familial colorectal cancer or adenomatous polyposis patients by systematically performing yeast functional assays and applying the ACMG criteria for missense variant classification. Based on a group of ten families carrying eight different POLE exonuclease domain variants classified as pathogenic or likely pathogenic, we show that penetrance at 30, 50, and 70 years is 11.1% (95% CI: 4.2–17.5), 48.5% (33.2–60.3), and 74% (51.6–86.1) for colorectal cancers.
In the only other study of penetrance published so far, which was mainly based on an analysis of data from the literature, Buchanan et al. found somewhat higher cumulative risks of colorectal cancer to age 70 years (97% [85–99%] for males and 92% [75–99%] for females) in carriers of the undoubtedly pathogenic POLE L424V variant6. However, they found much lower risks [40% (26–57%) for males and 32% (20–47%) for females] when they included all of the reported rare germline variants predicted to be pathogenic by multiple commonly used in silico tools. While this discrepancy might be explained by different levels of pathogenicity between variants, it could also be due to the inclusion of variants of uncertain significance in that study. Indeed, whereas some of their 15 POLE variants had enough data available to be classified as pathogenic or likely pathogenic based on good evidence for cosegregation with the disease (L424V1, N363K26, Y458F27), yeast functional studies (L424V28), and somatic evidence of driver variants on the same residue (L424, D368, Y458)3, all other variants would have been classified as variants of unknown pathogenicity by the ACMG guidelines. We thus think that our data, based on a set of eight different deleterious POLE variants, are the most accurate evaluation of colorectal cancer risk in PPAP to date. Our results put colorectal cancer risk levels in PPAP syndrome on a par with those associated with Lynch syndrome29. It seems, therefore, reasonable to implement in PPAP colorectal cancer screening guidelines similar to those applied in Lynch syndrome, that is, colonoscopy every 1–2 years beginning at age 20–25 years or 2–5 years before the youngest age at diagnosis of colorectal cancer in the family before age 25 years.
Management of carriers of pathogenic POLE variants requires reliable information not only about colorectal cancer risk but also about extracolonic cancers. These other cancers increase the global burden of cancer in PPAP to still higher levels, as cumulative incidence for any cancer at 30, 50, and 70 years was 21.3% (12.1–29.5), 61% (47.3–71.2), and 89% (72.6–95.6). Among extracolonic cancers, we found a strikingly high number of brain tumors, mostly giant-cell glioblastomas. We estimated cumulative risks of brain tumors to age 70 years to be 18.7% (3.2–25.8). Although confidence margins were wide, these results show that brain tumors belong to the tumor spectrum of PPAP and raise the question of whether a surveillance program for the early detection of brain tumors is justified. Tumors of the central nervous system have been associated with several hereditary syndromes, including Li–Fraumeni syndrome, neurofibromatosis, nevoid basal cell carcinoma syndrome, tuberous sclerosis, von Hippel–Lindau disease, familial adenomatous polyposis, and Lynch syndrome. It has been shown that long-term compliance with a comprehensive surveillance protocol for early tumor detection in individuals with Li–Fraumeni syndrome, a condition associated with a high risk of glioblastomas in children and young adults, is feasible and that early tumor detection through surveillance is associated with improved long-term survival30. On the other hand, it is not recommended to screen for brain tumors in conditions such as Lynch syndrome31,32 that are estimated to confer a much lower risk of brain tumors (0.5–3.7% by age 70 years)31,33,34,35. Brain tumor penetrance in PPAP clearly falls somewhere between these two situations; it is higher than in Lynch syndrome but still much lower than in Li–Fraumeni syndrome. Our estimates clearly need to be refined before deciding whether prospective studies of early detection programs that may include magnetic resonance imaging or computerized tomography scan should be conducted.
We also found several endometrial and ovarian tumors in female carriers of the variants. These observations tally well with the recently published comprehensive analysis of human cancer that found somatic POLE and POLD1 driver variants in a restricted set of cancer types (brain, colorectal, endometrial, and ovarian)3 that overlaps with the main tumor types we found in carriers of POLE germline variants. These data suggest that, contrary to the initial description of PPAP, extracolonic cancers occur in POLE as well as in POLD1 variant carriers.
We performed yeast functional assays primarily to help in classifying POLE exonuclease domain variants found in our patients. Interestingly, these assays revealed that, while some variants increased mutagenesis to a similar extent as the exonuclease-deficient strain, other variants, in particular those predicted to inhibit DNA binding (P286L and N363K), had a mutator effect that significantly exceeded that of the exonuclease-deficient strain, by as much as one order of magnitude. It has been assumed that the ultramutated phenotype of POLE mutant-carrying tumors was a direct consequence of exonuclease deficiency, but these results suggest that variants that inhibit DNA binding must impact replication fidelity in additional ways. Strikingly high levels of mutagenicity had already been shown for POLE P286R, the most frequently recurring cancer-associated variant, in similar yeast assays36. Interestingly, P286R is also predicted to inhibit DNA binding. To explain this hypermutagenic phenotype, a model has recently been proposed wherein limited access of the 3’-terminus to the exonuclease site might promote binding at the polymerase site, thus stimulating polymerization and extension of mismatched primer termini, thereby dramatically increasing variant rates37,38. The fact that different variants at the same (P286L) and other residues (N363K) that inhibit DNA binding are also associated with very high variant rates lends weight to that model.
This study has potential limitations. Although fluctuation analysis is an established test for the analysis of genetic instability, and human POLE and yeast POL2 exonuclease domains are 69% identical at the protein level, it cannot be ruled out that identical amino acid substitutions might result in different functional consequences in human and yeast cells, as illustrated by a recent publication in which the cancer variant hotspot V411L did not significantly elevate mutagenesis in a similar yeast assay28. Besides, whereas ultrahypermutagenic variants such as P286L and N363K can be confidently classified as deleterious, the level at which a cut-off of hypermutagenicity should be set for clinical use of the test is not known. In this study, we decided to classify as deleterious variants that significantly increased mutagenicity levels compared with the wild-type strain. However, that might not be stringent enough; some variants with mildly increased mutagenicity levels might not cause disease or might result in a disease with reduced penetrance. Finally, family data were obtained from genealogical trees and medical records compiled by clinical geneticists and genetic counselors. Given the large size of some families and the recording of their medical history across several generations, systematic confirmation of family history through medical records was not feasible and information given about the older generations might be unreliable.
In summary, CRC risk levels in POLE variant carriers warrant screening and management guidelines similar to those currently recommended for people with Lynch syndrome. POLE (and most probably POLD1) should be included in multigene panel testing for hereditary colorectal cancer and adenomatous polyposis as recently recommended by the UK Cancer Genetics Group39. Further refinement of our brain tumor estimates is needed to inform genetic counseling and management. Our results, and those of other groups, showing strikingly different mutagenic effects of the various POLE exonuclease domain variants raise the question of whether these differences might correlate with cancer penetrance and localization, an endeavor that will need data from a larger group of affected families.
References
Palles C, Cazier J-B, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144.
Rayner E, van Gool IC, Palles C, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81.
Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056.e10.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337.
Heitzer E, Tomlinson I. Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev. 2014;24:107–113.
Buchanan DD, Stewart JR, Clendenning M, et al. Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med. 2018;20:890–895.
Umar A, Risinger JI, Hawk ET, Barrett JC. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat Rev Cancer. 2004;4:153–158.
Institut National Du Cancer. La polypose associée aux mutations bi-alléliques du gène MUTYH—Ref: POLYPMUTMUT11. http://www.e-cancer.fr/Expertises-et-publications/Catalogue-des-publications/La-polypose-associee-aux-mutations-bi-alleliques-du-gene-MUTYH. Accessed 29 January 2018.
Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B. 1977;39:1–22.
Koller D, Friedman N. Probabilistic graphical models: principles and techniques. Cambridge, MA: MIT Press; 2009.
Totir LR, Fernando RL, Abraham J. An efficient algorithm to compute marginal posterior genotype probabilities for every member of a pedigree with loops. Genet Sel Evol. 2009;41:52.
Alarcon F, Bourgain C, Gauthier-Villars M, Planté-Bordeneuve V, Stoppa-Lyonnet D, Bonaïti-Pellié C. PEL: an unbiased method for estimating age-dependent genetic disease risk from pedigree data unselected for family history. Genet Epidemiol. 2009;33:379–385.
Alarcon F, Planté-Bordeneuve V, Olsson M, Nuel G. Non-parametric estimation of survival in age-dependent genetic disease and application to the transthyretin-related hereditary amyloidosis. PLoS One. 2018;13:e0203860.
Jin YH, Clark AB, Slebos RJC, et al. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat Genet. 2003;34:326–329.
Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34.
Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82.
Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179.
Hall BM, Ma C-X, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565.
Jain R, Rajashankar KR, Buku A, et al. Crystal structure of yeast DNA polymerase ε catalytic domain. PLoS One. 2014;9:e94835.
Gouge J, Ralec C, Henneke G, Delarue M. Molecular recognition of canonical and deaminated bases by P. abyssi family B DNA polymerase. J Mol Biol. 2012;423:315–336.
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–388.
Shevelev IV, Hübscher U. The 3’ 5’ exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–376.
Vande Perre P, Siegfried A, Corsini C, et al. Germline mutation p.N363K in POLE is associated with an increased risk of colorectal cancer and giant cell glioblastoma. Fam Cancer. 2019;18:173–178.
Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424.
Rohlin A, Zagoras T, Nilsson S, et al. A mutation in POLE predisposing to a multi-tumour phenotype. Int J Oncol. 2014;45:77–81.
Hansen MF, Johansen J, Bjørnevoll I, et al. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer. 2015;14:437–448.
Barbari SR, Kane DP, Moore EA, Shcherbakova PV. Functional analysis of cancer-associated DNA polymerase ε variants in Saccharomyces cerevisiae. G3 (Bethesda). 2018;8:1019–1029.
Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on Colorectal Cancer. Am J Gastroenterol. 2014;109:1159–1179.
Villani A, Shore A, Wasserman JD, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li–Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17:1295–1305.
Vasen HF, Sanders EA, Taal BG, et al. The risk of brain tumours in hereditary non-polyposis colorectal cancer (HNPCC). Int J Cancer. 1996;65:422–425.
Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456.
Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218.
Barrow E, Robinson L, Alduaij W, et al. Cumulative lifetime incidence of extracolonic cancers in Lynch syndrome: a report of 121 families with proven mutations. Clin Genet. 2009;75:141–149.
Therkildsen C, Ladelund S, Rambech E, Persson A, Petersen A, Nilbert M. Glioblastomas, astrocytomas and oligodendrogliomas linked to Lynch syndrome. Eur J Neurol. 2015;22:717–724.
Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901.
Parkash V, Kulkarni Y, Ter Beek J, Shcherbakova PV, Kamerlin SCL, Johansson E. Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase ε. Nat Commun. 2019;10:373.
Xing X, Kane DP, Bulock CR, et al. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat Commun. 2019;10:374.
Taylor A, Brady AF, Frayling IM, et al. Consensus for genes to be included on cancer panel tests offered by UK genetics services: guidelines of the UK Cancer Genetics Group. J Med Genet. 2018;55:372–377.
Acknowledgements
The authors thank Dmitry Gordenin for providing the yeast strain, Irmine Ferreira for technical assistance, and Sophie Grandjouan and Xavier Jeunemaître for useful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hamzaoui, N., Alarcon, F., Leulliot, N. et al. Genetic, structural, and functional characterization of POLE polymerase proofreading variants allows cancer risk prediction. Genet Med 22, 1533–1541 (2020). https://doi.org/10.1038/s41436-020-0828-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0828-z
Keywords
This article is cited by
-
Multiple duodenal epithelial tumors in a patient with polymerase proofreading-associated polyposis in POLE variant
Clinical Journal of Gastroenterology (2024)
-
Characterization of POLE c.1373A > T p.(Tyr458Phe), causing high cancer risk
Molecular Genetics and Genomics (2023)
-
Histomorphological and molecular genetic characterization of different intratumoral regions and matched metastatic lymph nodes of colorectal cancer with heterogenous mismatch repair protein expression
Journal of Cancer Research and Clinical Oncology (2023)
-
Solving the enigma of POLD1 p.V295M as a potential cause of increased cancer risk
European Journal of Human Genetics (2022)
-
POLE, POLD1, and NTHL1: the last but not the least hereditary cancer-predisposing genes
Oncogene (2021)