Abstract
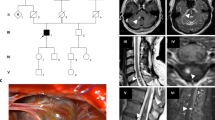
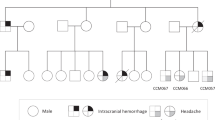
Loss-of-function variants in CCM1/KRIT1, CCM2/MGC4607, and CCM3/PDCD10 genes are identified in the vast majority of familial cases with multiple cerebral cavernous malformations. However, genomic DNA sequencing combined with large rearrangement screening fails to detect a pathogenic variant in 5% of the patients. We report a family with two affected members harboring multiple CCM lesions, one with severe hemorrhages and one asymptomatic. No causative variant was detected using DNA sequencing of the three CCM genes, CNV detection analysis, and RNA sequencing. However, a loss of heterozygosity in CCM2 was observed on cDNA sequences in one of the two affected members, which strongly suggested that this locus might be involved. Whole genome sequencing (WGS) identified a balanced structural variant on chromosome 7 with a breakpoint interrupting the CCM2 gene, preventing normal mRNA synthesis. These data underline the importance of WGS in undiagnosed patients with typical multiple CCM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The datasets generated during the current study are available from the corresponding author.
References
Labauge P, Denier C, Bergametti F, Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6:237–44.
Riant F, Bergametti F, Ayrignac X, Boulday G, Tournier-Lasserve E. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277:1070–5.
Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, et al. Genotype–phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60:550–6.
Spiegler S, Najm J, Liu J, Gkalympoudis S, Schröder W, Borck G, et al. High mutation detection rates in cerebral cavernous malformation upon stringent inclusion criteria: one-third of probands are minors. Mol Genet Genom Med. 2014;2:176–85.
Riant F, Cecillon M, Saugier-Veber P, Tournier-Lasserve E. CCM molecular screening in a diagnosis context: novel unclassified variants leading to abnormal splicing and importance of large deletions. Neurogenetics. 2013;14:133–41.
Spiegler S, Rath M, Hoffjan S, Dammann P, Sure U, Pagenstecher A, et al. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics. 2018;19:55–9.
Pilz RA, Schwefel K, Weise A, Liehr T, Demmer P, Spuler A, et al. First interchromosomal insertion in a patient with cerebral and spinal cavernous malformations. Sci Rep. 2020;10:6306.
Zhang ZD, Du J, Lam H, Abyzov A, Urban AE, Snyder M, et al. Identification of genomic indels and structural variations using split reads. BMC Genomics. 2011;12:375.
Acknowledgements
We thank the family for participating in this study. This research was made possible through access to the data generated by the France Genomic Medicine Plan 2025.
Funding
No financial assistance was received in support of the study.
Author information
Authors and Affiliations
Contributions
FR designed the work that led to the submission, acquired data, and played an important role in interpreting the results. AC, PL, and XA acquired data and revised the manuscript. TG acquired the data. NC played an important role in interpreting the results and revised the manuscript. ETL revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Study participants or legal representatives gave written informed consent for clinical testing, research use and publication. The analyses were performed in accordance with French regulations and the principles of the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaussenot, A., Ayrignac, X., Chatron, N. et al. Loss of heterozygosity in CCM2 cDNA revealing a structural variant causing multiple cerebral cavernous malformations. Eur J Hum Genet (2024). https://doi.org/10.1038/s41431-024-01626-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-024-01626-7