Abstract
Purpose
We aimed to assess the biological and clinical significance of the human cysteine protease inhibitor cystatin M/E, encoded by the CTS6 gene, in diseases of human hair and skin.
Methods
Exome and Sanger sequencing was performed to reveal the genetic cause in two related patients with hypotrichosis. Immunohistochemical, biophysical, and biochemical measurements were performed on patient skin and 3D-reconstructed skin from patient-derived keratinocytes.
Results
We identified a homozygous variant c.361C>T (p.Gln121*), resulting in a premature stop codon in exon 2 of CST6 associated with hypotrichosis, eczema, blepharitis, photophobia and impaired sweating. Enzyme assays using recombinant mutant cystatin M/E protein, generated by site-directed mutagenesis, revealed that this p.Gln121* variant was unable to inhibit any of its three target proteases (legumain and cathepsins L and V). Three-dimensional protein structure prediction confirmed the disturbance of the protease/inhibitor binding sites of legumain and cathepsins L and V in the p.Gln121* variant.
Conclusion
The herein characterized autosomal recessive hypotrichosis syndrome indicates an important role of human cystatin M/E in epidermal homeostasis and hair follicle morphogenesis.
Similar content being viewed by others
INTRODUCTION
The importance of regulated proteolysis in epithelia and execution of protease-mediated processes1,2,3,4 is well illustrated by the identification of pathogenic variants in proteases and their inhibitors in a variety of human cornification/hair diseases (Supplementary Table S1 online).5,6,7,8,9,10,11 These include a number of cysteine proteases of the lysosomal cathepsin family and their cognate inhibitors of the cystatin family. In addition, studies in mice have shown that misregulation of proteolysis disturbs epidermal homeostasis, as witnessed by for example cathepsin L deficiency,12,13 targeted ablation of cathepsin D,14 and cystatin M/E deficiency.15
The protein encoded by the human CST6 gene (MIM 601891) was originally discovered independently by two groups and named cystatin M or cystatin E (here further referred to as cystatin M/E).16,17 Cystatin M/E is an atypical cystatin in that it was found to inhibit both lysosomal cysteine proteases such as cathepsin L (CTSL) and cathepsin V (CTSV), and the asparaginyl endopeptidase legumain (LGMN). We showed that cystatin M/E had an unusually tissue-specific expression pattern with high expression levels restricted to the skin.18 Our research on the function of this protein led to the discovery that the phenotype of the spontaneous ichq mouse mutant was caused by homozygosity for Cst6 null alleles. The resulting cystatin M/E deficiency in these mice causes disturbed epidermal cornification, impaired barrier function, and neonatal lethality because of excessive transepidermal water loss.15,19,20 Biochemical evidence indicated that human and mouse cystatin M/E controls the activity of several proteases and may indirectly regulate activity of epidermal transglutaminases, which are key enzymes in skin cornification.21 Using double knockout models in mice, we showed that a tightly regulated balance between cathepsin L and cystatin M/E is essential not only for tissue integrity of the epidermis, but also for maintenance of healthy hair follicles (HFs) and corneal epithelium.22 Our most recent data, in which we rescued the lethal skin phenotype of cystatin M/E–deficient mice by transgenic, epidermis-specific, re-expression of cystatin M/E under control of the human involucrin promoter, showed that cystatin M/E is not only a key molecule in a biochemical pathway that controls stratum corneum (SC) homeostasis but additionally plays an important role in maintaining HF integrity.23
In the current study, we report for the first time a loss-of-function (LOF) variant in the human CST6 gene, which causes a novel autosomal recessive hypotrichosis syndrome.
MATERIALS AND METHODS
Patients
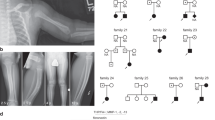
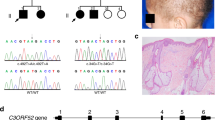
We ascertained a consanguineous Turkish family with hypotrichosis, including three affected and four unaffected individuals (Fig. 1a). All patients (PII:1, PII:2, and PIII:2) were offspring of parents who were first cousins (in generation I and generation II). Saliva samples for DNA extraction were collected from all individuals. Biophysical measurements were performed on the right upper ventral forearm skin of PII:2 (father) and two healthy age-matched male controls. Skin biopsies from these individuals were taken from the same body location. Photographs were taken from PII:2 (father) and PIII:2 (daughter, index patient). The parents did not give permission to publish frontal facial photographs nor was it allowed to take biopsies from the index patient (PIII:2). We obtained informed consent from all subjects using protocols approved by the local ethics committee (CMO area Arnhem-Nijmegen), which comply with the Declaration of Helsinki principles.
Pedigree, clinical features, and schematic representation of CST6 loss-of-function (LOF) variant. (a) Pedigree and genotypes of the family described. The homozygous LOF variant c.361C>T (p.Gln121*) was identified in individuals II:1, II:2, and III:2 (index patient), who were all born to consanguineous parents. Index patient is indicated by the arrowhead. Squares, males; circles, females; black filled symbols, affected; the half-filled symbols indicate heterozygous carriers of the CST6 variant. (b) The index patient (PIII:2) and (c) her father (PII:2) showed hypotrichosis due to progressive hair loss from three years of age. (d,e) The father developed eczematous lesions later in life. (f) The homozygous nonsense variant c.361C>T (p.Gln121*) results in a premature stop codon in exon 2 of the CST6 gene. The cystatin M/E (CST6) protein is 149 amino acids in size and the positions of the α-helix, the β-sheets, and the cathepsin and legumain inhibitory domains are indicated.
Infinium_CytoSNP_850K genotyping array analysis
DNA amplification, tagging, and hybridization were performed according to the manufacturer’s protocol (Illumina). Details of the procedure can be found in Supplementary Materials and Methods.
Exome sequencing
Exome sequencing was performed on genomic DNA extracted from the blood of the index patient (PIII:2). Details of the procedure can be found in Supplementary Materials and Methods.
Sequencing of the candidate gene
The pathogenic variant was confirmed in the patient by Sanger sequencing (BigDye terminator v1.1 sequencing kit, 3730 DNA analyzer, Applied Biosystems) using primers flanking exon 2 of CST6 (sense: 5’-AAGTGAGGATCAAGGGTCAAGAC-3’, antisense: 5’-CAGTTCAGGATGGACAACCT-3’). Sequence data was visualized with Mutation Surveyor (version 4.0.7, Softgenetics). Segregation analysis of the pathogenic variant in all available family members was performed.
Biophysical measurements
Details of the biophysical measurements (transepidermal water loss, subcutaneous [SC] hydration, skin redness and pigmentation) can be found in Supplementary Materials and Methods online.
Development of 3D human epidermal equivalents
Primary human keratinocytes were isolated from biopsies of family members PII:2 and II:3 and six healthy controls (wild-type CST6, CST6+/+) and expanded according to the Rheinwald–Green protocol.24 Passage 1–2 keratinocytes were used to generate 3D epidermal equivalents as described previously.25 The quality and phenotype of the epidermal equivalents was investigated by morphology, protein expression (immunohistochemistry), and gene expression (quantitative polymerase chain reaction [qPCR]). The reconstructed epidermis was used to generate cell extracts to study protease activity (enzymology) and cystatin M/E protein expression (western blotting).
RNA isolation and qPCR analysis
A brief description of the procedure can be found in Supplementary Materials and Methods. An overview of the qPCR primer pairs used is shown in Table S2.
Morphological and immunohistochemical analysis
A brief description of the procedure can be found in Supplementary Materials and Methods. An overview of the antibodies used is shown in Table S3.
Protein extraction from epidermal equivalents
Proteins were extracted from the 3D epidermal equivalents. A brief description of the procedure can be found in Supplementary Materials and Methods.
Western blotting
Proteins were separated by SDS-PAGE using the NuPAGE electrophoresis system and precast 12% Bis-Tris polyacrylamide gels under reducing conditions (Invitrogen). A brief description of the procedure can be found in Supplementary Materials and Methods.
Production of recombinant cystatin M/E variant p.Gln121*
Cystatin M/E variant p.Gln121* was obtained using the recombinant plasmid pGEX2T-cystatin M/E and Quick-change site-directed mutagenesis kit (Stratagene). Primers used for the generation of the p.Gln121* variant were 5’-GGCAGCAGGGGCGCAGTAGGAGAAGCTGCGC-3’ (sense) and 5’- GCGCAGCTTCTCCTACTGCGCCCCTGCTGCC -3’ (antisense). Sequence analysis was performed to check for the desired pathogenic variants in the complementary DNA (cDNA). Production and purification of recombinant wild-type and mutant forms of human cystatin M/E were performed as described previously.21 pGEX2T plasmids containing the wild-type or mutated cystatin M/E cDNAs were transformed into Escherichia coli BL21 Star (DE3) bacteria (Invitrogen). Cultures were grown from one single colony overnight at 37 °C in LB medium with ampicillin, followed by induction with isopropyl-1-thio-β-D-galactopyranoside (Roche) at a final concentration of 0.5 mM for 4 hours at 30 °C. Cells were lysed by four cycles of freezing in liquid nitrogen and thawing. Glutathione Sepharose 4B beads (Amersham Biosciences) were used for purification of glutathione S-transferase fusion protein from the lysate. Cleavage of the fusion protein into glutathione S-transferase and cystatin M/E occurred overnight at 4 °C using thrombin (Sigma) at a thrombin to fusion protein ratio of 1:500 in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2.5 mM CaCl2. Recombinant cystatin M/E was purified from glutathione S-transferase using glutathione Sepharose 4B beads and dialyzed against phosphate-buffered saline with a YM3 filter (Millipore). Protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce).
Fluorimetric enzyme assays
Protease inhibitor activity of recombinant human wild-type cystatin M/E and the p.Gln121* variant against recombinant human legumain,26 human CTSV, and human CTSL (both from R&D Systems) was determined by measuring inhibition of the proteases using fluorogenic synthetic substrates Z-Ala-Ala-Asn-MCA (Peptides International) for legumain, and Z-Leu-Arg-AMC (R&D Systems) for CTSV and CTSL. Protease inhibition was measured after an incubation period of 5 minutes (CTSL) or 10 minutes (legumain and CTSV) at room temperature. The buffer that was used in the legumain assay contained 0.1 M phosphate (pH 5.7), 2 mM EDTA, 1 mM dithiothreitol, 2.67 mM L-cysteine, and 5 µg/µl bovine serum albumin. CTSV and CTSL assays were performed in a buffer (pH 5.5) containing 0.1 M acetate, 1 mM ethylenediaminetetraacetate (EDTA), 2 mM dithiothreitol, and 5 µg/µL bovine serum albumin. Cathepsin and legumain protease activity was determined in protein extracts by measuring the hydrolysis of fluorescent substrates as described above.
Structural modeling
The structure of human cystatin M/E in complex with legumain has been solved experimentally and can be found in Protein Data Bank (PDB) file 4N6O.27 This PDB file was superposed on the structure of cystatin A in complex with cathepsin V, which can be found in PDB file 3KFQ (not yet published). Superposition of the assemblies and subsequent analysis was performed using the MOTIF-alignment option in the YASARA & WHAT IF Twinset.28,29
RESULTS
Clinical evaluation and genetic analysis
A 9-year-old girl (individual III:2 in Fig. 1a, index patient) was referred to the clinical geneticist by a dermatologist because of progressive hair loss resulting in hypotrichosis. Her father (PII:2) and her uncle (II:1) showed great similarities with regard to the clinical phenotype. The girl’s mother (II:3) and 6-year-old sister (III:1) were unaffected. The girl’s parents and her paternal grandparents had consanguineous marriages. The paternal grandparents (I:1 and I:2) did not have any symptoms of hypotrichosis. The clinical features of the index patient and her father are summarized in Table S4. From birth both suffered from a dry skin, blepharitis, and photophobia. After three years the clinical phenotype of hypotrichosis appeared (Fig. 1b, c). Overall body hair was sparse, very slowly growing, and both patients had only a few eyelashes (not shown as no permission was given to publish frontal facial photographs). Later in life the father developed eczematous lesions on hands (Fig. 1d, e) and feet (not shown). Both patients were not able to sweat, except after extreme physical exercise.
High-resolution single-nucleotide polymorphism (SNP) array did not show any pathogenic copy-number variants. After pretest genetic counseling and consent of the index patient’s parents, we performed exome sequencing. This analysis revealed a homozygous nonsense pathogenic variant c.361C>T (p.Gln121*) resulting in a premature stop codon in exon 2 of the CST6 gene (Fig. 1f). We confirmed this pathogenic variant with Sanger sequencing in the index patient and other family members. The results were consistent with autosomal recessive inheritance of c.361C>T as the causative pathogenic variant. The father of the index patient, who had very similar symptoms, was also found to be homozygous for the variant. All clinically unaffected family members were heterozygous.
Epidermal acanthosis and abnormal distribution of stratum corneum water content
To investigate the biology underlying the phenotype of cystatin M/E deficiency in this family, we first performed several noninvasive biophysical measurements on the midventral forearm skin of patient PII:2 (father) to detect possible abnormalities. Transepidermal water loss measurements in this patient were comparable with healthy controls, indicating no difference of inside-out skin barrier function (Table S5). Interestingly, spectrophotometric determination of skin redness and pigmentation showed that the a* value was increased in the patient, demonstrating the presence of erythema. Pigmentation (b* value) was normal. In vivo reflectance confocal microscopy (RCM) measurements showed a thickened epidermis (acanthosis) as well as a thickened SC in the cystatin M/E–deficient patient (Table S5). The water content of the patient’s SC was comparable with healthy controls (Fig. 2a). However, the water content pattern of the patient was more heterogeneous compared with the controls (Fig. 2b, c). This finding indicates a different distribution of water content in the SC, which can explain the dry skin.
Heterogeneous stratum corneum hydration in patient skin. (a) Permittivity measurements (bars represent mean +/-SD) of the patient and the healthy controls, demonstrating no overall quantitative difference between the patient and healthy controls. (b) Epsilon subcutaneous (SC) hydration image of the patient skin, showing a more heterogeneous water content pattern depicted by the larger black spaces (low SC hydration, representing the absence of water) between the green islets (high SC hydration, indicating the presence of water). (c) Epsilon SC hydration image of a healthy control, with small black areas between the homogeneous green islets, indicating a homogeneous distribution of the water content in the SC.
Abnormal epidermal proliferation and differentiation, and atypical dermal collagen fiber density
For in-depth phenotypical characterization we microscopically analyzed the skin biopsies. Hematoxylin and eosin (H&E) staining confirmed the RCM findings that the epidermal and SC thickness of the patient was increased as compared with six healthy controls (Fig. 3a, b). H&E staining also showed stronger eosinophilic collagen fibers in the papillary dermis of the patient’s dermis compared with healthy control dermis. This was even more prominent in an Elastica van Gieson stained section, which showed densely packed collagen fibers in the patient’s papillary dermis (Fig. 3c, d). Protein expression analysis revealed the presence of cystatin M/E in the upper granular layer of the epidermis (Fig. 3e, f) suggesting that the truncated protein was expressed. The increased number of proliferating cells, represented by Ki67 staining (Fig. 3g, h), was consistent with the increased epidermal thickness. We observed an increased protein expression of epidermal differentiation genes like filaggrin (FLG), loricrin (LOR), and involucrin (IVL) in the patient’s skin (Fig. 3i–n). The expression of transglutaminase 1 (TGM1), an enzyme responsible for crosslinking structural proteins of the cornified envelope in epidermal keratinocytes, was clearly induced in the upper epidermal layers of the patient (Fig. 3o, p). The increased expression levels of Ki67, TGM1, and the three differentiation genes were confirmed at the messenger RNA (mRNA) level (Supplementary Figure S1). To explain the observed increased redness (erythema) of the patient’s skin, we evaluated CD31 expression (endothelial cell marker) as a marker for angiogenesis but no differences were found when compared with healthy controls (data not shown).
Morphological and histological analysis of CST6 loss-of-function (LOF) skin. Midventral forearm skin biopsies obtained from patient PII:2 (1st and 3rd column) and two healthy controls (2nd and 4th column) were examined for morphology and protein expression. Hematoxylin and eosin staining showed acanthosis and dense collagen fibers in the papillary dermis (a, b), which was confirmed by Elastica van Gieson staining (c, d). Cystatin M/E expression was present in the stratum granulosum (e, f). Induced expression was found in the patient skin for the proliferation marker Ki67 (g, h), epidermal differentiation proteins filaggrin (i, j), loricrin (k, l) and involucrin (m, n), and the crosslinking enzyme transglutaminase 1 (o, p). Scale bar represents 400 µm.
Disrupted protease binding sites in cystatin M/E p.Gln121*
The presence of cystatin M/E protein in the epidermis of the patient (Fig. 3e, f) indicated that the c.361 C>T pathogenic variant did not preclude translation of a (truncated) protein. Based on the known protease binding sites of cystatin M/E (Fig. 1f), we hypothesized that truncation of the protein would lead to a compromised inhibitory activity of cystatin M/E against cathepsins L and V. To study the functional consequences of the c.361 C>T pathogenic variant, we generated recombinant truncated cystatin M/E (p.Gln121*) by site-directed mutagenesis. Wild-type and mutant cystatin M/E were tested for inhibitory activity against several cathepsins and legumain. Ki values were determined using an Easson–Stedman plot as previously described.21 When the Ki could not be determined in case of very low affinity, the ratio of protease activity in the presence (Vi) and absence (V0) of inhibitor was determined, and the remaining protease activity at a fixed high inhibitor (I) concentration (300 nM) was calculated. Ki values of wild-type human cystatin M/E and the p.Gln121* variant for interaction with human cathepsins and human legumain are shown in Table S6. Wild-type cystatin M/E is a high affinity inhibitor for cathepsin L, cathepsin V, and legumain as witnessed by their respective Ki values of 1.78 nM, 0.47 nM, and 0.25 nM,21 whereas cathepsin B is not inhibited by human cystatin M/E (vi/v0 > 0.71 at [I] = 300 nM). The Ki value of the p.Gln121* variant for interaction with cathepsins L and V could not be determined (vi/v0 > 0.86 and vi/v0 > 0.98 respectively at [I] = 300 nM), which indicates that the recombinant mutated variant was not able to inhibit any of these cathepsins. As the legumain inhibitory domain is positioned in the first exon (Fig. 1f) we anticipated that legumain inhibitory activity would be retained. This, however, was not the case as no inhibition was observed by the p.Gln121* variant (vi/v0 > 1.09 at [I] = 300 nM). As a control we used heat-inactivated wild-type recombinant cystatin M/E showing that this denaturated protein was not able to inhibit both cathepsins and legumain.
Based on the position of the pathogenic variant (Fig. 1f) at the proximal end of exon 2 and just before two of the four β-sheets, we hypothesized that the protein structure of the truncated protein had changed, resulting in an inactive molecule. Indeed, 3D-modeling using the published X-ray crystal structures of the cystatin M/E–legumain (PDB file 4N6O) and cystatin A–cathepsin V (PDB file 3KFQ) complexes indicates that the p.Gln121* pathogenic variant will severely affect the protein structure and function (Fig. 4). According to the created in silico model of the complex, legumain and cathepsin V both seem to bind to their own distinct sites on cystatin M/E. However, both sites require interactions with residues located in the last two β-strands (direct), or with residues that are correctly positioned by those residues in the last two β-strands (indirect). As a result, the deletion of these β-strands caused by pathogenic variant p.Gln121* will affect these binding sites and thereby affect the interaction with other proteins.
3D visualization of the cystatin M/E–legumain–cathepsin V complex. The visualization was made by superposing Protein Data Bank (PDB) files 4N6O and 3KFQ. Legumain and cathepsin V are shown in purple and magenta respectively. Cystatin M/E is colored by element (β-strands = red, ɑ-helix = blue, loops = cyan/green). The residues deleted after truncation of the protein are colored gray. (a) Overview of the complex in ribbon presentation. (b) Overview of the complex with protein surface shown. (c) Close-up of the protein–protein interaction sites with side chains of the residues shown.
Free legumain activity in cystatin M/E–deficient 3D skin equivalents
Because cystatin M/E is known to target three proteases—cathepsin L, cathepsin V, and legumain, all of which are expressed in human epidermis and HFs—we tried to assess the contribution of unrestricted activity of these proteases due to the p.Gln121* variant. Therefore, we generated 3D epidermal equivalents from the patient’s keratinocytes (PII:2, homozygous CST6 p.Gln121*), keratinocytes from the unaffected mother (II:3, heterozygous CST6 p.Gln121*), and keratinocytes from healthy individuals (N = 6, homozygous CST6 wild type). Similar to the in vivo biopsy material of PII:2, we observed cystatin M/E protein expression in the stratum granulosum of the homozygous CST6 p.Gln121* epidermal equivalents (Fig. 5a). Western blot analysis revealed the expression of a truncated protein of approximately 10 kDa in cell lysates of the p.Gln121* epidermal equivalent (Fig. 5b). The heterozygous p.Gln121* epidermal equivalent expressed both truncated and wild-type cystatin M/E as indicated by two bands, one at 12 kDa and one at 10 kDa (Fig. 5b). The 16-kDa bands indicate the presence of glycosylated cystatin M/E in the 3D epidermal equivalents. Clearly, the truncated cystatin M/E is not glycosylated as the presumed glycosylation site carrying an N-linked carbohydrate chain at position 137Asn is deleted due to the p.Gln121* variant.17 In contrast to in vivo skin we could not detect differences in the expression of specific genes/proteins involved in differentiation and proliferation (not shown). Protease activity measurements in the cell extracts of these epidermal equivalents indicated that cathepsin L and cathepsin V activity was low to undetectable in all epidermal equivalents (not shown). Strikingly, we found a high level of legumain activity in the patient-derived CST6 p.Gln121* epidermal equivalent compared with six control CTS6 wild-type epidermal equivalents (Fig. 5c). Although this finding did not allow unequivocal identification of the relevant protease responsible for the pathological alterations, it is consistent with the observation that legumain inhibitory activity is lost in the p.Gln121* variant.
Analysis of CST6 loss-of-function (LOF) 3D epidermal equivalents. (a) Hematoxylin and eosin (H&E) staining showed no specific morphological differences between epidermal constructs generated with primary keratinocytes that harbor a homozygous or heterozygous CST6 p.Gln121* variant compared with the wild-type control(s). Cystatin M/E protein is expressed in the stratum granulosum of all constructs. Scale bar represents 400 µm. (b) Western blot analysis of cell extracts derived from normal healthy skin biopsies (lane 1) and from epidermal equivalents from healthy controls (lanes 2 and 3), the patient (PII:2, father) with the homozygous CST6 p.Gln121* LOF variant (lane 4), and the healthy mother (II:3) who is heterozygous for the variant (lane 5). CST6trunc = truncated cystatin M/E, CST6glyc = glycosylated cystatin M/E, CST6wt = wild-type cystatin M/E. (c) Conversion of the fluorogenic z-A-A-N-AMC substrate. Cell extract of the patient-derived construct contains more free legumain activity compared with controls.
DISCUSSION
Our previous genetic studies in mice and in vitro biochemical studies using human enzymes and inhibitors have provided information on the role of cystatin M/E in epidermal cornification (Figure S2A). Based on the phenotype of the cystatin M/E–deficient ichq mice, and the biochemical and cell biological properties of human cystatin M/E, we anticipated that deficiency in humans would cause a severe ichthyosis-like phenotype rather than a hair phenotype. For this reason we have previously sequenced the CST6 gene in a large number (>100) of patients with autosomal recessive congenital ichthyosis who were negative for variants in known ichthyosis-associated genes. In none of these patients did we detect variants in the coding sequences of the CST6 gene.30 Although the patients described here do have a mild epidermal phenotype, their hypotrichosis is the most prominent and serious clinical feature, and led us to classify this syndrome as a heritable hair disease rather than an ichthyosis.
The clinical skin phenotype of the two patients was less severe than we expected based on our mouse studies and in vitro knockdown experiments.15,22,23,30 The severe and lethal skin phenotype in ichq mice, where the protein is completely absent, might be due to larger quantities of body hair, compared with humans, because neonatal keratinization of the HFs and subsequent dehydration is a hallmark of these mice. In the current study we show that the patient’s keratinocytes still produce a significant amount of truncated protein, which is unable to inhibit protease activity. However, this truncated protein may still have some hitherto unknown functions that are important for a proper development of the epidermis, as complete short hairpin RNA (shRNA)-mediated knockdown of cystatin M/E in a human skin equivalent showed a more severe phenotype.30
We herein show abnormal epidermal proliferation and differentiation, and atypical dermal collagen fiber density in skin biopsies of the patient, features that we did not or could not observe in the in vitro epidermal equivalents generated with the patient’s keratinocytes. These differences between the in vivo and in vitro situation are likely due the fact that the 3D skin model used here only contains keratinocytes in a sterile environment, thereby lacking interactions with skin microbes, fibroblasts, and/or other dermal and epidermal cells. Potential microbe–host or immune cell–mediated interactions may cause the phenotypical characteristics seen in our patient. Of note, there is currently no in vitro model available to grow hairs from epidermal keratinocytes, which precluded investigation of the hair phenotype in vitro.
Despite the fact that skin biopsies were taken from macroscopically healthy skin, histopathology and immunostaining revealed epidermal hyperproliferation and mildly abnormal differentiation compared with normal skin from healthy volunteers taken from the same anatomical location. Obviously it has to be taken into account that we could only study histology of one single patient (the father but not the daughter), and the observed abnormalities may be within the normal range. The dry skin observed in the two patients and the distribution of the SC water content were clearly abnormal compared with what is observed in healthy controls. It has to be noted that both patients had atopic eczema-like lesions, which may be mechanistically related to their dry skin. Speculatively, cystatin M/E deficiency may yet be another example of an epidermal defect that predisposes to atopic eczema, as has been reported for variants in filaggrin and genes that lead to a disturbed protease/inhibitor balance resulting in an aberrant skin barrier function.31,32,33 Observations in rare skin barrier diseases, for which there are now a few examples, will lend further support to initiatives to prevent atopic disease by strengthening skin barrier function in early life.
We demonstrated that due to the c.361 C>T LOF npathogenic variant in the CST6 gene, the resulting truncated cystatin M/E protein is unable to inhibit legumain and the cathepsins L and V. As all three proteases are expressed in the human HF and also colocalize with cystatin M/E in the distal part and infundibulum of the HF34 (see Figure S2B), it is likely that unrestricted activity of these proteases is the cause of hypotrichosis in patients. Our previous animal studies in double knockout mice and transgenic mice revealed that unrestricted cathepsin L activity is responsible for ichthyosis and lethality in the ichq mouse, but not for the persistent alopecia and fibrotic changes.22,23 However, it should be noted that the cystatin M/E controlled biochemical pathway in mice is distinct from humans. Mice do not have a cathepsin V orthologue, and we showed that mouse cystatin M/E differs from the human protein at amino acid position 64 (Asn instead of Asp), which renders it inactive against legumain. Ablation of legumain in mice with a cystatin M/E–deficient background (Cst6−/−Lgmn−/−) did not allow survival beyond the neonatal phase and mice displayed exactly the same phenotype as ichq mice. Human cystatin M/E, however, is a specific and high affinity inhibitor of human legumain. In the current study we found free legumain activity in an extract of the epidermal construct generated from patient keratinocytes, which was more than fourfold higher than in control keratinocytes (Fig. 5c). This suggests that cystatin M/E is a major inhibitor of epidermal legumain activity. We were unable to quantitatively assess the effect of cystatin M/E deficiency on cathepsin L and V activities because no activity could be detected in mutant or wild-type cell extracts. We surmise that activity of these enzymes is masked by the presence of other inhibitors in the extracts, such as hurpin and the cystatins A, B, and C.21,35 Due to the lack of material from the patient (e.g., biopsy material including HFs) and the limitations of our 3D model (no hair growth) we are, at this point, not able to dissect the mechanism in more detail or identify the key protease that causes the hypotrichosis.
Our previous animal work has also shown evidence that free cathepsin L activity is responsible for the eye pathology in cystatin M/E–deficient mice.22 These mice developed severe keratitis, thickening of the corneal stroma, and squamous metaplasia of the corneal epithelium. The patients presented in the current study also have clinically relevant eye problems, although milder than in the mice, such as dry eyes, blepharitis, and photophobia. If these problems are also caused by unrestrained cathepsin L activity this might provide therapeutic opportunities.
In conclusion, we have identified a new autosomal recessive hypotrichosis syndrome caused by a LOF pathogenic variant in the CST6 gene. These findings provide another example of the importance of protease–antiprotease balance in the regulation of epidermal and hair follicle homeostasis.
References
Zeeuwen PL. Epidermal differentiation: the role of proteases and their inhibitors. Eur J Cell Biol. 2004;83:761–773.
Zeeuwen PL, Cheng T, Schalkwijk J. The biology of cystatin M/E and its cognate target proteases. J Invest Dermatol. 2009;129:1327–1338.
Meyer-Hoffert U. Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Arch Immunol Ther Exp. 2009;57:345–354.
de Veer SJ, Furio L, Harris JM, Hovnanian A. Proteases: common culprits in human skin disorders. Trends Mol Med. 2014;20:166–178.
Chavanas S, Bodemer C, Rochat A, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:141–142.
Toomes C, James J, Wood AJ, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23:421–424.
Basel-Vanagaite L, Attia R, Ishida-Yamamoto A, et al. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. Am J Hum Genet. 2007;80:467–477.
Oeffner F, Fischer G, Happle R, et al. IFAP syndrome is caused by deficiency in MBTPS2, an intramembrane zinc metalloprotease essential for cholesterol homeostasis and ER stress response. Am J Hum Genet. 2009;84:459–467.
Blaydon DC, Nitoiu D, Eckl KM, et al. Mutations in CSTA, encoding cystatin A, underlie exfoliative ichthyosis and reveal a role for this protease inhibitor in cell-cell adhesion. Am J Hum Genet. 2011;89:564–571.
Pigors M, Sarig O, Heinz L, et al. Loss-of-function mutations in SERPINB8 linked to exfoliative ichthyosis with impaired mechanical stability of intercellular adhesions. Am J Hum Genet. 2016;99:430–436.
Ngcungcu T, Oti M, Sitek JC, et al. Duplicated enhancer region increases expression of CTSB and segregates with keratolytic winter erythema in South African and Norwegian families. Am J Hum Genet. 2017;100:737–750.
Roth W, Deussing J, Botchkarev VA, et al. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000;14:2075–2086.
Tobin DJ, Foitzik K, Reinheckel T, et al. The lysosomal protease cathepsin L is an important regulator of keratinocyte and melanocyte differentiation during hair follicle morphogenesis and cycling. Am J Pathol. 2002;160:1807–1821.
Egberts F, Heinrich M, Jensen JM, et al. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J Cell Sci. 2004;117 Pt 11:2295–2307.
Zeeuwen PL, Vlijmen-Willems IM, Hendriks W, Merkx GF, Schalkwijk J. A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum Mol Genet. 2002;11:2867–2875.
Sotiropoulou G, Anisowicz A, Sager R. Identification, cloning, and characterization of cystatin M, a novel cysteine proteinase inhibitor, down-regulated in breast cancer. J Biol Chem. 1997;272:903–910.
Ni J, Abrahamson M, Zhang M, et al. Cystatin E is a novel human cysteine proteinase inhibitor with structural resemblance to family 2 cystatins. J Biol Chem. 1997;272:10853–10858.
Zeeuwen PL, Vlijmen-Willems IM, Jansen BJ, et al. Cystatin M/E expression is restricted to differentiated epidermal keratinocytes and sweat glands: a new skin-specific proteinase inhibitor that is a target for cross-linking by transglutaminase. J Invest Dermatol. 2001;116:693–701.
Zeeuwen PL, Vlijmen-Willems IM, Olthuis D, et al. Evidence that unrestricted legumain activity is involved in disturbed epidermal cornification in cystatin M/E deficient mice. Hum Mol Genet. 2004;13:1069–1079.
Sundberg JP, Boggess D, Hogan ME, et al. Harlequin ichthyosis (ichq): a juvenile lethal mouse mutation with ichthyosiform dermatitis. Am J Pathol. 1997;151:293–310.
Cheng T, Hitomi K, van Vlijmen-Willems IM, et al. Cystatin M/E is a high affinity inhibitor of cathepsin V and cathepsin L by a reactive site that is distinct from the legumain-binding site. A novel clue for the role of cystatin M/E in epidermal cornification. J Biol Chem. 2006;281:15893–15899.
Zeeuwen PL, van Vlijmen-Willems IM, Cheng T, et al. The cystatin M/E-cathepsin L balance is essential for tissue homeostasis in epidermis, hair follicles, and cornea. FASEB J. 2010;24:3744–3755.
Oortveld MAW, van Vlijmen-Willems I, Kersten FFJ, et al. Cathepsin B as a potential cystatin M/E target in the mouse hair follicle. FASEB J. 2017;31:4286–4294.
Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343.
Niehues H, Schalkwijk J, van Vlijmen-Willems I, et al. Epidermal equivalents of filaggrin null keratinocytes do not show impaired skin barrier function. J Allergy Clin Immunol. 2017;139:1979–1981 e1913.
Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J Biol Chem. 2003;278:38980–38990.
Dall E, Fegg JC, Briza P, Brandstetter H. Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew Chem. 2015;54:2917–2921.
Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402.
Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. 29
Jansen PA, van den Bogaard EH, Kersten FF, et al. Cystatin M/E knockdown by lentiviral delivery of shRNA impairs epidermal morphogenesis of human skin equivalents. Exp Dermatol. 2012;21:889–891.
Palmer CN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446.
Cork MJ, Danby SG, Vasilopoulos Y, et al. Epidermal barrier dysfunction in atopic dermatitis. J Invest Dermatol. 2009;129:1892–1908.
Wang XW, Wang JJ, Gutowska-Owsiak D, et al. Deficiency of filaggrin regulates endogenous cysteine protease activity, leading to impaired skin barrier function. Clin Exp Dermatol. 2017;42:622–631.
Cheng T, van Vlijmen-Willems IM, Hitomi K, et al. Colocalization of cystatin M/E and its target proteases suggests a role in terminal differentiation of human hair follicle and nail. J Invest Dermatol. 2009;129:1232–1242.
Welss T, Sun J, Irving JA, et al. Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry. 2003;42:7381–7389.
Acknowledgements
We are grateful to the family who participated in this study. This study was funded by the Dutch Organization for Scientific Research (NWO-ALW, project number 821.02.013). E.v.d.B. was funded by a Veni grant from the Dutch Organization for Scientific Research (project number 91616054). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
van den Bogaard, E.H.J., van Geel, M., van Vlijmen-Willems, I.M.J.J. et al. Deficiency of the human cysteine protease inhibitor cystatin M/E causes hypotrichosis and dry skin. Genet Med 21, 1559–1567 (2019). https://doi.org/10.1038/s41436-018-0355-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0355-3