Abstract
Purpose
To investigate the effectiveness of phenotype-based search approaches using publicly available online databases.
Methods
We included consecutively solved cases from our exome database. For each case, the combination of Human Phenotype Ontology terms reported by the referring clinician was used to perform a search in three commonly used databases: OMIM (first 300 results), Phenolyzer (first 300 results), and Mendelian (all 100 results).
Results
One hundred cases were included (43 females; mean age: 10 years). The actual molecular diagnosis identified through exome sequencing was not included in the search results of any of the queried databases in 33% of cases. In 85% of cases it was not found within the top five search results. When included, its median rank was 61 (range: 1–295), 21 (1–270), and 29 (1–92) in OMIM, Phenolyzer and Mendelian, respectively.
Conclusion
This study demonstrates that, in most cases, phenotype-based search approaches using public online databases is ineffective in providing a probable diagnosis for Mendelian conditions. Genotype-first approach through molecular-guided diagnostics with backward phenotyping may be a more appropriate approach for these disorders, unless a specific diagnosis is considered a priori based on highly unique phenotypic features or a specific facial gestalt.
Similar content being viewed by others
INTRODUCTION
The traditional diagnostic approach in medical genetics utilizes the observation and cataloging of the combination of phenotypic features in a patient to search for a probable genetic diagnosis. Nevertheless, the growing number of gene–disease associations as well as overlapping clinical features of different genetic disorders complicate the differential diagnosis, especially in clinically and genetically heterogeneous disorders. Variable expression and pleiotropic phenotypic effects further add to this complexity.1 Moreover, different clinical features may have varying degrees of specificity, and some of them may even be unrelated to the patient’s primary disease.2 These factors may result in a time-consuming, cumbersome search for a probable genetic diagnosis. Phenotype-based search tools have been developed to integrate available heterogeneous phenotypic data and aid in this complex diagnostic process. These are used routinely by medical geneticists. Nevertheless, the effectiveness and expected yield of phenotype-based search tools in the diagnostics of Mendelian disorders is not clear. The need to investigate their effectiveness is further emphasized as next-generation sequencing–based testing becomes more available and genomic sequencing is performed more routinely in clinical care, further revealing the complexity of gene–disease associations. In this study, we investigated the diagnostic yield of the phenotype-based search approach using publicly available online databases.
MATERIALS AND METHODS
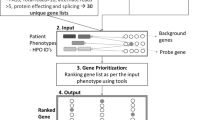
We retrospectively included consecutive probands examined at the Recanati Genetic Institute in the Rabin Medical Center, for whom exome sequencing and analysis yielded a molecular diagnosis during the period between November 2017 and August 2019. Both clinical and research exome cases were included. Fetal exome sequencing cases were not included. Exome sequence analysis is described in the Supplementary data. For each case, the combination of Human Phenotype Ontology (HPO) terms reported by the referring medical geneticist was used to perform a search in three commonly used online databases for a probable genetic diagnosis: OMIM (https://www.omim.org/)3, Phenolyzer (http://phenolyzer.wglab.org/),4 and Mendelian (http://www.mendelian.co/). We examined whether the actual molecular diagnosis (AMD) identified through exome sequencing analyses was included in the first 300 search results in OMIM and in Phenolyzer, and in all 100 search results in the Mendelian database.
RESULTS
We included 100 probands (57 males, 43 females; 67 clinical exomes, 33 research exomes). The mean age of the patients included was 10 years (median age: 5 years and 7 months; range: 1 month–47 years). Most cases (95/100) were trios of a proband and two parents, either with or without additional siblings. The most common main indication for exome sequencing was cognitive abnormalities, intellectual disability or developmental delay with or without additional neurological abnormalities, or multiple congenital anomalies. The characteristics of the patients included are summarized in Table 1.
As previously described, we performed search queries for each of the cases in three different public databases, using HPO terms reported by the referring medical geneticist. The search results did not include the AMD in any of the three databases (OMIM, Phenolyzer, and Mendelian) in 33.0% of cases. The AMD was included in the search results in one, two, or all three databases in 30.0%, 20.0%, and 17.0% of cases, respectively. The proportion of cases for which the AMD was included in at least one of the three databases did not differ markedly between the subgroup of patients with the most common main indication for exome sequencing (45/71, 63.0%) and those with other main indications (22/29, 76.0%) (Table 2).
Analysis of the search results demonstrated a low search yield in all three databases (Table 3), as the AMD was included in the search results in only 58.0%, 28.0%, and 35.0% of cases in OMIM, Phenolyzer, and Mendelian, respectively. In cases in which the AMD was included in the search results, its median rank in the results list was 61 in OMIM (range: 1–295), 21 in Phenolyzer (1–270), and 29 in Mendelian (1–92). It was ranked first in the results list in only 3/100 cases in OMIM, and in a single case in both Phenolyzer and Mendelian. Moreover, it was included in the top five search results in only 9.0%, 6.0%, 7.0%, and 15.0%, and in the first ten search results in 10.0%, 10.0%, 9.0%, and 21.0% of cases, in OMIM, Phenolyzer, Mendelian, and any of the three databases, respectively, further demonstrating the low search yield in all these databases. Using the clinical synopsis filter in OMIM did not change these results significantly (Table 3).
We searched for potential disorder-related factors that may have contributed to the low search yield. To this end, we further investigated the 79 cases for which the AMD was not included in the first ten search results in any of the three databases. We identified four potential disorder-related factors that may have affected the results and divided these 79 cases according to the factor that we assumed contributed the most to the low search yield in each case, as shown in Fig. 1. Not surprisingly, the low search yield was most commonly attributed to clinical overlap among different disorders (49/79, 62.0%). Another factor that may have affected the results in a subset of cases (12/79, 15.0%) was blended phenotype, that is, cases in which either not all of the phenotypic features reported by the clinician could be attributed to the AMD (11/12 cases) or where exome sequencing revealed two AMDs, each of which explained part of the clinical phenotype (a proven double molecular diagnosis; 1/12 cases). Additional factors identified were mild or partial manifestation of a known syndrome in 10/79 cases (13.0%) and a new gene–disease association or a very rare disease (<4 unrelated cases reported in the literature) in 8/79 cases (10.0%).
DISCUSSION
Phenotypic matching between a patient’s clinical presentation and disorders reported in the literature has motivated the study of phenotypic similarity-based algorithms in the field of genetic diagnostics,5,6,7,8,9 and constitutes a major subject of algorithmic development for the improvement of the diagnostic process of Mendelian disorders. In this study, we investigated the diagnostic yield of phenotype-based search in publicly available online databases. We found a low search yield, indicating that this search strategy is ineffective in the diagnostics of Mendelian disorders. We propose several potential explanations for this extremely low yield, which include factors related to the disorders, the clinical evaluation, or the databases used.
Disorder-related limitations in phenotype-based search
As demonstrated in this study, disorder-related factors that may affect the phenotype-based search results include clinical overlap shared by different syndromes, mild or partial manifestation of a known syndrome, instances of a new or very rare disorder, as well as investigation of blended phenotypes. Overlapping phenotypes is a well-recognized limitation in the investigation of gene–disease association.10,11 Search tools may assign a higher weight to unique clinical features than to common ones. Yet, phenotypic overlap still remains an important aspect to consider and it is not surprising that clinical overlap among different disorders was the disorder-related factor to which low search yield was most commonly attributed in our study. Lack of specificity of clinical features for clinically and genetically heterogeneous disorders can also affect phenotype search approaches for diagnosis, as certain phenotypic terms such as “developmental delay,” “intellectual disability,” “hypotonia,” and many others can be associated with a multitude of genetic disorders.
Mild or partial manifestation of a known syndrome may stem from variable expression of the disease. In light of the growing understanding of nonlinearity of genotype–phenotype associations, it became apparent that individuals with the same genetic disorder or even the same pathogenic variant may manifest a wide spectrum of disease severity.12 With the increased availability and application of clinical genomic sequencing, it is expected that more individuals will be diagnosed with mild or partial manifestations of known syndromes. It is also possible that in cases in which exome sequencing was performed early on during infancy or early childhood, age-dependent penetrance of some of the known disease manifestations may result in a reported set of HPO terms apparently representing a milder or partial manifestation of the AMD, impeding its inclusion in the phenotype-based search results.
Since its introduction, exome sequencing has brought to the identification of about 160 new disease-associated genes annually.13,14 As demonstrated in our study, the ongoing identification of novel gene–disease associations may impose limitations on phenotype-based search for a probable diagnosis in online databases, as this search strategy depends on the frequency at which these databases are updated and include the most recent published evidence.
We defined a blended phenotype in cases for which only part of the patient’s phenotype, as represented by the HPO terms used, was explained by a single AMD. This may occur in cases with an established multiple Mendelian diagnoses in a single patient, which is not as rare as previously thought and accounts for an average of 4.3% of cases diagnosed by exome sequencing (range 1.4–7.2%).15,16 Alternatively, a blended phenotype may be found in cases in which genomic sequencing reveals a single AMD that explains only part of the clinical features and does not uncover additional genetic causes to explain the rest of the patient’s phenotype. This may occur in cases of multiple Mendelian diagnoses that cannot be all revealed by exome sequencing, since they are related to additional gene–disease associations yet uncovered or due to exome sequencing limitations in revealing certain types of pathogenic variants, such as polynucleotide repeat expansions, mitochondrial DNA variants, or noncoding variants. It may also occur when a Mendelian disorder and a multifactorial condition coexist in the same individual. Moreover, some cases could represent instances with less typical or new, yet unrecognized, manifestations of the AMD due to phenotypic expansion of a known disorder and/or limited phenotypic characterization of a new disease association. This is relevant as continued characterization and deep phenotyping of patients with newly reported disorders help to better understand these novel conditions and delineate the clinical spectrum beyond the first gene–disease association report.17,18 In all of these scenarios, an unspecific phenotype is expected to negatively impact the search yield, as it creates “phenotypic noise” limiting the search for a single probable diagnosis that can explain the patient’s phenotype.
Clinical evaluation–related limitations in phenotype-based search
Clinical evaluation–related limitations of phenotype-based search can be attributed to the HPO term selection strategy. These include both the number of HPO terms used and their content, which directly impact the informativeness of the query. Concerning the number of HPO terms used, on one hand, it is expected that many of the probands selected for exome sequencing would not have a specific phenotype that allows to focus on a specific gene or a specific group of disorders or genes. Therefore, clinical overlap and the complexity of the differential diagnosis in these cases may not enable the clinician to choose only some phenotypic features for the analysis, which results in an apparently blended phenotype represented by the combination of selected HPO terms that include also phenotypic features unrelated to the patient’s AMD. On the other hand, an attempt to use a smaller number of HPO terms may result in partial representation of the patient’s AMD, which, again, may negatively affect the search yield. One recent study has suggested that using 5 well-chosen key HPO terms is as successful as using 10 or 15 HPO terms for the interpretation of genomic sequencing results.19 Nevertheless, it is unclear whether this suggested approach would be as effective in the case of phenotype-based search for a probable diagnosis in online databases prior to exome sequencing analyses.
Concerning the content of HPO terms used, it was previously suggested that when coming to use HPO terminology, ideally the most specific HPO terms should be used.20 It might be the case that some of the algorithms automatically expand the query from a more specific term to a more general term; nevertheless, it is not determined whether specific terms are superior to general terms in all cases and for all types of disorders. For instance, it is not clear whether in all cases the inclusion of separate HPO terms specifying all of the patient’s dysmorphic features will necessarily yield better search results compared with the inclusion of the general HPO term “abnormal facial shape” (HP:0001999). Moreover, at least in some cases, a meticulous query to obtain the most specific HPO terms may depend on the clinical expertise of the referring physician, as well as on ancillary test results. Therefore, in certain cases it may require evaluation by other medical specialists and performance of additional tests, prolonging the diagnostic odyssey. The issue of selecting specific versus general HPO terms for diagnostic purposes in different cases and in different types of disorders is an important subject to be continuously evaluated and optimized. A recently published article suggests how to optimize the set of HPO terms used.20 The selection of relevant HPO terms depends, nevertheless, mainly on clinical judgment, making it an important source of variability that imposes limitations when using phenotype-based search tools.
Database-related limitations in phenotype-based search
Some of the inherent features of online phenotype-based search tools may negatively affect the search results. One of these factors may stem from the limitations of their algorithms that are used to prioritize a list of probable diagnoses according to a phenotype query. A major limitation of publicly available phenotype databases/catalogs and online search tools can be attributed to their manual curation. This may result in a lag between publication of new gene–disease associations and database updates, as well as outdated information in these databases of the most recently known disease manifestations for a given disorder.
Potential implications
Medical geneticists are facing the challenge of a time-consuming evaluation in each case and increasing demands related to advanced genetic and genomic testing.21 Furthermore, a recent study that investigated the current conditions in medical genetics practice found that wait times and average new patient caseloads have increased over time, while the number of geneticists has not.22 These changes require adaptations that will keep cost-effectiveness and shorten turnaround times for analyses and results without compromising the quality of patient care. To investigate potential adaptations, we focused in this study on phenotype-based search for a differential diagnosis in online databases. This is a key component in the practice of medical genetics. Our results demonstrate that phenotype-based search approaches using public online databases is ineffective in diagnostics of Mendelian conditions in the era of advanced genomic testing. They suggest that better phenotype-based search tools are needed to improve the diagnostic process. In addition, they emphasize the need to consider a more structured strategy for HPO term selection. Finally, these results suggest that a genotype-first approach with backward phenotyping may be more efficient, except for patients who present pathognomonic or extremely rare or unique phenotypic features or a specific facial gestalt, for whom a specific diagnosis can be considered a priori. This suggested shift toward molecular diagnostics through a genotype-first approach should be further investigated in the future for better differential refinement of its potential use in different types of disorders. The proposed changes in phenotypic evaluation are summarized in Fig. 2.
Limitations
This study has several limitations. First, it did not include “all-comer” patients from our genetics clinic, including those with a specific phenotypic gestalt that may allow reaching a diagnosis by gene-specific pathogenic variant testing or gene panel testing and therefore not requiring exome sequencing. These cases would probably have shown a better diagnostic yield in phenotype-based search tools. Nevertheless, the methodological structure of this study aimed to represent the real-life experience of Mendelian disorder diagnostics in cases for which exome sequencing is necessary and indicated.
Second, most cases in this study (71.0%) had the same main clinical indication for exome sequencing of intellectual disability or developmental delay with or without additional neurological features, or multiple congenital anomalies. It is possible that in cases with different indications for exome sequencing the phenotype-based search would return a different yield. Yet, this should be tested separately for specific groups of disorders in future studies.
Third, in this study, due to practical considerations, we investigated the results in three commonly used publicly available online phenotype-based catalogs and tools, and did not attempt to do an exhaustive evaluation of all online databases available for that purpose. Nevertheless, considering the very low yield we obtained from our study and the limitations of phenotype-based search tools discussed above, other additional tools are unlikely to yield significantly different results. However, this could be further investigated in additional studies in the future.
Conclusion
This study demonstrates that phenotype-based search using publicly available online databases and catalogs is ineffective in streamlining the diagnoses of Mendelian conditions. Our results suggest that molecular diagnostics through a genotype-first approach with deep backward phenotyping should be considered as a central diagnostic strategy in medical genetics, with added modifications on a case by case basis. These suggested measures are aimed at improving the diagnostics of Mendelian disorders in the era of advanced genomic testing. They should be further investigated in future studies to refine their differential applicability in diverse groups of Mendelian conditions.
Data availability
The de-identified data set used in this study is available upon reasonable request from the corresponding author.
References
Chakravorty, S. & Hegde, M. Gene and variant annotation for mendelian disorders in the era of advanced sequencing technologies. Annu. Rev. Genomics Hum. Genet. 18, 229–256 (2017).
Köhler, S. et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 85, 457–464 (2009).
Online Mendelian Inheritance in Man (OMIM). https://omim.org/ (2020).
Yang, H., Robinson, P. N. & Wang, K. Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 12, 841–843 (2015).
Jia, J. et al. RDAD: a machine learning system to support phenotype-based rare disease diagnosis. Front. Genet. 9, 587 (2018).
Saklatvala, J. R., Dand, N. & Simpson, M. A. Text-mined phenotype annotation and vector-based similarity to Improve identification of similar phenotypes and causative genes in monogenic disease patients. Hum. Mutat. 39, 643–652 (2018).
Li, Q., Zhao, K., Bustamante, C. D., Ma, X. & Wong, W. H. Xrare: a machine learning method jointly modeling phenotypes and genetic evidence for rare disease diagnosis. Genet. Med. 21, 2126–2134 (2019).
Chen, J. et al. Novel phenotype-disease matching tool for rare genetic diseases. Genet. Med. 21, 339–346 (2019).
James, R. A. et al. A visual and curatorial approach to clinical variant prioritization and disease gene discovery in genome-wide diagnostics. Genome Med. 8, 13 (2016).
van Driel, M. A., Bruggeman, J., Vriend, G., Brunner, H. G. & Leunissen, J. A. M. A text mining analysis of the human phenome. Eur. J. Hum. Genet. 14, 535–542 (2006).
Yu, H. & Zhang, V. W. Precision medicine for continuing phenotype expansion of human genetic diseases. Biomed. Res. Int. 2015, 745043 (2015).
Yadav, A., Vidal, M. & Luck, K. Precision medicine—networks to the rescue. Curr. Opin. Biotechnol. 63, 177–189 (2020).
Boycott, K. M. et al. International cooperation to enable the diagnosis of all rare genetic diseases. Am. J. Hum. Genet. 100, 695–705 (2017).
Bamshad, M. J., Nickerson, D. A. & Chong, J. X. Mendelian gene discovery: fast and furious with no end in sight. Am. J. Hum. Genet. 105, 448–455 (2019).
Balci, T. B. et al. Debunking Occam’s razor: diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin. Genet. 92, 281–289 (2017).
Posey, J. E. et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 376, 21–31 (2017).
Bend, R. et al. Phenotype and mutation expansion of the PTPN23 associated disorder characterized by neurodevelopmental delay and structural brain abnormalities. Eur. J. Hum. Genet. 28, 76–87 (2020).
Zhang, L. X. et al. Further delineation of the clinical spectrum of KAT6B disorders and allelic series of pathogenic variants. Genet. Med. 22, 1338–1347 (2020).
Kernohan, K. D., Hartley, T., Alirezaie, N., Care4Rare Canada Consortium, Robinson, P. N., Dyment, D. A. & Boycott, K. M. Evaluation of exome filtering techniques for the analysis of clinically relevant genes. Hum. Mutat. 39, 197–201 (2018).
Köhler, S. et al. Encoding clinical data with the human phenotype ontology for computational differential diagnostics. Curr. Protoc. Hum. Genet. 103, e92 (2019).
Sukenik-Halevy, R., Ludman, M. D., Ben-Shachar, S. & Raas-Rothschild, A. The time-consuming demands of the practice of medical genetics in the era of advanced genomic testing. Genet. Med. 18, 372–377 (2016).
Maiese, D. R., Keehn, A., Lyon, M., Flannery, D. & Watson, M. Current conditions in medical genetics practice. Genet. Med. 21, 1874–1877 (2019).
Author information
Authors and Affiliations
Contributions
Conceptualization: A.F., L.B.-S. Data curation: all authors. Formal analysis: A.F., N.R.-S., G.L., A.R.S., C.G.-J., L.B., L.B.-S. Investigation: A.F., N.R.-S., N.O., G.L., A.R.S., C.G.-J., L.B., L.B.-S. Methodology: A.F., L.B.-S. Supervision: L.B.-S. Writing—original draft: A.F., L.B.-S. Writing—review & editing: all authors.
Corresponding author
Ethics declarations
Ethics Declaration
This study was approved by the Institutional Review Board (IRB) of Rabin Medical Center. All patients and family members included in this study were consented for genetic and genomic studies.
Competing interests
A.F. has received speaker honoraria from Pfizer Pharmaceuticals. A.R.S. is an employee of Regeneron Pharmaceuticals Inc. and receives compensation for his employment. C.G.-J. is a full-time employee of the Regeneron Genetics Center from Regeneron Pharmaceuticals Inc. and receives salary and stock options as compensation. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Fellner, A., Ruhrman-Shahar, N., Orenstein, N. et al. The role of phenotype-based search approaches using public online databases in diagnostics of Mendelian disorders. Genet Med 23, 1095–1100 (2021). https://doi.org/10.1038/s41436-020-01085-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-01085-7
This article is cited by
-
Intellectual disability genomics: current state, pitfalls and future challenges
BMC Genomics (2021)