Abstract
Genomic healthcare programmes, both in a research and clinical context, have demonstrated a pivotal opportunity to prevent, diagnose, and treat rare diseases. However, implementation factors could increase overall costs and affect uptake. As well, uncertainties remain regarding effective training, guidelines and legislation. The purpose of this rapid evidence review was to draw together the available global evidence on the implementation of genomic testing programmes, particularly on population-based screening and diagnostic programmes implemented at the national level, to understand the range of factors influencing implementation. This review involved a search of terms related to genomics, implementation and health care. The search was limited to peer-reviewed articles published between 2017–2022 and found in five databases. The review included thirty articles drawing on sixteen countries. A wide range of factors was cited as critical to the successful implementation of genomics programmes. These included having policy frameworks, regulations, guidelines; clinical decision support tools; access to genetic counselling; and education and training for healthcare staff. The high costs of implementing and integrating genomics into healthcare were also often barriers to stakeholders. National genomics programmes are complex and require the generation of evidence and addressing implementation challenges. The findings from this review highlight that there is a strong emphasis on addressing genomic education and engagement among varied stakeholders, including the general public, policymakers, and governments. Articles also emphasised the development of appropriate policies and regulatory frameworks to govern genomic healthcare, with a focus on legislation that regulates the collection, storage, and sharing of personal genomic data.
Similar content being viewed by others
Introduction
There has been considerable progress in the field of genomic medicine with the increasing application of genomic technologies in healthcare to screen, diagnose, and treat diseases [1, 2]. Despite the rapidly evolving field of genomic research, its integration and implementation into healthcare has been slow and varies drastically between and within countries [3, 4]. The provision of genomic medicine has thus far been hindered by inconclusive evidence of the clinical utility of genomic information and the insufficient implementation science evidence [1, 2].
Implementation science enables us to understand the factors shaping the adoption of genomics and to identify tools that support its implementation into healthcare. While healthcare providers broadly support the adoption of genomic medicine, they are limited in their ability to do so due to the lack of genomics education and training, healthcare fragmentation and the scarcity of digital tools for genomics integrated with electronic health records (EHR). Moreover, there remains a lack of research focusing on macro-level factors such as health systems, health policies, financing, and generalisability of genomics programmes [5]. In order to identify and overcome the barriers to the adoption of genomics and ensure its effective and timely translation into clinical settings, implementation frameworks and outcome measures should be applied from the initial research stages through to the development and integration of genomic medicine. Once genomics programmes are integrated into healthcare settings, they should continue to be evaluated and adapted and adjusted as necessary to ensure sustainable implementation [5].
A number of countries have adopted genomic healthcare programmes at a national or regional level. The United Kingdom has government support to lead developments in genomic medicine, delivered through a national health service (NHS) [6]. Following the delivery of the 100,000 Genomes Project, whole genome sequencing (WGS) is now available along with other clinical genomic tests for rare diseases and cancers with equitable access through a national NHS Genomic Medicine Service [7]. This includes linking clinical care with a national de-identified research database of genomic and health data (with patient consent) to promote a learning healthcare system. Genomics England (GEL), in partnership with the NHS, is now designing and implementing a research study to explore the potential benefits and challenges of WGS in newborns [8].
The Australian Genomics Health Alliance is a national network of state-level genomic initiatives collaborating to translate approaches to genomics into standardised practice through clinical and laboratory research. This includes a focus on health policy, health economics, education and implementation science [9].
In France, The Plan France Médecine Génomique 2025 (PFMG 2025) aims to develop a national framework for big-genomic data. Leading French public research organisations participate, while France Génomique manages the programme’s technological aspects, including sequencing platforms and data analysis infrastructure. Patients’ electronic records are standardised, and regulatory frameworks are being developed. The PFMG 2025 aims include developing an economic model and processes for harmonising protocols and methods to support implementation into healthcare.
There are understandably variable approaches to genomics programmes in different countries due to differences in funding, infrastructure and variable approaches to evaluating their implementation. This can present a challenge to harmonizing the enablers and barriers to implementation, including common factors that may be relevant internationally.
While a variety of examples of national or regional genomic healthcare programmes exist, there is a recently emerging body of evidence regarding evaluation and implementation of these programmes, including factors acting as barriers and enablers with variability in the number, type and duration of these studies in different countries internationally. In order to inform the effective planning and design of future programmes, it is helpful to understand these implementation factors up-front. This evidence is crucial to bridging the gap between research findings and the systematic and sustainable uptake of genomics into clinical care. This rapid evidence review aims to identify these factors and discuss their implications for programme planning, practice, and policy and support programme design by considering these implementation factors up-front. We chose a rapid review approach because it best supported Genomic England need for a capture of the current knowledge, trends and gaps in the fast-moving field of genomics, to inform its future programmes.
Methods
The review design was informed by guidance for rapid evidence reviews developed by Tricco et al. [10]. The review followed a phased approach, beginning with a broad search strategy and subsequently expanding with each search round. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement to guide the review design and report the methods and findings [11]. Due to the rapid nature of the review (10 weeks), the questions and search strategy were targeted to identify relevant articles that could be analysed within the review timeframe. A protocol was developed before searching and is registered in the Open Science Framework (OSF) platform (https://osf.io/hkywe/).
Search strategy
We identified search terms using a combination of free-text and controlled terms building on previous work and suggestions from employees at GEL, as this review was commissioned to inform implementation approaches for programmes and services developed by GEL and delivered within the UK NHS.
We tested and refined the terms by running exploratory searches in principal databases. We assessed a provisional search strategy for sensitivity versus breadth on PubMed, using different combinations of Boolean operators and search strings (see Appendix 1 for the complete search strategy).
The search was limited to articles published between 2017 and 2022 due to the rapidly changing nature of genomic technologies and to keep the review scope manageable for a rapid timeframe. However, there was no language or location limitation. The search strategy focused on three categories (genomics, implementation science, and healthcare). Final searches were conducted in February 2022 on five databases (Web of Science, Medline, PubMed, CINAHL and EMBASE).
Document selection
The search results were imported into Rayyan, which is a validated tool with semi-automated features enabling the detection of duplicated publications from the different databases [12]. The software also displays citation details, titles, and abstracts of each publication, facilitating screening.
The initial title and abstract screening for eligibility were conducted in unison. Following the initial screening at the title/abstract level, two researchers cross-checked 10% of exclusions against the inclusion criteria. The remaining publications that met the inclusion criteria were organised and allocated randomly to continue full-text screening for eligibility. In this phase, 100% of included and 25% of excluded papers were reviewed by a different reviewer (CV-P). Both screening at title and abstract and full-text level were performed by 4 reviewers (GA-G, TM, ND, FR). Detailed inclusion and exclusion criteria are depicted in (Table 1).
Data extraction
Data extraction was conducted using an extraction form on REDCap software to organise the review process. The extraction form was first piloted, and necessary amendments were made before extracting data from the included documents. Data were extracted by four reviewers and checked by a different team member.
Data synthesis
Data were synthesised using framework analysis [13]. The analysis focused on developing themes that can accurately represent the data. The categories for the framework were based on the research questions guiding the review and the information emerging from the documents.
Quality assessment
The methodological quality of the empirical articles was critically appraised in parallel to data synthesis using the Mixed Methods Appraisal Tool (MMAT) [14, 15]. The MMAT was developed to allow reviewers to assess the methodological quality of diverse study designs, including qualitative, quantitative, and mixed methods. Selected studies catalogued as reviews were not assessed using the MMAT since this tool is only indicated for quantitative, qualitative and mixed-methods studies. Overall, papers had an average score of 4.4 of 5.0. Lowest scores were related to insufficient interpretation based on the data and inconsistencies in using methods [16, 17]. The results from the assessments can be found in Appendix 2.
Results
Article selection
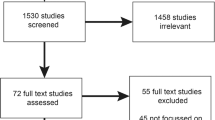
The initial search yielded 5027 articles (after duplicates were removed). In total, 4958 articles were excluded as these did not meet the inclusion criteria outlined above. We reviewed 62 articles at the full-text level and excluded 32 because they did not describe genomic programmes, did not include implementation at a national or regional level or were focused on specific conditions. The final review included 30 articles (see Fig. 1 for the PRISMA Flow Diagram).
Article characteristics
The main article characteristics are summarised in Table 2. Seven articles were from Australia, six from the US, four from France, three from the UK, one from Canada, one from the Netherlands, and eight were global in scope, including countries such as Pakistan, Estonia, Nigeria, Malaysia, Slovenia, Cyprus, Israel, Saudi Arabia, Belgium and Sweden. Sixteen articles described programmes implemented at the national level, four at the regional level, and eight described multi-sited programmes but did not specify if these were regional or national in scale. Fourteen articles focused on the use of genomics in screening, three focused on diagnostic genomic testing, and thirteen addressed both. Twenty-one articles were non-empirical reports in the form of commentaries, while the remaining nine articles were empirical in nature, of which seven were qualitative, one was quantitative, and one was mixed methods.
Implementation processes for programmes at a national scale
Most of the genomic programmes implemented at a national scale were in the pilot phase. The articles focussed on clinical settings [16, 18,19,20,21,22,23,24,25], governmental perspectives on genomic programmes [20, 21, 26,27,28,29,30,31], and, to a lesser extent, on the patients’ or participants’ experiences of these programmes [22, 27].
The cost-effectiveness and the funding of genomic programmes were recurrent concerns in implementing sustainable systems. It was frequently mentioned that the short-term funding of genomic programmes could hamper long-term continuity [26, 29, 32], and without palpable high-quality evidence, some governments could not justify creating ongoing publicly funded programmes [28]. Authors also frequently mentioned the need for an infrastructure capable of storing a large amount of confidential data and keeping up with the rapidly evolving technological landscape of genomics [16, 20, 21, 24, 27, 32,33,34,35]. Many articles also emphasised the importance of setting up an evaluation framework to obtain feedback and make changes [18, 27, 36,37,38].
Genetic counselling services were also regularly mentioned as a key means to supply information to patients, families, and the general public [22, 25, 27, 28, 32, 37, 39,40,41]. Some articles also highlighted that having transparent and standardised clinical pathways facilitated implementation [18, 24], but this needs to be accompanied by training programmes for healthcare providers to be effective [3, 20, 22, 25, 30, 37, 42,43,44]. Finally, topics such as culturally appropriate communication for patients and families, time efficiency in the delivery of services and interdisciplinarity of the staff involved were also mentioned as integral parts of programme implementation [9, 20, 21, 23, 25, 27, 32, 34, 35, 41]. Additional information can be found below in (Table 3).
Implementation barriers and enablers
Factors acting as barriers to implementation
Various barriers to implementing genomic programmes were mentioned across the 30 articles, many of which were interrelated. Nineteen articles identified the lack of genomic education and literacy and the need for significant upskilling of the healthcare workforce as a barrier. Fourteen articles identified the significant costs of enacting system-wide change as a barrier to implementation, with many pointing to the costs involved, not only for running tests and analysing them, but also the additional time and staff needed to provide adequate education, training, consent, and counselling, and the costs involved in integrating genomics information and technology into the healthcare system. Nine articles also perceived a lack of guidelines, regulations, and standards as a significant barrier, which may be related to the perceived lack of government and policy-making support for genomics. Six articles identified the lack of integration between Electronic Health Records (EHR) and genomics data as a barrier to successful implementation, particularly considering healthcare fragmentation and the need for EHRs to be accessible to patients to share across different healthcare organisations [22, 35]. Two articles identified that genomic medicine implementation is hindered by the quantity and types of evidence required for healthcare payers to justify reimbursement, being unwilling to reimburse for new diagnostics that do not meet their thresholds for clinical validity and utility [37, 45]. One article also identified parents’ emotional, psychological and time costs as a barrier to rapid genomic sequencing in intensive care, compounded by counselling in stressful hospital intensive care settings [41].
Factors acting as enablers in implementation
Ten articles discussed the need to incorporate genomics into medical and nursing education programmes and provide training to existing healthcare workers in conjunction with enhanced regulation, guidelines, and standards [44]. Ten articles identified a combination of the increasing accessibility and affordability of genomic technology (e.g., rGS and NGS) and increasing data storage capacities as factors facilitating the implementation of genomic programmes. Eight articles highlighted the value of lesson-sharing and collaborative communication between hospitals, clinicians and countries. Four articles discussed the need for health services to accommodate genomics at an organisational level, including integrating genomic data and EHRs and embedding programmes into clinical portfolios, such as cancer and infectious diseases [24]. Eight articles discussed the role of genetic counselling, the use of decision aids, patient engagement and education in facilitating implementation. Finally, two articles discussed the need for modelling to demonstrate implementation at a larger scale, with a focus on integration and employing adaptive management philosophies that allow for flexibility to facilitate changes in response to learning and external factors [18, 24]. Further details about the main barriers and enablers discussed in the articles can be found in Appendix 3.
Discussion
This rapid evidence review synthesized implementation barriers and enablers across several countries and identified challenges and opportunities to consider for implementing genomics diagnostic or screening programmes. As such, this section will discuss some key areas and themes from the findings. It is important to bear in mind that these enablers and barriers to implementation vary across geographical areas and countries. As such, they should be considered general barriers and enablers that will affect different countries and areas differently. It is also important to note that addressing any one of these factors would not be sufficient to guarantee implementation; a holistic approach must be taken to address micro and macro-level implementation challenges.
Genomic education and training of healthcare staff
In line with the rapid advancement of genomics across the fields of medicine, science, technology, ethics and legislation, there is a need for continuous education and the exchange of knowledge between different stakeholders to keep up with the pace of progress [46]. The successful integration of genomics into healthcare fundamentally depends on healthcare professionals with the appropriate genomic knowledge and skills, and this was the most cited factor in the articles in this review. For the promise of genomic medicine to be translated into improved patient outcomes, it is necessary to establish quality evidence-based education with clear objectives for learning [47]. This would require integrating genomics into training for the upcoming workforce (e.g., though undergraduate, graduate, or specialist professional training), as well as initiatives to target the existing health care workforce who would not have had any prior education in genomics or who need to refresh their education to reflect more recent developments. For these interventions to be successful, there is a need for reporting standards which entail consistent descriptions enabling replication, comparisons and helping those developing interventions to learn lessons from past efforts. An example of this is a program logic model and reporting standards for genomics education and evaluation, proposed following international collaboration and consensus [47, 48]. Of note, there is no overarching governing body to publish or mandate the use of reporting standards for education interventions for genomics, but global and national bodies can facilitate awareness and implementation.
It is also essential to develop online learning tools that improve accessibility internationally and facilitate sharing educational efforts [27]. Most healthcare professionals prefer education that demonstrates clinical utility via workshops, lectures, conferences, or online materials [44]. Indeed, it is noted that the motivation to learn new skills is tied to the applicability of the information [49].
Other approaches include developing cross-professional genomics competency frameworks in [50,51,52]; utilizing techniques such as the ‘flipped classroom’ blended learning approach [49]; and embedding trained genomic specialists within mainstream clinical settings to provide on-hand education and support [16]. These strategies can provide greater consistency, encourage multidisciplinary team working, and lead to more effective practice through more flexible and constructive use of healthcare professionals’ time, and focusing in-person interactions on case-based learning or interactive discussion [17, 49,50,51,52]. Hands-on learning or experiential learning approaches are recognized ways to support the development of skills and confidence in genomics [17, 52]. Clinical Decision Support (CDS) tools can also help clinicians with limited knowledge by providing just-in-time information with links to evidence [53].
While education is not the sole factor guaranteeing the success of genomic programmes, it is clearly integral to provide a foundation for implementation. Crellin et al., (2019) highlight that adults are most likely to be receptive to educational interventions when they recognize a need to learn, and the benefits of the knowledge acquired. Many specialists continue to hold concerns about genomics including a lack of clinical guidelines, the often-uncertain nature of genomic results, and the potential to cause psychological harm to patients; nevertheless, most acknowledge that integrating genomics into healthcare is inevitable [54]. It is important to ensure that education is not addressed in isolation, but considered in the context of other factors such as the ability to enable multidisciplinary team working and coordinated care pathways, patient and public perspectives, and ethical issues surrounding genomics.
Public support and patient education and engagement
The successful implementation of genomics will also depend on the culturally appropriate engagement of the general public, for which public health education and genomics promotion programs are necessary [27, 53]. Such programmes should draw on behavioural economics and deliberative public engagement methods [27]. Common approaches to engaging the general public include educational events, online platforms, mass media and social media engagement [16, 46]. In Finland, the government plans to educate its citizens on making informed decisions regarding genetic testing and study participation. It intends to implement guidelines on using genomic data, such as the educational genome portal and virtual health services, to enable people to engage with and learn to use their genomic data [46]. The UK, France, Canada, Denmark and Finland have all started integrating genomic education into the primary and secondary education curricula, using online platforms for education, holding educational symposiums and providing specialised training to teachers [46, 55]. In New England, the professional development programme Teaching the Genome Generation (TtGG) provides teachers with the necessary tools to educate students on genomics through biology classes [56].
Patient engagement and education is crucial as it is the stakeholder group most impacted by genomics, and strategies to address this must go hand-in-hand with approaches to educating healthcare professionals as discussed above. This can be provided via workshops, community outreach programmes, interactive and multimedia e-learning tools and decision aids to support consent [22, 46, 57, 58]. Actively involving diverse groups of patients in implementation, for example through patient advisory boards or through co-design of information materials, has also shown to enhance success of implementation [16].
Communication and counselling
Communication between various stakeholders is crucial to developing sustainable genomic programmes. For instance, interactions between and within teams in hospital settings are essential to long-term success [27, 35, 42]. Tools such as webinars, conference calls, and presentations between the teams involved in the programmes encourage collaboration and enable feedback and eventual modifications while also unifying the different strands of the project [17, 35]. Team members of the American-based IGNITE projects thought this strategy was critical to engagement and buy-in by physicians and hospital management – which ultimately facilitated the incorporation of genetic medicine into routine care [35]. For Australian Genomics, a study measuring collective ties between clinicians showed that “hands-on learning” and “making group decisions” were the most potent influences on their genomic practice. This was achieved by strategically building a genomic learning community by creating boundary-spanning roles [17].
Although genetic counselling is considered best practice, several barriers exist to its successful implementation. Genetic counsellors are a highly specialized resource with limited numbers, and therefore difficult to scale. Models to facilitate large-scale delivery could include mainstreaming genetic counselling skills among non-genetic experts (such as nurses, pharmacists and general practitioners), or using digital tools such as telehealth or chatbots to help triage patients and support decision making [22, 27, 59].
Challenges also exist with regards to patient communication. In some circumstances, such as critical care settings, patients are required to understand information quickly in high-stress environments [41]. Information must therefore be sufficiently tailored to support patents’ diverse decision-making needs. Social factors such as discouragement by family members, health insurance coverage, ability to pay, and lack of awareness of available genetic services from patients and clinicians have also been identified as barriers to accessing genetic counselling [22, 59]. As well as public, patient and healthcare professional education, successful strategies to overcome these barriers include delivering genetic counselling in more accessible locations (such as primary care facilities) or providing this service remotely [32]. Genomic counselling seeks to help patients “understand and adapt to the medical, psychological and familial implications of genetic contributions to disease” [57] and is an important facilitator to information about genomic technologies.
Advocacy actions and information campaigns improve public awareness of genetic technologies. They are likely to make access to genomics information easier for clinicians and patients and, in turn, facilitate the implementation of genomics into routine care [20]. Nevertheless, these public interventions must be culturally appropriate and accessible to be effective [24, 27]. However, there is a gap in the literature regarding the public perception of genomic technologies, their needs and expectations, and their experience with these techniques. This sociological information would help all stakeholders by helping orient research perspectives and improving the guidance and support offered to patients.
Regulatory frameworks
As the field of genomics progresses rapidly, there remains the problem of insufficient clinical evidence combined with a lack of ethical, legal and social regulations [30]. Indeed, fully realising genomic medicine’s potential requires a multi-pronged research, clinical, policy and regulatory agendas. As genomics evolves, so does the regulatory landscape around it, albeit in an often uncertain and ad hoc manner. As well, there are limitations to understanding genetic links between health and disease, particularly for rare conditions, when data is scarce or siloed. For its potential to be truly harnessed, genomic research and healthcare data need to be integrated and accessible at a large scale [60].
Regulatory approaches to genomics in research, clinical care and public health vary with regards to issues of liability, consent, quality assurance, and data and privacy protection. Increased availability of direct-to-consumer (DTC) testing creates additional challenges as this is accessed outside of health care and research initiatives and is typically unregulated [60, 61]. The current approach to research, clinical care, public health, and DTC testing is siloed and fails to recognise how genomics data is produced and used across these areas [61]. Policymakers and legislators need to regulate the return of results, confidentiality and privacy while creating policies that promote genomic education and economic incentives that bring together the interests of different stakeholders [62]. The convoluted ecosystem of stakeholders necessitates technical standards that are neutral and adaptable for diverse purposes and relevant to the wide-ranging set of clinical, research, commercial and public users [63].
It is widely acknowledged that the potential of genomics-enabled research and clinical care is proportional to the amount of data that can be accessed and analysed and the implications shared [62]. As such, regulatory frameworks for genomics need to be perceptive of the connections blurring the boundaries between research and clinical care, such as when sequencing activity should be considered research or care and how findings with clinical implications should be managed and by whom [33]. Pooling data can also help increase sample sizes to a level that makes investigating every rare condition feasible. In 2018, 21 European Union (EU) member states signed a joint declaration to enable the cross-border sharing of human genomes by 2022 [60]. However, genomic data may allow the re-identification of donors, which has privacy implications and is also sensitive regarding fundamental rights. The 2016 General Data Protection Regulation (GDPR) establishes the rules for personal data protection and sharing by individuals, companies, and organisations at the EU level. However, the reality has not lived up to hopes, with the member states using their discretion to interpret and implement the regulation differently [64]. Challenges also exist within countries, where health care is administered at a provincial or state level; for example, differences in legislation and common law interpretations between Australian states create challenges for researchers and clinicians, leading to uncertain expectations of how institutions will protect data [65]. Shared guidelines or codes of conduct effectively provide practical guidance and procedural clarity to avoid inconsistencies. Researchers and scientists can participate in their development, thereby increasing the chances that data protection will be addressed in line with the relevant sector and reinforcing the code’s factual and scientific legitimacy [64].
Ultimately, codes of conduct ease responsibility issues by clarifying the appropriate safeguards for genomic data protection. They promote the coordinated application of rules and, over time, they directly influence the laws governing data sharing by enabling improved engagement with data protection principles and an enhanced understanding of obligations [64].
Differences in legislation and common law interpretations between Australian states create challenges for researchers and clinicians, leading to uncertain expectations of how institutions will protect data [65]. Creating more precise expectations and encouraging public trust would be better promoted by having a more harmonised regulatory approach. Public trust is vital to collecting, storing, and sharing genomic data [66]. Regulations promoting accountable and transparent collaboration and standardised data collection and reporting are critical to fostering such trust.
A federated approach is seen as a viable strategy in cases where data cannot be pooled for legal or practical reasons [60]. Under this approach, independent bodies host the data in a secure processing environment governed by technical standards that enable large-scale analysis [63].
Although challenging, international collaboration is crucial to addressing regulatory issues relating to genomics in research and health care, and maximising the value of data. Consortia such as the Global Alliance for Genomics and Health (GA4GH), including researchers, data scientists, healthcare professionals and patient advocacy groups, are working towards consistent standards in regulation, policy and data management to enable more effective comparison across countries [62, 63]. Even at the level of electronic patient health records, EHR software vendors are highly proprietary, creating complications for standardisation and harmonisation with research systems [63]. Standard methods for collecting, storing, sharing, accessing and analysing genomics data need to be agreed upon if the benefits of genomics medicine are to benefit the population at large.
Legal differences across domains should be harmonised to reduce confusion and increase compliance. For instance, regulatory differences between research and clinical care cause unnecessary confusion concerning data identifiability, deidentification, and re-identification [61]. There is a need for stakeholders to have clarity and education about regulations governing the different domains of genomics to ensure transparency and accountability.
In the absence of widely accessible genomics services via national healthcare systems, DTC companies have generated a market and grown rapidly, leading to decreased costs and increased access to genetic tests [67]. However, governments today remain unable to monitor or quantify DTC testing effectively and are limited in their ability to regulate it [27]. DTC tests are often delivered without oversight from a healthcare practitioner, results may be inaccurate or misleading, and some operate outside of regulatory frameworks entirely if they are based offshore and deliver services via the internet [68]. DTC testing may be addressing a gap in public interest and need in the absence of sufficient access to genomic testing in health care, but risks causing greater complications with regards to the barriers mentioned in this review including regulation and public trust [68, 69]. As such, policymakers need to develop to regulation and medical coverage approaches that enables genomics tests to be the most effective pathway to clinical care [62].
Ethical considerations
There are a number of ethical issues raised by genomics in research and health care which can significantly influence implementation of genomics programmes. Should the genomic test be regarded as research or as clinical care? What are the responsibilities of the clinician and/or researcher to feedback research findings to participants? Should relevant results be communicated to family members as well? How can genomic programmes be designed and delivered in an equitable manner? The answers to these questions have variable implications for implementation of genomic programmes, including workforce capacity; regulatory frameworks;as well as public and patient engagement and trust.
It is generally acknowledged that patients should be kept informed about the type of information they may receive throughout the genomic testing process (whether clinical or research-based), and be respectful of their choices regarding the types of results they wish to receive. There are recognized challenges to facilitating valid consent while addressing the fact that genomic results can be complex and uncertain [33]. To enable this, patients must be empowered to actively participate in decisions about their care, which includes having awareness of the ethical, legal and social implications of genomics [46].
Recommendations summary
In discussing the barriers and enablers to the implementation of genomics programmes many of the articles reviewed also highlighted recommendations for moving forward which are summarised here.
It is recommended that genomic education needs to be improved and made more accessible for all stakeholders including the public, patients, healthcare providers, policymakers and governments [17, 27, 37, 44, 46, 49, 53, 54]. For such education interventions to be successful reporting standards comprising consistent descriptions which enable replication, comparisons and lessons learned are advocated [47, 48].
With regard to communication, it is recommended to establish genomic counselling services with communication and information tool guidelines to maintain good communication with patients and their families and enable informed decision-making [22, 27, 32, 33, 41, 46, 59]. It is also key to involve patients, participants and the public in decision making processes, and to incorporate their experience of implementations in it as well. Moreover, multidisciplinary engagement and communication across a range of conditions and expertise is advised [17, 18, 37, 43, 70].
Finally, to enable the successful implementation of genomics in healthcare articles advise developing appropriate policies and regulatory frameworks to govern genomics, with a particular focus on harmonising regulation across the domains of research and clinical care and establishing legislation regulating the collection, storage, and sharing of personal genomic data [33, 60,61,62,63,64].
Limitation and strengths
The review used a rapid design, meaning that only a limited number of databases and websites were accessed, which facilitated timely results. It is possible that specific subject headings, keyword terms and synonyms have been missed. The review was strengthened by having a multidisciplinary team with five reviewers searching for peer-reviewed articles and cross-checking their relevance, following robust systematic research guidelines.
Conclusion
This review has outlined the implementation process for genomic screening or diagnostic programmes on a national scale and the interrelated factors that act as barriers and facilitators to their implementation. It is essential to acknowledge the differences in policy, funding, etc within and between countries, and the extent to which they are affected by those barriers or enablers. Across the articles reviewed, most of the programmes were still in pilot phases and early stages. Genomic healthcare has grown over the past years, but a range of interlinked factors must be addressed for these programmes to thrive and translate into national-scale implementation. As is often the case, investment in research is not yet matched by clear financial and policy commitments to widespread implementation and workforce education, with many programmes only operating on a small scale with limited accessibility to the public and within a complex and convoluted international landscape. Taken together these findings can help to inform the design of future programmes, as well as future research on the factors influencing the implementation of population-based genomic programmes. Hopefully, the implementation of the programmes and initiatives discussed above will lead to durable national programmes and enable increased and equitable access to genomics in our healthcare systems.
References
Roberts MC, Kennedy AE, Chambers DA, Khoury MJ. The current state of implementation science in genomic medicine: opportunities for improvement. Genet Med. 2017;19:858–63.
Taylor N, Best S, Martyn M, Long JC, North KN, Braithwaite J, et al. A transformative translational change programme to introduce genomics into healthcare: a complexity and implementation science study protocol. BMJ Open. 2019;9:e024681.
Best S, Long JC, Gaff C, Braithwaite J, Taylor N. Organizational perspectives on implementing complex health interventions: clinical genomics in Australia. J Health Organ Manag. 2021;35:825–45.
Best S, Vidic N, An K, Collins F, White SM. A systematic review of geographical inequities for accessing clinical genomic and genetic services for non-cancer related rare disease. Eur J Hum Genet. 2022;30:645–52. https://doi.org/10.1038/s41431-021-01022-5
Bangash H, Kullo IJ. Implementation Science to Increase Adoption of Genomic Medicine: An Urgent Need. J Pers Med [Internet]. 2020;11:19. https://www.mdpi.com/2075-4426/11/1/19
HM Government. Genome UK: the future of healthcare [Internet]. Genome UK: the future of healthcare. London; 2020 Sep [cited 2022 Aug 11]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/920378/Genome_UK_-_the_future_of_healthcare.pdf
NHS England. NHS England » NHS Genomic Medicine Service [Internet]. Genomics. 2022 [cited 2022 Aug 11]. Available from: https://www.england.nhs.uk/genomics/nhs-genomic-med-service/
Pichini A, Ahmed A, Patch C, Bick D, Leblond M, Kasperaviciute D, et al. Developing a National Newborn Genomes Program: An Approach Driven by Ethics, Engagement and Co-design. Front Genet [Internet]. 2022 May;13. Available from: https://www.frontiersin.org/articles/10.3389/fgene.2022.866168/full
Best S, Long J, Theodorou T, Hatem S, Lake R, Archibald A, et al. Health practitioners’ perceptions of the barriers and enablers to the implementation of reproductive genetic carrier screening: a systematic review. Prenat Diagn [Internet]. 2021;41:708–19. http://onlinelibrary.wiley.com/doi/abs/10.1002/pd.5914
Tricco AC, Langlois EV, Straus SE Rapid reviews to strengthen health policy and systems: a practical guide [Internet]. Alliance for Health Policy and Systems Research, & World Health Organization. (2017). Rapid reviews to strengthen health policy and systems: A practical guide. 2017 [cited 2022 Apr 20]. Available from: https://apps.who.int/iris/handle/10665/258698
Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ [Internet]. 2021 Mar;n160. Available from: https://doi.org/10.1136/bmj.n160
Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst Rev 2016 51 [Internet]. 2016;5:1–10. https://systematicreviewsjournal.biomedcentral.com/articles/10.1186/s13643-016-0384-4
Gale NK, Heath G, Cameron E, Rashid S, Redwood S. Using the framework method for the analysis of qualitative data in multi-disciplinary health research. BMC Med Res Methodol [Internet]. 2013;13:117. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/1471-2288-13-117
Pace R, Pluye P, Bartlett G, Macaulay AC, Salsberg J, Jagosh J, et al. Testing the reliability and efficiency of the pilot Mixed Methods Appraisal Tool (MMAT) for systematic mixed studies review. Int J Nurs Stud [Internet]. 2012;49:47–53. https://pubmed.ncbi.nlm.nih.gov/21835406/
Souto RQ, Khanassov V, Hong QN, Bush PL, Vedel I, Pluye P. Systematic mixed studies reviews: Updating results on the reliability and efficiency of the mixed methods appraisal tool. Int J Nurs Stud [Internet]. 2015;52:500–1. https://pubmed.ncbi.nlm.nih.gov/25241931/
Sperber NR, Carpenter JS, Cavallari LH, Damschroder JL, Cooper-DeHoff RM, Denny JC, et al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med Genomics [Internet]. 2017;10:35. https://bmcmedgenomics.biomedcentral.com/articles/10.1186/s12920-017-0273-2
Long JC, Pomare C, Best S, Boughtwood T, North K, Ellis LA, et al. Building a learning community of Australian clinical genomics: a social network study of the Australian Genomic Health Alliance. BMC Med [Internet]. 2019;17:44. https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1274-0
Gaff CL, Winship MI, Forrest MS, Hansen PD, Clark J, Waring MP, et al. Preparing for genomic medicine: a real world demonstration of health system change. npj Genom Med [Internet]. 2017;2:16. http://www.nature.com/articles/s41525-017-0017-4
Bertier G, Joly Y. Clinical exome sequencing in France and Quebec: what are the challenges? What does the future hold? Life Sci Soc Policy [Internet]. 2018;14:17. http://www.proquest.com/docview/2080558781/abstract/77B7389E1C6F4178PQ/1
Laviolle B, Denèfle P, Gueyffier F, Bégué É, Bilbault P, Espérou H, et al. The contribution of genomics in the medicine of tomorrow, clinical applications and issues. Therapies [Internet]. 2019;74:9–15. https://linkinghub.elsevier.com/retrieve/pii/S0040595718302622
Nadauld LD, Ford JM, Pritchard D, Brown T. Strategies for clinical implementation: precision oncology at three distinct institutions. Health Aff [Internet]. 2018;37(May):751–6. http://www.healthaffairs.org/doi/10.1377/hlthaff.2017.1575
Snir M, Nazareth S, Simmons E, Hayward L, Ashcraft K, Bristow SL, et al. Democratizing genomics: Leveraging software to make genetics an integral part of routine care. Am J Med Genet Part C Semin Med Genet [Internet]. 2021;187:14–27. https://onlinelibrary.wiley.com/doi/10.1002/ajmg.c.31866
Tonkin E, Calzone KA, Badzek L, Benjamin C, Middleton A, Patch C, et al. A roadmap for global acceleration of genomics integration across nursing. J Nurs Scholarsh [Internet]. 2020;52:329–38. https://onlinelibrary.wiley.com/doi/10.1111/jnu.12552
Vidgen ME, Williamson D, Cutler K, McCafferty C, Ward RL, McNeil K, et al. Queensland Genomics: an adaptive approach for integrating genomics into a public healthcare system. npj Genom Med [Internet]. 2021;6:71. https://www.nature.com/articles/s41525-021-00234-4
Vinkšel M, Writzl K, Maver A, Peterlin B. Improving diagnostics of rare genetic diseases with NGS approaches. J Community Genet [Internet]. 2021;12:247–56. https://link.springer.com/10.1007/s12687-020-00500-5
Spackman E, Hinde S, Bojke L, Payne K, Sculpher M. Using cost-effectiveness analysis to quantify the value of genomic-based diagnostic tests: recommendations for practice and research. Genet Test Mol Biomark [Internet]. 2017;21:705–16. http://www.liebertpub.com/doi/10.1089/gtmb.2017.0105
Burns BL, Bilkey GA, Coles EP, Bowman FL, Beilby JP, Pachter NS, et al. Healthcare System Priorities for Successful Integration of Genomics: An Australian Focus. Front Public Heal [Internet]. 2019;7:358. https://www.mdpi.com/2075-4426/11/5/358
Rowe CA, Wright CF. Expanded universal carrier screening and its implementation within a publicly funded healthcare service. J Community Genet [Internet]. 2020;11:21–38. http://link.springer.com/10.1007/s12687-019-00443-6
Denommé-Pichon A-S, Vitobello A, Olaso R, Ziegler A, Jeanne M, Tran Mau-Them F, et al. Accelerated genome sequencing with controlled costs for infants in intensive care units: a feasibility study in a French hospital network. Eur J Hum Genet [Internet]. 2022;30:567–76. https://www.nature.com/articles/s41431-021-00998-4
Traversi D, Pulliero A, Izzotti A, Franchitti E, Iacoviello L, Gianfagna F, et al. Precision medicine and public health: new challenges for effective and sustainable health. J Pers Med [Internet]. 2021;11:135. https://www.mdpi.com/2075-4426/11/2/135
Vidgen ME, Fowles LF, Istiko SN, Evans E, Cutler K, Sullivan K, et al. Evaluation of a Genetics Education Program for Health Interpreters: A Pilot Study. Front Genet [Internet]. 2022;12. Available from: https://www.frontiersin.org/articles/10.3389/fgene.2021.771892/full
Pearce C, Goettke E, Hallowell N, McCormack P, Flinter F, McKevitt C. Delivering genomic medicine in the United Kingdom National Health Service: a systematic review and narrative synthesis. Genet Med [Internet]. 2019;21:2667–75. https://www.sciencedirect.com/science/article/pii/S1098360021012168
Gaille M, Horn R. The ethics of genomic medicine: redefining values and norms in the UK and France. Eur J Hum Genet [Internet]. 2021;29:780–8. https://www.nature.com/articles/s41431-020-00798-2
Prins BP, Leitsalu L, Pärna K, Fischer K, Metspalu A, Haller T, et al. Advances in genomic discovery and implications for personalized prevention and medicine: Estonia as example. J Pers Med. [Internet]. 2021;11:358. https://www.mdpi.com/2075-4426/11/5/358
Zebrowski AM, Ellis DE, Barg FK, Sperber NR, Bernhardt BA, Denny JC, et al. Qualitative study of system-level factors related to genomic implementation. Genet Med [Internet]. 2019;21:1534–40. https://linkinghub.elsevier.com/retrieve/pii/S1098360021016920
Elsink K, Huibers MMH, Hollink IHIM, Simons A, Zonneveld-Huijssoon E, van der Veken LT, et al. Implementation of early next-generation sequencing for inborn errors of immunity: a prospective observational cohort study of diagnostic yield and clinical implications in Dutch genome diagnostic centers. Front Immunol. 2021;12:1–11.
Levy KD, Blake K, Fletcher-Hoppe C, Franciosi J, Goto D, Hicks JK, et al. Opportunities to implement a sustainable genomic medicine program: lessons learned from the IGNITE Network. Genet Med [Internet]. 2019;21:743–7. http://www.proquest.com/docview/2473249946/abstract/F7D4F383C2FE44D0PQ/1
Stark Z, Boughtwood T, Phillips P, Christodoulou J, Hansen DP, Braithwaite J, et al. Australian genomics: a federated model for integrating genomics into healthcare. Am J Hum Genet. 2019;105:7–14.
Balasopoulou A, Mooy F-M, Baker DJ, Mitropoulou C, Skoufas E, Bulgiba A, et al. Advancing global precision medicine: an overview of genomic testing and counseling services in Malaysia. Omi A J Integr Biol [Internet] 2017;21:733–40. http://www.liebertpub.com/doi/10.1089/omi.2017.0136
Delatycki MB, Alkuraya F, Archibald A, Castellani C, Cornel M, Grody WW, et al. International perspectives on the implementation of reproductive carrier screening. Prenat Diagn [Internet]. 2020;40:301–10. https://onlinelibrary.wiley.com/doi/10.1002/pd.5611
Lynch F, Nisselle A, Stark Z, Gaff CL, McClaren B. Parents’ experiences of decision making for rapid genomic sequencing in intensive care. Eur J Hum Genet [Internet]. 2021;29(May):1804–10. https://www.nature.com/articles/s41431-021-00950-6
Abimiku AG, Croxton T, Ozumba PJ, Agala N, Balogun O, Jonathan E, et al. Blueprint for building a biorepository in a resource-limited setting that follows international best practices. Afr J Lab Med [Internet]. 2019;8. Available from: http://www.ajlmonline.org/index.php/AJLM/article/view/722
Tonkin E, Calzone KA, Badzek L, Benjamin C, Middleton A, Patch C, et al. A maturity matrix for nurse leaders to facilitate and benchmark progress in genomic healthcare policy, infrastructure, education, and delivery. J Nurs Scholarsh [Internet]. 2020;52:583–92. https://onlinelibrary.wiley.com/doi/abs/10.1111/jnu.12586
White S, Jacobs C, Phillips J. Mainstreaming genetics and genomics: a systematic review of the barriers and facilitators for nurses and physicians in secondary and tertiary care. Genet Med [Internet]. 2020;22:1149–55. https://linkinghub.elsevier.com/retrieve/pii/S1098360021011813
Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, Caulfied MJ, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet [Internet]. 2019;104(May):13–20. https://linkinghub.elsevier.com/retrieve/pii/S0002929718304221
Zimani AN, Peterlin B, Kovanda A. Increasing Genomic Literacy Through National Genomic Projects. Front Genet [Internet]. 2021;12. https://www.frontiersin.org/articles/10.3389/fgene.2021.693253/full
Nisselle A, Janinski M, Martyn M, McClaren B, Kaunein N, Maguire J, et al. Ensuring best practice in genomics education and evaluation: reporting item standards for education and its evaluation in genomics (RISE2 Genomics). Genet Med. 2021;23:1356–65.
Nisselle A, Martyn M, Jordan H, Kaunein N, McEwen A, Patel C, et al. Ensuring best practice in genomic education and evaluation: a program logic approach. Front Genet [Internet]. 2019;10. Available from: https://www.frontiersin.org/article/10.3389/fgene.2019.01057
Fee-Schroeder KC, Nelson DM. Flipped classroom strategy: an accessible, application-driven approach to genomics education. Clin J Oncol Nurs. 2019;23:145–8.
Pichini A, Bishop M A nationally agreed cross-professional competency framework to facilitate genomic testing. Genet Med [Internet]. 2022;24:1743–52. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1098360022007535
Simpson S, Seller A, Bishop M Using the findings of a national survey to inform the work of england’s genomics education programme. Front Genet [Internet]. 2019;10. Available from: https://www.frontiersin.org/article/10.3389/fgene.2019.01265/full
McClaren BJ, Crellin E, Janinski M, Nisselle AE, Ng L, Metcalfe SA, et al. Preparing medical specialists for genomic medicine: continuing education should include opportunities for experiential learning. Front Genet [Internet]. 2020;11. Available from: https://www.frontiersin.org/article/10.3389/fgene.2020.00151/full
Klein ME, Parvez MM, Shin J-G. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J Pharm Sci. 2017;106:2368–79.
Crellin E, McClaren B, Nisselle A, Best S, Gaff C, Metcalfe S. Preparing Medical Specialists to Practice Genomic Medicine: Education an Essential Part of a Broader Strategy. Front Genet [Internet]. 2019;10. https://www.frontiersin.org/article/10.3389/fgene.2019.00789/full
Whitley KV, Tueller JA, Weber KS. Genomics education in the era of personal genomics: academic, professional, and public considerations. Int J Mol Sci. 2020;21:768.
LaRue KM, McKernan MmP, Bass KM, Wray CG. Teaching the Genome Generation: Bringing Modern Human Genetics into the Classroom Through Teacher Professional Development. J STEM Outreach [Internet]. 2018;1. https://www.jstemoutreach.org/article/3680-teaching-the-genome-generation-bringing-modern-human-genetics-into-the-classroom-through-teacher-professional-development
Metcalfe SA. Genetic counselling, patient education, and informed decision-making in the genomic era. Semin Fetal Neonatal Med. 2018;23:142–9.
Lyon GJ, Segal JP. Practical, ethical and regulatory considerations for the evolving medical and research genomics landscape. Appl Transl Genomics. 2013;2:34–40.
Riaz M, Tiller J, Ajmal M, Azam M, Qamar R, Lacaze P. Implementation of public health genomics in Pakistan. Eur J Hum Genet [Internet]. 2019;27:1485–92. http://www.nature.com/articles/s41431-019-0428-z
Saunders G, Baudis M, Becker R, Beltran S, Béroud C, Birney E, et al. Leveraging European infrastructures to access 1 million human genomes by 2022. Nat Rev Genet. 2019;20:693–701.
Wolf SM, Ossorio PN, Berry SA, Greely HT, McGuire AL, Penny MA, et al. Integrating rules for genomic research, clinical care, public health screening and DTC testing: creating translational law for translational genomics. J Law, Med Ethics. 2020;48:69–86.
Ginsburg GS, Phillips KA. Precision medicine: from science to value. Health Aff. 2018;37:694–701.
Rehm HL, Page AJH, Smith L, Adams JB, Alterovitz G, Babb LJ, et al. GA4GH: International policies and standards for data sharing across genomic research and healthcare. Cell Genomics [Internet]. 2021;1:100029. https://linkinghub.elsevier.com/retrieve/pii/S2666979X21000367
Molnár‐Gábor F, Korbel JO. Genomic data sharing in Europe is stumbling—Could a code of conduct prevent its fall? EMBO Mol Med [Internet]. 2020;12. https://onlinelibrary.wiley.com/doi/10.15252/emmm.201911421
Eckstein L, Chalmers D, Critchley C, Jeanneret R, McWhirter R, Nielsen J, et al. Australia: regulating genomic data sharing to promote public trust. Hum Genet. 2018;137:583–91.
Milne R, Morley KI, Almarri MA, Anwer S, Atutornu J, Baranova EE, et al. Demonstrating trustworthiness when collecting and sharing genomic data: public views across 22 countries. Genome Med. 2021;13:92.
McGeveran W, Schmitz C. General-purpose privacy regulation and translational genomics. J Law, Med Ethics. 2020;48:142–50.
Tiller J, Lacaze P. Regulation of Internet-based Genetic Testing: Challenges for Australia and Other Jurisdictions. Front Public Heal [Internet]. 2018;6. http://journal.frontiersin.org/article/10.3389/fpubh.2018.00024/full
Molster CM, Bowman FL, Bilkey GA, Cho AS, Burns BL, Nowak KJ, et al. The evolution of public health genomics: exploring its past, present, and future. Front Public Heal. 2018;6
Long JC, Gul H, McPherson E, Best S, Augustsson H, Churruca K, et al. A dynamic systems view of clinical genomics: a rich picture of the landscape in Australia using a complexity science lens. BMC Med Genomics [Internet]. 2021;14:63. https://bmcmedgenomics.biomedcentral.com/articles/10.1186/s12920-021-00910-5
Funding
This work was commissioned by GEL to inform implementation approaches for programmes and services developed by GEL and delivered within the UK NHS. This work was not funded by any particular grant or award.
Author information
Authors and Affiliations
Contributions
GA-G, TM, ND, FR, AP and CV-P made substantial contributions to the conception and design of the work. GA-G, TM, ND, and FR analysed and interpreted the data. GA-G, TM, ND, FR, AP and CV-P drafted the work and substantively revised it. CV-P and GA-G were responsible for supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
AP is employed by Genomics England. The authors declare no competing interests.
Ethical approval
For this work ethical approval was not required considering its methodological nature.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alarcón Garavito, G.A., Moniz, T., Déom, N. et al. The implementation of large-scale genomic screening or diagnostic programmes: A rapid evidence review. Eur J Hum Genet 31, 282–295 (2023). https://doi.org/10.1038/s41431-022-01259-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01259-8
This article is cited by
-
Precision public health in the era of genomics and big data
Nature Medicine (2024)
-
2023 in the European Journal of Human Genetics
European Journal of Human Genetics (2024)
-
Use of a multi-phased approach to identify and address facilitators and barriers to the implementation of a population-wide genomic screening program
Implementation Science Communications (2023)
-
Investigating genomic medicine practice and perceptions amongst Australian non-genetics physicians to inform education and implementation
npj Genomic Medicine (2023)
-
Genes=disease (?)
European Journal of Human Genetics (2023)