Abstract
Purpose
Kaposiform lymphangiomatosis (KLA) is a rare, frequently aggressive, systemic disorder of the lymphatic vasculature, occurring primarily in children. Even with multimodal treatments, KLA has a poor prognosis and high mortality rate secondary to coagulopathy, effusions, and systemic involvement. We hypothesized that, as has recently been found for other vascular anomalies, KLA may be caused by somatic mosaic variants affecting vascular development.
Methods
We performed exome sequencing of tumor samples from five individuals with KLA, along with samples from uninvolved control tissue in three of the five. We used digital polymerase chain reaction (dPCR) to validate the exome findings and to screen KLA samples from six other individuals.
Results
We identified a somatic activating NRAS variant (c.182 A>G, p.Q61R) in lesional tissue from 10/11 individuals, at levels ranging from 1% to 28%, that was absent from the tested control tissues.
Conclusion
The activating NRAS p.Q61R variant is a known “hotspot” variant, frequently identified in several types of human cancer, especially melanoma. KLA, therefore, joins a growing group of vascular malformations and tumors caused by somatic activating variants in the RAS/PI3K/mTOR signaling pathways. This discovery will expand treatment options for these high-risk patients as there is potential for use of targeted RAS pathway inhibitors.
Similar content being viewed by others
INTRODUCTION
Lymphatic anomalies encompass a spectrum of diseases with marked heterogeneity. In 2014, the International Society for the Study of Vascular Anomalies (ISSVA) reclassified lymphatic anomalies into simple and complex.1 Complex lymphatic anomalies, which include generalized lymphatic anomaly (GLA), Gorham–Stout disease (GSD), and channel type lymphatic malformation (CTLM), have more systemic involvement (soft tissue, bone, spleen, other viscera, as well as effusions) and have significant impact on morbidity, mortality, and overall quality of life. Kaposiform lymphangiomatosis (KLA) is a more recently characterized systemic and frequently aggressive lymphatic anomaly, typically presenting in childhood or early adulthood (Fig. 1) (refs. 2,3,4,5). KLA is clinically heterogeneous. Frequently there is thoracic involvement, and the abdomen can also be diffusely involved, as well as the spleen, gastrointestinal tract, bone, and other organs. Thoracic involvement in KLA is more extensive than in GLA, and includes mediastinal, pleural, and parenchymal pulmonary disease, often with recurrent pleural and/or pericardial effusions. Hemostatic anomalies, marked by coagulopathy and intralesional hemorrhage, are common and can be rapidly progressive and lethal. A defining feature of KLA is the presence of variably abundant foci of spindled lymphatic endothelial cells with incomplete canalization accompanying abnormal lymphatic channels of varying size. Although similar spindled endothelial cells are also found in kaposiform hemangioendothelioma (KHE), KHE is a localized vascular tumor with a distinct histopathology commonly seen in infants and toddlers and often associated with coagulopathy (Kasabach–Merritt phenomenon).6 A genetic cause for these complex lymphatic anomalies has not yet been identified, although a somatic activating NRAS variant (p.Q61R) was recently identified in a single case of GLA, and shown to have a pathogenic effect on the lymphatic endothelium.7 Despite multimodal therapy, the overall prognosis of KLA remains poor, with an overall survival of 34% (refs. 3,4). Identification of a causative variant would enable more efficacious targeted therapies for this aggressive disease.
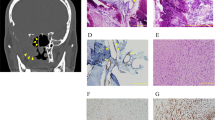
Clinical pathologic features of kaposiform lymphangiomatosis (KLA). (a–d) Participant KLA2. (a) Extensive cutaneous and subcutaneous involvement of trunk and thigh accompanied by hemorrhage and ascites. (b–d) Magnetic resonance imaging. (b) Coronal T2 fat-saturated image. Hyperintense infiltrative tissue in retroperitoneum and posterior mediastinum (arrow) with heterogeneous high signal of adjacent vertebral bodies. Ascites (asterisk) and multiple cystic lesions in the spleen (curved arrow). (c) Axial T2 fat-saturated image. High signal soft tissue in mediastinum (arrow), extending along bronchovascular bundles. Similar soft tissue abnormalities axillae. (d) Axial T1 contrast enhanced image. Intense enhancement of the abnormal soft tissue. (e–f) Participant KLA3. (e) Thoracoscopic image with extensive visceral pleural involvement. Parietal pleural adhesions also present (not shown). (f) Photomicrograph of pleural surface lesion with thin-walled anastomotic lesional channels with adjacent spindled endothelial cell component and erythrocyte extravasation. (g–j) Participant KLA6. (g) Swollen thigh and perineum with cutaneous telangiectasias and brown discoloration. (h–i) Photomicrographs of skin biopsy with (h) dilated dermal lymphatic channels (asterisk) and small cellular clusters (circle). Higher-magnification of circled area (i) shows complex abnormal lymphatic vessel adjacent to cluster of spindled hemosiderotic cells. (j) D2-40 immunopositivity in spindled cells similar to those in adjacent lymphatic channel. Focal separation of spindled cells suggestive of nascent lumens. (k–n) Participant KLA5. (k) Intraoperative image of anterior mediastinal fibrofatty mass with dilated blood-filled lymphatic channels. (l–n) Photomicrographs of lesional tissue demonstrating (l) several large, often blood-filled lymphatic vessels and anastomosing cellular cords containing interspersed red cells (circle). Higher-magnification of circled area (m) with clusters/ribbons of lymphatic endothelial cells, seemingly canalized and with luminal red cells. (n) Ribbons composed of D2-40 immunopositive lymphatic endothelial cells adjacent to abnormal, dilated lymphatic channels. Although no discernible histopathologic differences were found between participants, this type of rudimentary canalization was more prevalent in tissue sampled at autopsy.
Recently, several vascular anomalies have been shown to be caused by somatic activating variants in the MAPK and PI3K/AKT pathways (Table S1). We hypothesized that this might also be the case for KLA. Although we had previously identified hotspot variants in PIK3CA as a cause of localized lymphatic anomalies and combined vascular malformations with a lymphatic component,8,9 we did not find similar variants in KLA. Hence, we performed exome sequencing of tissue specimens—both lesional and uninvolved—from individuals with KLA. The University of Calgary Institutional Review Board and the Committee on Clinical Investigation of Boston Children’s Hospital approved this study. All participants provided informed consent to participate, including, for those whose photos are used, consent to publish photos.
MATERIALS AND METHODS
Exome sequencing and analysis
We performed exome sequencing of five KLA tumor samples as well as uninvolved tissue from three of those five individuals. Thin sections were scraped from diagnostic formalin-fixed paraffin-embedded (FFPE) tissue blocks, which had been obtained either at biopsy (2/5 cases), or at autopsy (3/5 cases). DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). For one tumor sample, we also recovered messenger RNA (mRNA) using the RNeasy Mini Kit (Qiagen). DNA libraries, and a complementary DNA (cDNA) library from the mRNA sample, were prepared using a TruSeq DNA Exome Kit and the TruSeq RNA Access Library Prep Kit (Illumina Corp, San Diego, CA, USA), respectively, and exome enrichment was performed with the SureSelectXT Human All Exon V5 capture kit (Agilent Technologies, Santa Clara, CA, USA). Each library was sequenced over a single lane of an Illumina HiSeq 2500, generating approximately 1 billion 100-bp paired-end read pairs per library.
We analyzed the exome sequences based on the Genome Analysis Toolkit (GATK) Best Practices for Somatic Short Variant Discovery.10 Briefly, sequence reads were aligned to the reference human genome (GRCh37) using the Burrows–Wheeler Aligner MEM algorithm (BWA-MEM),11 duplicates were marked using Picard, and base quality scores were recalibrated using GATK (see Supplementary Methods for all commands and arguments used). For each exome, 79–99% of target bases were covered at least 15-fold, and 42–92% of target bases were covered at least 50-fold, by unique reads with a minimum base quality score of 20 (Table S2).
We used Mutect2 in GATK4 to identify somatic variants, with a “panel of normals” created from 11 1000 Genomes participants,12 and using the gnomAD database13 as a “germline-resource.” For the three paired samples (KLA1, KLA2, and KLA3), we used the default Mutect2 parameters, with the exception of adjusting the “–af-of-alleles-not-in-resource” to 0.0000025, as recommended in the GATK4 Mutect2 Tutorial provided by the Broad Institute. We included in our analysis all somatic variant calls passing the standard Mutect2 filters. For the two unpaired tumor samples (KLA4 and KLA5), we ran Mutect2 in “tumor-only” mode, and ignored the “germline_risk” filter, including in our analysis any calls in which that was the only filter applied. The resulting somatic variant calls were then annotated using ANNOVAR.14 See Supplementary Methods for all commands used.
Digital PCR
FFPE tissue blocks or unstained glass slides of diagnostic specimens, collected previously during biopsy, were obtained from six additional individuals. Thin sections were scraped from the blocks or slides (for the five tumor samples included in the exome analysis, new thin sections were scraped from the same FFPE tissue blocks used for exome analysis), and DNA was extracted using a GeneRead FFPE DNA Extraction Kit (Qiagen), with the only modification to the kit protocol being that the Proteinase K digestion was extended overnight. Digital polymerase chain reaction (dPCR) was performed using a fully validated TaqMan NRAS p.Q61R assay (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA; assay ID: C_44193858_10). Genomic DNA was diluted so that the concentration of target DNA was 200–2000 copies/uL. PCR reaction mix was prepared per manufacturer’s protocol and 14.5 uL was loaded on each QuantStudio™ 3D Digital PCR chip v2 (Thermo Fisher Scientific). Digital chips were run on a ProFlex™ 2× Flat PCR thermocycler (Applied Biosystems by Thermo Fisher Scientific) with the following cycling conditions: 96 °C 10 minutes, then 39 cycles of 60 °C for 2 minutes, followed by 98 °C 30 seconds, then a final elongation step at 60 °C 2 minutes and 10 °C hold. Digital imaging was performed on the QuantStudio™ 3D Digital PCR Instrument and results were analyzed using the QuantStudio™ 3D Analysis Suite Software version 3.1.2 (Applied Biosystems by Thermo Fisher Scientific).
Additional analyses in KLA2
We did not identify the NRAS p.Q61R variant in one individual, KLA2, despite testing two independent lesional tissue samples, and despite the fact that no clinical or histopathological differences were apparent between this individual and the other ten participants. We compared b-allele frequencies between the lesional and control samples to look for regions of loss of heterozygosity, and used Lumpy15 to look for large structural variants or copy-number variations (Figure S1; commands in Supplementary Methods). We searched the exome data of KLA2 for other somatic hotspot variants,16 and for rare (i.e., frequency <0.001 in gnomAD database), nonsynonymous variants in other genes that have previously been implicated in vascular anomalies (Table S1). For this search, we loosened the stringency of variant calling, looking at all the raw calls output by Mutect2 (i.e., without filtering), and then investigating any calls of interest in the Integrative Genomics Viewer (IGV) to exclude mapping errors or sequencing artifacts. Additionally, for this search, we looked at germline variants in the lesional tissue sample, called using the HaplotypeCaller from GATK4, following their recommended best practices.10
RESULTS
Identification of consensus NRAS variant
We identified a single gene—NRAS— in which three or more of the KLA participants had a novel (i.e., not previously identified in the gnomAD database), nonsynonymous, somatic variant identified in their lesional tissue exome. A single somatic NRAS variant—c.182A>G, p.Q61R—was called in three KLA lesional exomes. This variant was present at levels ranging from 3% to 14%, with 6 to 26 sequence reads supporting the variant call (Table 1, Fig. 2). Visual inspection in IGV17 revealed that it was also present in a fourth lesional sample at 4% (though with only three sequence reads supporting the variant call, which is likely why it was not identified by the variant calling software) (Table 1). This variant was also present on 20/105 reads (19%) from the KLA5 RNA sequencing data. No variant reads were identified in any of the three uninvolved tissue samples.
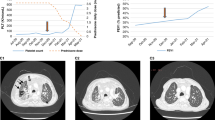
Exome sequencing and digital polymerase chain reaction (dPCR) of NRAS c.182A>G, p.Q61R allele in participant KLA4. (a) Integrative Genomics Viewer (IGV) screenshot of sequencing reads from lesional tissue exome of participant KLA4 supporting the variant allele. Variant is on 26/188 reads (14%). (b) dPCR results from lesional tissue of participant KLA4, demonstrating amplification of variant allele (14%). (c) dPCR results from an unrelated unaffected control, demonstrating no amplification of variant allele.
We performed digital PCR (dPCR) (and, in one case, a targeted high-throughput sequencing panel) to validate that this was a true variant (as opposed to a sequencing error), and to look for the variant in additional KLA samples. This sensitive variant detection method confirmed the presence of the NRAS p.Q61R variant in all four positive exome tumors, and confirmed the absence of the variant in the single negative exome tumor (KLA2). In addition, the variant was identified in all six new KLA tumor samples, at levels ranging from 1% to 8% (Table 1, Fig. 2).
NRAS p.Q61R screen in kaposiform hemangioendothelioma
We also used dPCR to screen for the NRAS p.Q61R variant in kaposiform hemangioendothelioma (KHE). We tested six KHE tumor samples for the NRAS p.Q61R variant, and all were negative (Table S3).
Additional analyses in KLA2
We did not identify the NRAS p.Q61R variant in one individual, KLA2, despite testing two independent lesional tissue samples. Using Lumpy,15 we also excluded large structural variants, copy-number variations (100 bp–10 kbp in size), or loss of heterozygosity in the exome data of this individual (Figure S1; commands in Supplementary Methods). No somatic or germline known hotspot variants were found, but from the list of vascular anomaly–associated genes (Table S1), we identified a missense variant in GNAQ (c.175 A>C, p.M59L), and two variants—a missense and a nonsense—in TEK (c.508G>C, p.V66L and c.340C>T, p.R114X).
The GNAQ variant identified in KLA2 (c.175A>C, p.M59L) has not previously been reported in the gnomAD or ClinVar databases, and appears only once in the COSMIC database (COSM6304069). However, it was recently reported in five cases of acral melanoma.18 Though the variant was present on 25 sequencing reads in the KLA2 lesional exome, there were several potential problems with the call (Figure S2). A BLAST search of the reads carrying the putative variant revealed that they could also map to an intergenic region of chromosome 2, with a single mismatch corresponding to a common polymorphism (rs3730153; minor allele frequency 22%). It is therefore likely that participant KLA2 carries the chromosome 2 germline polymorphism, and that the GNAQ variant represents a low-level artifact in both the lesional and uninvolved exomes due to mismapping of a small proportion of reads.
DISCUSSION
Our finding of a somatic NRAS p.Q61R variant in the lesional tissue in 10 of 11 individuals with KLA strongly implicates this variant in the pathogenesis of the disease, and suggests an important role for NRAS signaling in lymphatic vessel development and proliferation. NRAS is a proto-oncogene that encodes a small GTPase that normally functions to regulate cell proliferation via the MAPK and PI3K/AKT signaling pathways19,20,21—consistent with the pathways in which somatic variants in other vascular anomalies have so far been identified (Table S1). It is a member of the Ras gene family, which also includes HRAS and KRAS, and which are frequently altered in human cancers.19,20,21 In particular, NRAS variants are identified in more than 20% of melanomas.22 Three codons—12, 13, and 61—are variation hotspots (being variant in 74, 41, and 422 tumors, respectively, in a survey of 11,119 tumors16). Variants at each of these three sites result in a constitutively active NRAS protein and therefore, constitutive activation of downstream signaling pathways, resulting in unchecked cell proliferation.21
In addition to the prevalence of these variants in human cancers, somatic activating RAS variants have been identified in several benign cutaneous, skeletal, and vascular disorders (Table S4). Codon 61 NRAS variants, specifically, have been identified in epidermal nevi (MIM 162900) (ref. 23), multiple congenital melanocytic nevi (CMNS [MIM 137550]) and neurocutaneous melanosis (NCMS [MIM 249400]) (ref. 24), cutaneous skeletal hypophosphatemia syndrome (CSHS),25 and vascular anomalies including pyogenic granuloma.26,27 The presence of the NRAS p.Q61R variant in 10 of 11 tested KLA samples, therefore, adds KLA to the list of mosaic conditions linked to postzygotic activating NRAS variants, and to the even broader list of phenotypes associated with somatic variants in the RAS-MAPK and PI3K/AKT pathways. In general, the clinical and histopathologic similarities in all 11 patients seemed to justify a diagnosis of KLA. The single NRAS p.Q61R-negative individual therefore suggests the possibility of some locus or allelic heterogeneity within KLA.
Germline alterations in components of the RAS/MAPK pathway, including NRAS, cause a group of syndromes known as RASopathies, including neurofibromatosis type 1, Noonan syndrome, Costello syndrome, capillary malformation–arteriovenous malformation syndrome, and others.28 The NRAS p.Q61R variant has not been identified in any of these syndromes. Additionally, germline RASopathies are associated with craniofacial, neurodevelopmental, and other shared phenotypic features that are not observed in KLA or other somatic mosaic conditions linked to variants in the RAS-MAPK pathway, which are often collectively referred to as somatic mosaic RASopathies.
The NRAS p.Q61R variant has recently been reported in a single case of generalized lymphatic anomaly (GLA), formerly known as diffuse lymphangiomatosis.7 The distinction of KLA from GLA has only recently been made.3,4 While both are characterized by a generalized lymphatic anomaly of soft tissue that often includes viscera and bone, KLA is more commonly marked by significant coagulopathy, a more aggressive clinical course, and is pathologically distinct as defined by the presence of a spindled lymphatic endothelial component.3,4 It is of interest that the case of GLA with an NRAS p.Q61R variant reported by Manevitz-Mendelson et al. had histopathology that included dispersed spindle-like cells, which made the authors consider the possibility of KLA, although the lack of cutaneous involvement and consumptive coagulopathy prompted the authors to make a diagnosis of GLA.7 As we did not examine any cases of GLA for this study, it is possible that these two forms of generalized lymphatic disorders share a common genetic etiology. We would, therefore, recommend that genetic screening for this NRAS variant be performed in all individuals with aggressive and/or disseminated lymphatic anomalies.
Interestingly, in preliminary functional analyses by Manevitz-Mendelson et al., artificial overexpression of the NRAS p.Q61R variant allele in zebrafish actually impaired lymphatic development.7 This raises the possibility that a mosaic microenvironment with wild-type lymphatic endothelial cells may be necessary to cause a proliferative phenotype in vivo. Alternatively, the observed effects may be the result of artificial expression under a pan-vascular promoter, such as fli-1, and/or a dosage effect from NRAS overexpression.29 It is also unclear how this NRAS variant is associated with such divergent biologic behavior as seen in metastatic melanoma, KLA, and more banal anomalies. KLA is heterogeneous, both in disease progression and particularly in the response to treatment. While we did screen our exome data to look for potential permissive alleles and genetic modifiers, no obvious variants were detected in this small subset. Additional work, particularly in model organisms, will be needed to further explore these possibilities.
KLA shares some characteristics with another vascular anomaly, KHE. Though they differ clinically as KHE is a more localized, infiltrative tumor seen in infancy, they share the presence of spindled cells with abnormal lymphatic channels and a propensity for significant hemorrhagic complications.6 The cause of KHE is also not known, but recently, a GNA14 p.Q205L variant was identified in a single KHE and a related entity, tufted angioma (MIM 607859) (ref. 30). Our finding that the consensus KLA variant is absent from six KHE samples indicates that KLA and KHE are likely also genetically separate entities. Whether other NRAS variants occur in KHE, or whether the GNA14 variant is recurrent, remains to be determined.
In the single NRAS p.Q61R-negative individual (KLA2), we identified putative somatic variants in two genes associated with other vascular anomalies—GNAQ and TEK (Table S1). Though the GNAQ variant (c.175 A>C, p.M59L) was also reported in a study of acral melanoma,18 we showed that, in our data, it was likely a false positive variant call, representing the mismapping of a small number of reads spanning a polymorphism on chromosome 2 (see Results, Additional analyses in KLA2). We note that the previous report of this variant in acral melanoma18 identified the variant in 5 of 85 individuals by high-throughput sequencing of a panel of candidate genes, but that the authors did not validate the variant call by a second method. The single report of this variant in the COSMIC database was also not validated by a second method (see COSMIC study ID COSU660). It is, therefore, possible that these results were also confounded by the presence of this chromosome 2 polymorphism, suggesting that the GNAQ c.175A>C, p.M59L variant should be carefully scrutinized if it is identified in future sequencing studies. This highlights the importance of secondary validation of high-throughput sequencing results, especially for somatic variant calls that may be present on a low number of sequencing reads. In addition, given the high population frequency of the confounding chromosome 2 polymorphism, this false positive variant call would likely have been replicated in the panel of normals if a larger number of individuals had been included (we only included 11, while the recommended minimum is 40), emphasizing the benefit of using a large panel of normals in such analyses.
The two TEK variants identified in KLA2—c.508G>C, p.V66L and c.340C>T, p.R114X—are located in the extracellular domain and include a loss-of-function allele, whereas the TEK variants implicated in venous malformations are gain-of-function variants in the intracellular portion of the protein (amino acid positions 849, 897, 914) (refs. 31,32,33). However, TEK is likely to be haploinsufficient,13 and inherited heterozygous loss-of-function TEK variants cause primary congenital glaucoma (MIM 617272) by disrupting vascular development in the anterior chamber.34 It cannot be excluded that a very low-level somatic nonsense variant could have pathogenic effects on lymphatic endothelial organization, causing a spindled/kaposiform phenotype. The p.V66L and p.R114X variants were found at very low levels (2/94 and 8/739 sequencing reads, respectively) in the lesional exome, and were not seen in the control exome, but because they were not found in any other individuals, we did not pursue further validation. It, therefore, remains possible that these are merely false positive variant calls. Nevertheless, TEK represents a potential candidate for screening if additional NRAS p.Q61R-negative individuals with KLA are identified.
Treatment of KLA patients is challenging and the responses are variable. Multimodal treatment is directed toward minimizing disease progression, hemorrhagic complications, and lymphatic chylous effusions. Primary resection is not recommended because it can lead to rapid decompensation.3 Supportive therapy is essential in the treatment of KLA patients and this may require correcting the coagulopathy and draining of effusions (pleural, pericardial, and ascites) when clinically necessary. Despite these treatment efforts, the five-year survival rate was 51%, and the overall survival was just 34% in the initial series of 20 patients.3 Since the demonstration of the safety and efficacy of sirolimus therapy for complicated vascular anomalies,35 and our discovery of PIK3CA activating variants as a cause of simple and combined lymphatic malformations,8,9 many patients, including those with KLA, have regularly been treated with this mTOR inhibitor. The effect of mTOR inhibition in KLA is mixed, with many patients requiring concurrent antiproliferative therapies, including vincristine, methylprednisolone, thalidomide, and/or interferon.3,4 Studies are presently in preparation to update the survival rate following the introduction of sirolimus treatment.
Identification of the NRAS p.Q61R variant in KLA provides a means for a more targeted treatment approach. As RAS activation also activates PI3K/mTOR signaling, it is not surprising that patients would have some benefit from sirolimus therapy. However, current therapies do not effectively target RAS/MAPK signaling. Fortunately, many efforts are underway to develop novel treatments for NRAS-variant cancers such as melanoma. These have proven difficult to treat, presumably due to the high affinity of RAS for GTP.36,37,38 Nevertheless, progress is being made—for example, clinical trials of binimetinib, a MEK1 and MEK2 inhibitor, have shown small benefits in NRAS-variant melanoma cohorts,39 and may be more efficacious when combined with immunotherapy.40 Inhibitors of additional downstream components of NRAS signaling, including RAF and ERKs, are also being clinically tested.38
In conclusion, we have shown that somatic NRAS p.Q61R variants are recurrent in KLA, providing not only a more specific means for diagnosis, but also a new approach for more effective targeted treatments. We have shown that this variant is not found in KHE (at least not as commonly as in KLA), further distinguishing these two disorders. We have also potentially identified (though not validated) a novel missense (p.V66L) and a novel nonsense (p.R114X) variant in TEK, in the lone NRAS p.Q61R-negative patient in our cohort, which represents a candidate for consideration if additional p.Q61R-negative patients are found. Finally, we have highlighted a potentially recurrent false positive GNAQ variant (c.175 A>C, p.M59L) that should be carefully evaluated whenever it appears as a candidate in high-throughput sequencing studies.
References
Wassef M, Blei F, Adams D, et al. Vascular anomalies classification: recommendations from the International Society for the Study of Vascular Anomalies. Pediatrics. 2015;136:e203–e214.
Safi F, Gupta A, Adams D, Anandan V, McCormack FX, Assaly R. Kaposiform lymphangiomatosis, a newly characterized vascular anomaly presenting with hemoptysis in an adult woman. Ann Am Thorac Soc. 2014;11:92–95.
Croteau SE, Kozakewich HP, Perez-Atayde AR, et al. Kaposiform lymphangiomatosis: a distinct aggressive lymphatic anomaly. J Pediatr. 2014;164:383–388.
Fernandes VM, Fargo JH, Saini S, et al. Kaposiform lymphangiomatosis: unifying features of a heterogeneous disorder. Pediatr Blood Cancer. 2015;62:901–904.
Goyal P, Alomari AI, Kozakewich HP, et al. Imaging features of kaposiform lymphangiomatosis. Pediatr Radiol. 2016;46:1282–1290.
Zukerberg LR, Nickoloff BJ, Weiss SW. Kaposiform hemangioendothelioma of infancy and childhood. An aggressive neoplasm associated with Kasabach-Merritt syndrome and lymphangiomatosis. Am J Surg Pathol. 1993;17:321–328.
Manevitz-Mendelson E, Leichner GS, Barel O, et al. Somatic NRAS mutation in patient with generalized lymphatic anomaly. Angiogenesis. 2018;21:287–298.
Kurek KC, Luks VL, Ayturk UM, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115.
Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166:1048–1054.e510.
Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84.
Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163.
Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192.
Moon KR, Choi YD, Kim JM, et al. Genetic alterations in primary acral melanoma and acral melanocytic nevus in Korea: common mutated genes show distinct cytomorphological features. J Invest Dermatol. 2018;138:933–945.
Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22.
Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430.
Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689.
Jakob JA, Bassett RL Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–4023.
Hafner C, Toll A, Gantner S, et al. Keratinocytic epidermal nevi are associated with mosaic RAS mutations. J Med Genet. 2012;49:249–253.
Kinsler VA, Thomas AC, Ishida M, et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol. 2013;133:2229–2236.
Lim YH, Ovejero D, Sugarman JS, et al. Multilineage somatic activating mutations in HRAS and NRAS cause mosaic cutaneous and skeletal lesions, elevated FGF23 and hypophosphatemia. Hum Mol Genet. 2014;23:397–407.
Lim YH, Douglas SR, Ko CJ, et al. Somatic activating RAS mutations cause vascular tumors including pyogenic granuloma. J Invest Dermatol. 2015;135:1698–1700.
Groesser L, Peterhof E, Evert M, Landthaler M, Berneburg M, Hafner C. BRAF and RAS mutations in sporadic and secondary pyogenic granuloma. J Invest Dermatol. 2016;136:481–486.
Aoki Y, Niihori T, Inoue S, Matsubara Y. Recent advances in RASopathies. J Hum Genet. 2016;61:33–39.
Burd CE, Liu W, Huynh MV, et al. Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 2014;4:1418–1429.
Lim YH, Bacchiocchi A, Qiu J, et al. GNA14 somatic mutation causes congenital and sporadic vascular tumors by MAPK activation. Am J Hum Genet. 2016;99:443–450.
Limaye N, Wouters V, Uebelhoer M, et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat Genet. 2009;41:118–124.
Vikkula M, Boon LM, Carraway KL 3rd, et al. Vascular dysmorphogenesis caused by an activating mutation in the receptor tyrosine kinase TIE2. Cell. 1996;87:1181–1190.
Wouters V, Limaye N, Uebelhoer M, et al. Hereditary cutaneomucosal venous malformations are caused by TIE2 mutations with widely variable hyper-phosphorylating effects. Eur J Hum Genet. 2010;18:414–420.
Souma T, Tompson SW, Thomson BR, et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest. 2016;126:2575–2587.
Adams DM, Trenor CC 3rd, Hammill AM, et al. Efficacy and safety of sirolimus in the treatment of complicated vascular anomalies. Pediatrics. 2016;137:e20153257.
Mandala M, Merelli B, Massi D. Nras in melanoma: targeting the undruggable target. Crit Rev Oncol Hematol. 2014;92:107–122.
Johnson DB, Lovly CM, Flavin M, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res. 2015;3:288–295.
Boespflug A, Caramel J, Dalle S, Thomas L. Treatment of NRAS-mutated advanced or metastatic melanoma: rationale, current trials and evidence to date. Ther Adv Med Oncol. 2017;9:481–492.
Dummer R, Schadendorf D, Ascierto PA, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18:435–445.
Queirolo P, Spagnolo F. Binimetinib for the treatment of NRAS-mutant melanoma. Expert Rev Anticancer Ther. 2017;17:985–990.
Acknowledgements
We thank all who participated in this study, and their families. We thank the members of the Vascular Anomalies Center at Boston Children’s Hospital, for their ongoing support for patients and families affected by KLA, and the many benefactors who make this research possible. Research reported in this manuscript was supported by the National Institutes of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) AR-64231 (K.C.K.), the Wilkes Tumor Foundation (K.C.K.), and the Alberta Children’s Hospital Research Institute (K.C.K.). We acknowledge computing support from the Centre for Health Genomics and Informatics of the Alberta Children’s Hospital Research Institute. We are grateful for the GATK4 tutorials provided by the Broad Institute. Zaw Aung provided Python scripts for filtering variant lists. We thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Barclay, S.F., Inman, K.W., Luks, V.L. et al. A somatic activating NRAS variant associated with kaposiform lymphangiomatosis. Genet Med 21, 1517–1524 (2019). https://doi.org/10.1038/s41436-018-0390-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0390-0
Keywords
This article is cited by
-
Treatment of severe Kaposiform lymphangiomatosis positive for NRAS mutation by MEK inhibition
Pediatric Research (2023)
-
Genomic profiling informs diagnoses and treatment in vascular anomalies
Nature Medicine (2023)
-
NRASQ61R mutation in human endothelial cells causes vascular malformations
Angiogenesis (2022)
-
Sirolimus in the treatment of kaposiform lymphangiomatosis
Orphanet Journal of Rare Diseases (2021)
-
A review of mechanisms of disease across PIK3CA-related disorders with vascular manifestations
Orphanet Journal of Rare Diseases (2021)