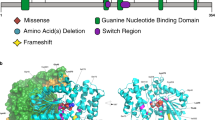
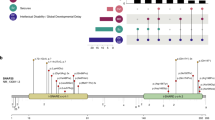
Abstract
Heterozygous PRRT2 variants are frequently implicated in Self-limited Infantile Epilepsy, whereas homozygous variants are so far linked to severe presentations including developmental and epileptic encephalopathy, movement disorders, and intellectual disability. In a study aiming to explore the genetics of epilepsy in the Sudanese population, we investigated several families including a consanguineous family with three siblings diagnosed with self-limited infantile epilepsy. We evaluated both dominant and recessive inheritance using whole exome sequencing and genomic arrays. We identified a pathogenic homozygous splice-site variant in the first intron of PRRT2 [NC_000016.10(NM_145239.3):c.-65-1G > A] that segregated with the phenotype in this family. This work taps into the genetics of epilepsy in an underrepresented African population and suggests that the phenotypes of homozygous PRRT2 variants may include milder epilepsy presentations without movement disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
The reported variants in family E11 were submitted to ClinVar (SCV002058115; SCV002059214). Exome and genomic array data are not publicly available. Additional details on the study that led to this manuscript, the clinical presentation of the individuals with PRRT2 variants, and genetic testing are available as Supplementary Methods.
References
Thomas RH, Berkovic SF. The hidden genetics of epilepsy—A clinically important new paradigm. Nat Rev Neurol. 2014;10:283–92. https://doi.org/10.1038/nrneurol.2014.62.
Ottman, R, Risch, N Genetic Epidemiology and Gene Discovery in Epilepsy. In: Noebels JL, Avoli M, Rogawski MA, et al. (eds). Jasper’s Basic Mechanisms of the Epilepsies. 4th ed. National Center for Biotechnology Information (US), Bethesda (MD), 2012. Available from: https://www.ncbi.nlm.nih.gov/books/NBK98200/.
Helbig I, Heinzen EL, Mefford HC, the ILAE Genetics Commission. Primer Part 1-The building blocks of epilepsy genetics. Epilepsia. 2016;57:861–8. https://doi.org/10.1111/epi.13381.
Sánchez Fernández I, Loddenkemper T, Gaínza-Lein M, Sheidley BR, Poduri A. Diagnostic yield of genetic tests in epilepsy: a meta-analysis and cost-effectiveness study. Neurology. 2019;92:e418–28. https://doi.org/10.1212/WNL.0000000000006850.
Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci. 2010;107:1779–86. https://doi.org/10.1073/pnas.0906079106.
Dobon B, Hassan HY, Laayouni H, Luisi P, Ricaño-Ponce I, Zhernakova A, et al. The genetics of East African populations: a Nilo-Saharan component in the African genetic landscape. Sci Rep. 2015;5:9996. https://doi.org/10.1038/srep09996.
Mohammed IN, Abdel Moneim M, Abdel Rahman A. The profile of childhood epilepsy in Sudan. Khartoum Med J. 2010;03:444–7.
Genomics England PanelApp. Genetic epilepsy syndromes panel (Version: 2.2). 2020. https://panelapp.genomicsengland.co.uk.
Wang J, Lin ZJ, Liu L, Xu HQ, Shi YW, Yi YH, et al. Epilepsy-associated genes. Seizure. 2017;44:11–20. https://doi.org/10.1016/j.seizure.2016.11.030.
Scheffer IE, Grinton BE, Heron SE, Kivity S, Afawi Z, Iona X, et al. PRRT2 phenotypic spectrum includes sporadic and fever-related infantile seizures. Neurology. 2012;79:2104–8. https://doi.org/10.1212/WNL.0b013e3182752c6c.
Schubert J, Paravidino R, Becker F, Berger A, Bebek N, Bianchi A, et al. PRRT2 mutations are the major cause of benign familial infantile seizures. Hum Mutat. 2012;33:1439–43. https://doi.org/10.1002/humu.22126.
Landolfi A, Barone P, Erro R. The spectrum of PRRT2-associated disorders: update on clinical features and pathophysiology. Front Neurol. 2021;12:629747. https://doi.org/10.3389/fneur.2021.629747.
Ebrahimi-Fakhari D, Saffari A, Westenberger A, Klein C. The evolving spectrum of PRRT2-associated paroxysmal diseases. Brain: A J Neurol. 2015;138:3476–3495. https://doi.org/10.1093/brain/awv317.
Döring JH, Saffari A, Bast T, Brockmann K, Ehrhardt L, Fazeli W, et al. The phenotypic spectrum of PRRT2-associated paroxysmal neurologic disorders in childhood. Biomedicines. 2020;8:E456. https://doi.org/10.3390/biomedicines8110456.
Chen Y, Chen D, Zhao S, Liu G, Li H, Wu ZY. Penetrance estimation of PRRT2 variants in paroxysmal kinesigenic dyskinesia and infantile convulsions. Front Med. 2021;15:877–86. https://doi.org/10.1007/s11684-021-0863-4.
Delcourt M, Riant F, Mancini J, Milh M, Navarro V, Roze E, et al. Severe phenotypic spectrum of biallelic mutations in PRRT2 gene. J Neurol Neurosurg Psychiatry. 2015;86:782–785. https://doi.org/10.1136/jnnp-2014-309025.
Raponi M, Baralle D. Alternative splicing: good and bad effects of translationally silent substitutions: alternative splicing. FEBS J. 2010;277:836–40. https://doi.org/10.1111/j.1742-4658.2009.07519.x.
Singer-Berk M, Gudmundsson S, Baxter S, Seaby EG, England E, Wood JC, et al. Advanced variant classification framework reduces the false positive rate of predicted loss-of-function variants in population sequencing data. Am J Hum Genet. 2023;110:1496–508. https://doi.org/10.1016/j.ajhg.2023.08.005.
Abramowicz A, Gos M. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J Appl Genet. 2018;59:253–68. https://doi.org/10.1007/s13353-018-0444-7.
Funding
This work was supported by grants from the ‘Deutsche Forschungsgemeinschaft’ (DFG Research Unit FOR2715; grants Le1030/16-1 and Le1030/16-2) and the ‘Stiftung no epilep’ to Holger Lerche. Mahmoud Koko was supported by a doctoral grant from the ‘Deutscher Akademischer Austauschdienst’ (DAAD program 57214224). LEOE was supported by Princess Nourah bint Abdulrahman University Researchers supporting project number (PNURSP2022R172), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. MAS was supported by King Saud University Researchers supporting project number (RSP-2020/38), King Saud University, Riyadh, Saudi Arabia. The funding source(s) had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Conception and Design: HL, LE, JS, AY, MK, MT, ME, IM, AH, MEI, MS. Primary clinical evaluation: ME, IM, AH, MS, AY. Recruitment and sampling of families: AY, RA, MK, AA, MA, IO, EA, EZ, FE, WA, EOA, EE, MOI, NH, HM, AB. Control datasets: LE, AY, MEI, MA, YB, MOI, MK. Exome Sequencing: JA, JS, MK, LE, AY, MA, YB, MEI, PN, HL. Array genotyping: MT, JS, MK, PN, HL. Data analysis and interpretation: MK, JS, HL. Writing - first draft: MK. Writing - critical revisions: MK, HL, ME, LE, AY. All authors revised and approved the final version of the manuscript. See the Supplementary for a detailed account of individual contributions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics and informed consent
Written informed consent for publication of their clinical details and/or clinical images was obtained from the participants/patients or their parents. All procedures in this study were performed following the standards the 1975 Helsinki declaration and its later amendments as well as the standards of the research committees of the Faculty of Medicine, University of Khartoum (Khartoum, Sudan).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Koko, M., Elseed, M.A., Mohammed, I.N. et al. Bi-allelic PRRT2 variants may predispose to Self-limited Familial Infantile Epilepsy. Eur J Hum Genet (2024). https://doi.org/10.1038/s41431-024-01541-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41431-024-01541-x