Abstract
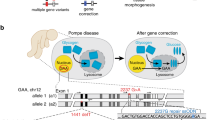
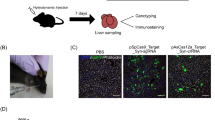
Gene editing for the cure of inborn errors of metabolism (IEMs) has been limited by inefficiency of adult hepatocyte targeting. Here, we demonstrate that in utero CRISPR/Cas9-mediated gene editing in a mouse model of hereditary tyrosinemia type 1 provides stable cure of the disease. Following this, we performed an extensive gene expression analysis to explore the inherent characteristics of fetal/neonatal hepatocytes that make them more susceptible to efficient gene editing than adult hepatocytes. We showed that fetal and neonatal livers are comprised of proliferative hepatocytes with abundant expression of genes involved in homology-directed repair (HDR) of DNA double-strand breaks (DSBs), key for efficient gene editing by CRISPR/Cas9. We demonstrated the same is true of hepatocytes after undergoing a regenerative stimulus (partial hepatectomy), where post-hepatectomy cells show a higher efficiency of HDR and correction. Specifically, we demonstrated that HDR-related genome correction is most effective in the replicative phase, or S-phase, of an actively proliferating cell. In conclusion, this study shows that taking advantage of or triggering cell proliferation, specifically DNA replication in S-phase, may serve as an important tool to improve efficiency of CRISPR/Cas9-mediated genome editing in the liver and provide a curative therapy for IEMs in both children and adults.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sanger sequencing data is being uploaded to a public online repository and accession numbers will be provided. The remainder of data are available in the main text and supplementary materials.
References
VanLith C, Guthman R, Nicolas CT, Allen K, Du Z, Joo DJ, et al. Curative ex vivo hepatocyte-directed gene editing in a mouse model of hereditary Tyrosinemia Type 1. Hum Gene Ther. 2018;29:1315–26.
Mingozzi F, High KA. Immune responses to AAV in clinical trials. Curr Gene Ther. 2011;11:321–30.
Mingozzi F, High KA. Immune responses to AAV vectors: Overcoming barriers to successful gene therapy. Blood. 2013;122:23–36.
Tran ND, Porada CD, Almeida-Porada G, Glimp HA, Anderson WF, Zanjani ED. Induction of stable prenatal tolerance to beta-galactosidase by in utero gene transfer into preimmune sheep fetuses. Blood. 2001;97:3417–23.
Porada CD, Park PJ, Almeida-Porada G, Liu W, Ozturk F, Glimp HA, et al. Gestational age of recipient determines pattern and level of transgene expression following in utero retroviral gene transfer. Mol Ther. 2005;11:284–93.
Flake AW. Genetic therapies for the fetus. Clin Obstet Gynecol. 2002;45:684–96.
Alapati D, Zacharias WJ, Hartman HA, Rossidis AC, Stratigis JD, Ahn NJ, et al. In utero gene editing for monogenic lung disease. Sci Transl Med. 2019;11:eaav8375.
Rossidis AC, Stratigis JD, Chadwick AC, Hartman HA, Ahn NJ, Li H, et al. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat Med. 2018;24:1513–8.
Lindblad B, Lindstedt S, Steen G. On the enzymic defects in hereditary tyrosinemia. Proc Natl Acad Sci USA. 1977;74:4641–5.
Endo F, Sun MS. Tyrosinaemia type I and apoptosis of hepatocytes and renal tubular cells. J Inherit Metab Dis. 2002;25:227–34.
Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis. 2001;21:563–71.
Gilgenkrantz H. Rodent models of liver repopulation. Methods Mol Biol. 2010;640:475–90.
Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–6.
VanLith CJ, Guthman RM, Nicolas CT, Allen KL, Liu Y, Chilton JA, et al. Ex vivo hepatocyte reprograming promotes homology-directed DNA repair to correct metabolic disease in mice after transplantation. Hepatol Commun. 2019;3:558–73.
Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–33.
Kreutz C, MacNelly S, Follo M, Waldin A, Binninger-Lacour P, Timmer J, et al. Hepatocyte ploidy is a diversity factor for liver homeostasis. Front Physiol. 2017;8:862.
Chen Y, Hata T, Rehman F, Kang L, Yang L, Kim BYS, et al. Visualization of hepatocellular regeneration in mice after partial hepatectomy. J Surg Res. 2019;235:494–500.
Tao Y, Wang M, Chen E, Tang H. Liver regeneration: Analysis of the main relevant signaling molecules. Mediators Inflamm. 2017;2017:4256352.
Iyama T, Wilson DM 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst). 2013;12:620–36.
Michalopoulos GK. Liver regeneration. J Cellular Physiol. 2007;213:286–300.
Cerone R, Holme E, Schiaffino MC, Caruso U, Maritano L, Romano C. Tyrosinemia type III: Diagnosis and ten-year follow-up. Acta Paediatr. 1997;86:1013–5.
Ruetschi U, Cerone R, Perez-Cerda C, Schiaffino MC, Standing S, Ugarte M, et al. Mutations in the 4-hydroxyphenylpyruvate dioxygenase gene (HPD) in patients with tyrosinemia type III. Hum Genet. 2000;106:654–62.
Chinsky JM, Singh R, Ficicioglu C, van Karnebeek CDM, Grompe M, Mitchell G, et al. Diagnosis and treatment of tyrosinemia type I: A US and Canadian consensus group review and recommendations. Genet Med. 2017;19:1380–95.
Vermeulen C, Geeven G, de Wit E, Verstegen M, Jansen RPM, van Kranenburg M, et al. Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am J Hum Genet. 2017;101:326–39.
Rafati M, Mohamadhashem F, Hoseini A, Ramandi SD, Ghaffari SR. Prenatal diagnosis of tyrosinemia type 1 using next generation sequencing. Fetal Pediatr Pathol. 2016;35:282–5.
David AL, Peebles D. Gene therapy for the fetus: Is there a future? Best Pract Res Clin Obstet Gynaecol. 2008;22:203–18.
Kim CH. Homeostatic and pathogenic extramedullary hematopoiesis. J Blood Med. 2010;1:13–9.
Dutta S, Sengupta P. Men and mice: Relating their ages. Life Sci. 2016;152:244–8.
Krishnan A, Samtani R, Dhanantwari P, Lee E, Yamada S, Shiota K, et al. A detailed comparison of mouse and human cardiac development. Pediatr Res. 2014;76:500–7.
Pressler R, Auvin S. Comparison of brain maturation among species: An example in translational research suggesting the possible use of bumetanide in newborn. Front Neurol. 2013;4:36.
Li N, Gou S, Wang J, Zhang Q, Huang X, Xie J, et al. CRISPR/Cas9-mediated gene correction in newborn rabbits with hereditary tyrosinemia type I. Mol Ther. 2020;29:1001–15.
Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766.
Jackman J, O’Connor PM. Methods for synchronizing cells at specific stages of the cell cycle. Curr Protoc Cell Biol. 2001;Chapter 8:Unit 8.3.
Thyagarajan B, Cruise JL, Campbell C. Elevated levels of homologous DNA recombination activity in the regenerating rat liver. Somat Cell Mol Genet. 1996;22:31–9.
Xiong W, MacColl Garfinkel AE, Li Y, Benowitz LI, Cepko CL. NRF2 promotes neuronal survival in neurodegeneration and acute nerve damage. J Clin Invest. 2015;125:1433–45.
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91.
Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG, et al. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–307.
Nijagal A, Le T, Wegorzewska M, Mackenzie TC. A mouse model of in utero transplantation. J Vis Exp. 2011;47:2303.
Waddington SN, Mitrophanous KA, Ellard FM, Buckley SM, Nivsarkar M, Lawrence L, et al. Long-term transgene expression by administration of a lentivirus-based vector to the fetal circulation of immuno-competent mice. Gene Ther. 2003;10:1234–40.
Wang X, Montini E, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. Kinetics of liver repopulation after bone marrow transplantation. Am J Pathol. 2002;161:565–74.
Bae S, Park J, Kim JS. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5.
Anderson EM, Haupt A, Schiel JA, Chou E, Machado HB, Strezoska Z, et al. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. J Biotechnol. 2015;211:56–65.
Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–70.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Acknowledgements
We thank Kari Allen for help with animal care. We thank Raymond Hickey for help with initial experiments. We thank LouAnn Gross and Tony Blahnik for histology and immunohistochemistry support. Children’s Hospital of Minnesota Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization: JBL, GM, and CTN. Methodology: JBL, GM, and CTN. Validation: RAK. Formal Analysis: CJV, GM, and DRO. Investigation: GM, CVL, and CTN. Resources: LH, WC, and BH. Writing-Original Draft: GM, CJV, CTN, and WST. Writing-Review & Editing: JBL and CTN. Visualization: CTN and JBL. Supervision: JBL and RAK. Project administration: RAK. Funding Acquisition: JBL.
Corresponding author
Ethics declarations
Competing interests
JBL is Chief Scientific Officer and RAK is Vice-President of Pre-Clinical Development of Castle Creek Biosciences, Inc. No funding or financial support for this work has been provided by Castle Creek Biosciences. The remaining authors have no competing interests.
Ethical approval
All animal experiments conducted in this study were approved by Mayo Clinic’s Institutional Animal Care and Use Committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mondal, G., VanLith, C.J., Nicolas, C.T. et al. Activation of homology-directed DNA repair plays key role in CRISPR-mediated genome correction. Gene Ther 30, 386–397 (2023). https://doi.org/10.1038/s41434-022-00369-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00369-8