Abstract
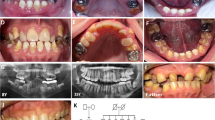
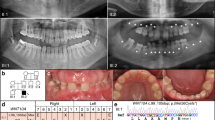
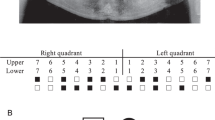
Amelogenesis imperfecta (AI) is a group of rare genetic conditions characterized by quantitative and/or qualitative tooth enamel alterations. AI can manifest as an isolated trait or as part of a syndrome. Recently, five biallelic disease-causing variants in the RELT gene were identified in 7 families with autosomal recessive amelogenesis imperfecta (ARAI). RELT encodes an orphan receptor in the tumor necrosis factor (TNFR) superfamily expressed during tooth development, with unknown function. Here, we report one Brazilian and two French families with ARAI and a distinctive hypomineralized phenotype with hypoplastic enamel, post-eruptive enamel loss, and occlusal attrition. Using Next Generation Sequencing (NGS), four novel RELT variants were identified (c.120+1G>A, p.(?); c.120+1G>T, p.(?); c.193T>C, p.(Cys65Arg) and c.1260_1263dup, p.(Arg422Glyfs*5)). Our findings extend the knowledge of ARAI dental phenotypes and expand the disease-causing variants spectrum of the RELT gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
RELT variants were submitted to the Leiden Open Variation Database (LOVD).Variants IDs are 0000917717; 0000917923;0000917924;0000917925.
Code availability
The data generated and analyzed during this study can be found in the published article and its supplementary files.
References
Smith CEL, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, et al. Amelogenesis imperfecta; genes, proteins, and pathways. Front Physiol. 2017;8:1–22.
Crawford PJ, Aldred M, Bloch-Zupan A. Amelogenesis imperfecta. Orphanet J Rare Dis. 2007;2:1–11.
de La Dure Molla M, Bournier BP, Manzanares MC, Acevedo AC, Hennekam RC, Friedlander L, et al. Elements of morphology: standard terminology for the teeth and classifying genetic dental disorders. Am J Med Genet A. 2019;179:1913–81.
Wright JT. Enamel phenotypes: genetic and environmental determinants. Genes. 2023;14:545–65.
Prasad MK, Geoffroy V, Vicaire S, Jost B, Dumas M, Le Gras S, et al. A targeted next-generation sequencing assay for the molecular diagnosis of genetic disorders with orodental involvement. J Med Genet. 2016;53:98–110.
Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, et al. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–83.
Kim J, Zhang H, Seymen F, Koruyucu M, Hu Y, Kang J, et al. Mutations in RELT cause autosomal recessive amelogenesis imperfecta. Clin Genet. 2018;95:375–83.
Nikolopoulos G, Smith CEL, Brookes SJ, El‐Asrag ME, Brown CJ, Patel A, et al. New missense variants in RELT causing hypomineralized amelogenesis imperfecta. Clin Genet. 2020;97:688–95.
Sica GL, Zhu G, Tamada K, Liu D, Ni J, Chen L. RELT, a new member of the tumor necrosis factor receptor superfamily, is selectively expressed in hematopoietic tissues and activates transcription factor NF-kappaB. Blood. 2001;97:2702–7.
Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26.
Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 2012;119:651–65.
Polek TC, Talpaz M, Spivak-Kroizman T. The TNF receptor, RELT, binds SPAK and uses it to mediate p38 and JNK activation. Biochem Biophys Res Commun. 2006;343:125–34.
Cusick JK, Mustian A, Goldberg K, Reyland ME. RELT induces cellular death in HEK 293 epithelial cells. Cell Immunol. 2010;261:1–8.
Cusick JK, Alhomsy Y, Wong S, Talbott G, Uversky VN, Hart C, et al. RELT stains prominently in B-cell lymphomas and binds the hematopoietic transcription factor MDFIC. Biochem Biophys Rep. 2020;24:100868.
Choi BK, Kim S-H, Kim YH, Lee DG, Oh HS, Han C, et al. RELT negatively regulates the early phase of the T-cell response in mice. Eur J Immunol. 2018;48:1739–49.
Gialluisi A, Pippucci T, Anikster Y, Ozbek U, Medlej-Hashim M, Mégarbané A, et al. Estimating the allele frequency of autosomal recessive disorders through mutational records and consanguinity: the Homozygosity Index (HI). Ann Hum Genet. 2012;76:159–67.
Ikeda A, Shahid S, Blumberg BR, Suzuki M, Bartlett JD. ADAM10 is expressed by ameloblasts, cleaves the RELT TNF receptor extracellular domain, and facilitates enamel development. Sci Rep. 2019;9:14086.
Acknowledgements
We thank Ms. Yorindel Cardozo for her assistance in the design of figure Appendix S6.
Funding
This study was supported by CAPES/COFECUB (Me918/2018) and the University of Brasilia (DPI/UnB, Brazil).
Author information
Authors and Affiliations
Contributions
KKMR: Contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. MCR: Contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. PMY: Contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. SR: Contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. LP: Contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. AB: Contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. RF: Contributed to analysis, and interpretation, drafted and critically revised the manuscript. VC-D: Contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. CG: Contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. ACA: Contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. MLD-M: Contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. “All authors gave their final approval and agree to be accountable for all aspects of the work”.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was developed in compliance with the Declaration of Helsinki and approved by the Ethics Committee of the Health Sciences Faculty, University of Brasília and by the Ethics Committee from Rothschild Hospital. Informed consent for publication of individual details and images was signed by all legal guardians and/or patients that agreed to participate in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Resende, K.K.M., Riou, M.C., Yamaguti, P.M. et al. Oro-dental phenotyping and report of three families with RELT-associated amelogenesis imperfecta. Eur J Hum Genet 31, 1337–1341 (2023). https://doi.org/10.1038/s41431-023-01440-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01440-7
This article is cited by
-
Deep phenotyping and population-level data can help resolve genomic variants
European Journal of Human Genetics (2023)