Abstract
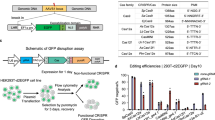
CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated endonuclease Cas9) nucleases have been widely applied for genome engineering. Staphylococcus aureus Cas9 (SaCas9) is compact, which can be packaged in AAV (adeno-associated virus) vector for in vivo gene editing. While, wild-type SaCas9 can induce unwanted off-target mutations and substantially limits the applications. So far, there are two reported SaCas9 variants with high-fidelity, including efSaCas9 from our previous study and SaCas9-HF. However, it remains unknown which one possessing the better fidelity and higher activity. Here, we performed a parallel comparison of efSaCas9 and SaCas9-HF in human cells through fluorescent reporter system and target deep sequencing, respectively. The results demonstrated that efSaCas9 possesses higher cleavage activity and fidelity than SaCas9-HF at the most endogenous sites in human cells. Collectively, our study provides insights for the rational selection of suitable SaCas9 for human genome editing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The deep sequencing data are available at the NCBI Sequence Read Archive (SRA) under PRJNA749863 (SRA accession number, SRR15253970 to SRR15253977, sample accession number, SAMN20423045).
References
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6.
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32.
Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6.
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97.
Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–83.
Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–9.
Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, et al. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–26.
Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–24.
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;155:479–80.
Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–84.
Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to Fokl nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–82.
Slaymaker IM, Gao LY, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88.
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–5.
Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–10.
Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9:3048.
Hu JH, Miller SM, Geurts MH, Tang WX, Chen LW, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63.
Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36:265–71.
Nishimasu H, Shi X, Ishiguro S, Gao LY, Hirano S, Okazaki S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–62.
Zinn E, Vandenberghe LH. Adeno-associated virus: fit to serve. Curr Opin Virol. 2014;8:90–97.
Truong DJJ, Kuhner K, Kuhn R, Werfel S, Engelhardt S, Wurst W, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–8.
Chew WL, Tabebordbar M, Cheng JKW, Mali P, Wu EY, Ng AHM, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13:868–74.
Moreno AM, Fu X, Zhu J, Katrekar D, Shih YRV, Marlett J, et al. In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol Ther. 2018;26:1818–27.
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–98.
Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25:229–33.
Gu X, Wang DQ, Xu ZJ, Wang JH, Guo L, Chai RJ, et al. Prevention of acquired sensorineural hearing loss in mice by in vivo Htra2 gene editing. Genome Biol. 2021;22:86.
Li Q, Su J, Liu Y, Jin X, Zhong XM, Mo L, et al. In vivo PCSK9 gene editing using an all-in-one self-cleavage AAV-CRISPR system. Mol Ther-Meth Clin D. 2021;20:652–9.
Xie HH, Ge XL, Yang FY, Wang B, Li S, Duan JZ, et al. High-fidelity SaCas9 identified by directional screening in human cells. Plos Biol. 2020;18:e3000747.
Tan YY, Chu AHY, Bao SY, Hoang DA, Kebede FT, Xiong WJ, et al. Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. P Natl Acad Sci USA. 2019;116:20969–76.
Zhang YL, Ge XL, Yang FY, Zhang LP, Zheng JY, Tan XF, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep-Uk. 2014;4:5405.
Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 2016;34:695–7.
Gao N, Zhang CD, Hu ZY, Li MM, Wei JJ, Wang YM, et al. Characterization of Brevibacillus laterosporus Cas9 (BlatCas9) for Mammalian Genome Editing. Front Cell Dev Biol. 2020;8:583164.
Karvelis T, Gasiunas G, Young JS, Bigelyte G, Silanskas A, Cigan M, et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253.
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–21.
Edraki A, Mir A, Ibraheim R, Gainetdinov I, Yoon Y, Song CQ, et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol Cell. 2019;73:714–26.
Hu ZY, Wang S, Zhang CD, Gao N, Li MM, Wang DQ, et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. Plos Biol. 2020;18:e3000686.
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500.
Xie HH, Tang LC, He XB, Liu XX, Zhou CC, Liu JJ, et al. SaCas9 Requires 5-NNGRRT-3 PAM for Sufficient Cleavage and Possesses Higher Cleavage Activity than SpCas9 or FnCpf1 in Human Cells. Biotechnol J. 2018;13:e1700561.
Tu MJ, Lin L, Cheng YL, He XB, Sun HH, Xie HH, et al. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017;45:11295–304.
Lin L, He XB, Zhao TY, Gu LK, Liu YQ, Liu XY, et al. Engineering the Direct Repeat Sequence of crRNA for Optimization of FnCpf1-Mediated Genome Editing in Human Cells. Mol Ther. 2018;26:2650–7.
Kim YH, Ramakrishna S, Kim H, Kim JS. Enrichment of cells with TALEN-induced mutations using surrogate reporters. Methods. 2014;69:108–17.
He XB, Wang YF, Yang FY, Wang B, Xie HH, Gu LK, et al. Boosting activity of high-fidelity CRISPR/Cas9 variants using a tRNA(Gln)-processing system in human cells. J Biol Chem. 2019;294:9308–15.
Hua K, Tao XP, Zhu JK. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J. 2019;17:499–504.
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–57.
Zhao DD, Li J, Li SW, Xin XQ, Hu MZ, Price MA, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol. 2021;39:35–40.
Kurt IC, Zhou RH, Iyer S, Garcia SP, Miller BR, Langner LM, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021;39:41–46.
Funding
This work was supported by grants from Basic research project of Henan Eye Hospital (20JCZD001), 23456 talent project of Henan Provincial People’s Hospital, National Natural Science Foundation of China (81201181), Natural Science Foundation of Zhejiang Province (2017C37176), Project of State Key Laboratory of Ophthalmology, Optometry and Visual Science, Wenzhou Medical University (J02-20190201) and Wenzhou City Grant, Zhejiang, China (2021Y1893).
Author information
Authors and Affiliations
Contributions
FG and ZS designed research, JL, HX, XL, YZ, JW, HC, and TY performed research and JL, J.J, JZ, ZS, and FG performed data analyses, and JL, FG, and ZS wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lv, J., Xi, H., Lv, X. et al. Two high-fidelity variants: efSaCas9 and SaCas9-HF, which one is better?. Gene Ther 29, 458–463 (2022). https://doi.org/10.1038/s41434-022-00319-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-022-00319-4