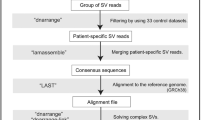
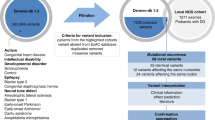
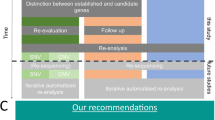
Abstract
About 0.3% of all variants are due to de novo mobile element insertions (MEIs). The massive development of next-generation sequencing has made it possible to identify MEIs on a large scale. We analyzed exome sequencing (ES) data from 3232 individuals (2410 probands) with developmental and/or neurological abnormalities, with MELT, a tool designed to identify MEIs. The results were filtered by frequency, impacted region and gene function. Following phenotype comparison, two candidates were identified in two unrelated probands. The first mobile element (ME) was found in a patient referred for poikilodermia. A homozygous insertion was identified in the FERMT1 gene involved in Kindler syndrome. RNA study confirmed its pathological impact on splicing. The second ME was a de novo Alu insertion in the GRIN2B gene involved in intellectual disability, and detected in a patient with a developmental disorder. The frequency of de novo exonic MEIs in our study is concordant with previous studies on ES data. This project, which aimed to identify pathological MEIs in the coding sequence of genes, confirms that including detection of MEs in the ES pipeline can increase the diagnostic rate. This work provides additional evidence that ES could be used alone as a diagnostic exam.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data are available on request from the corresponding author.
References
Finnegan DJ. Retrotransposons. Curr Biol. 2012;22:R432–7.
Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, et al. SVA Elements: A Hominid-specific Retroposon Family. J Mol Biol. 2005;354:994–1007.
Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40:1666–83.
Chenais B. Transposable Elements in Cancer and Other Human Diseases. Curr Cancer Drug Targets. 2015;15:227–42.
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82.
Feusier J, Watkins WS, Thomas J, Farrell A, Witherspoon DJ, Baird L, et al. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res. 2019;29:1567–77.
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703.
Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable Elements and Genome Organization: A Comprehensive Survey of Retrotransposons Revealed by the Complete Saccharomyces cerevisiae Genome Sequence. Genome Res. 1998;8:464–78.
Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L 1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6.
Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4859970/.
Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. Genom Med. 2018;3:1–10.
Goerner-Potvin P, Bourque G. Computational tools to unmask transposable elements. Nat Rev Genet. 2018;19:688–704.
Torene RI, Galens K, Liu S, Arvai K, Borroto C, Scuffins J, et al. Mobile element insertion detection in 89,874 clinical exomes. Genet Med. 2020;22:974–8.
Gardner EJ, Prigmore E, Gallone G, Danecek P, Samocha KE, Handsaker J, et al. Contribution of retrotransposition to developmental disorders. Nat Commun. 2019;10:4630.
Demidov G, Park J, Armeanu‐Ebinger S, Roggia C, Faust U, Cordts I, et al. Detection of mobile elements insertions for routine clinical diagnostics in targeted sequencing data. Mol Genet Genom Med. 2021;9:e1807.
Gardner EJ, Lam VK, Harris DN, Chuang NT, Scott EC, Pittard WS, et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–29.
Thevenon J, Duffourd Y, Masurel‐Paulet A, Lefebvre M, Feillet F, Chehadeh‐Djebbar SE, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–7.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34:3572–4.
Wu J, Lee W-P, Ward A, Walker JA, Konkel MK, Batzer MA, et al. Tangram: a comprehensive toolbox for mobile element insertion detection. BMC Genom. 2014;15:795.
Thung DT, de Ligt J, Vissers LE, Steehouwer M, Kroon M, de Vries P, et al. Mobster: accurate detection of mobile element insertions in next generation sequencing data. Genome Biol. 2014;15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4228151/.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Platzer K, Lemke JR. GRIN2B-Related Neurodevelopmental Disorder. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Mirzaa G, et al., editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993. http://www.ncbi.nlm.nih.gov/books/NBK501979/.
Guo Y, Dai Y, Yu H, Zhao S, Samuels DC, Shyr Y. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics. 2017;109:83–90.
Has C, Yordanova I, Balabanova M, Kazandjieva J, Herz C, Kohlhase J, et al. A novel large FERMT1 (KIND1) gene deletion in Kindler syndrome. J Dermatol Sci. 2008;52:209–12.
Has C, Wessagowit V, Pascucci M, Baer C, Didona B, Wilhelm C, et al. Molecular Basis of Kindler Syndrome in Italy: Novel and Recurrent Alu/Alu Recombination, Splice Site, Nonsense, and Frameshift Mutations in the KIND1 Gene. J Investig Dermatol. 2006;126:1776–83.
Youssefian L, Vahidnezhad H, Barzegar M, Li Q, Sotoudeh S, Yazdanfar A, et al. The Kindler Syndrome: A Spectrum of FERMT1 Mutations in Iranian Families. J Investig Dermatol. 2015;135:1447–50.
Zhou C, Song S, Zhang J. A novel 3017-bp deletion mutation in the FERMT1 (KIND1) gene in a Chinese family with Kindler syndrome. Br J Dermatol. 2009;160:1119–22.
Sawamura D, Nakano H, Matsuzaki Y. Overview of epidermolysis bullosa. J Dermatol. 2010;37:214–9.
Lai‐Cheong JE, Tanaka A, Hawche G, Emanuel P, Maari C, Taskesen M, et al. Kindler syndrome: a focal adhesion genodermatosis. Br J Dermatol. 2009;160:233–42.
Bruel A-L, Nambot S, Quéré V, Vitobello A, Thevenon J, Assoum M, et al. Increased diagnostic and new genes identification outcome using research reanalysis of singleton exome sequencing. Eur J Hum Genet. 2019;27:1519–31.
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
1000 Genomes Project Consortium. A map of human genome variation from population scale sequencing. Nature. 2010;467:1061–73.
Acknowledgements
We thank the probands and their families for their participation; and the Centre De Calcul (CCuB) at the University of Burgundy for providing technical support and management of the informatics core facility.
Funding
This work was supported by grants from the Regional Council of Burgundy (to C.T.‐R.), the FEDER 2017, PARI 2017, and CIFRE (ANRT) between Laboratoire Cerba and Regional Council of Burgundy for the doctoral work at Laboratoire Cerba and GAD.
Author information
Authors and Affiliations
Contributions
PG, MC and YD designed the study. PG, MC, AnV, SV, ET, CP, ALB, FTMT, HS, CTR, LF and YD analyzed the data. PG and MC performed PCR and MiSeq sequencing. MC performed cell cultures and experiments. AlV, PV, OP, ASDP, CTR, LF performed clinical analysis. All authors contributed to read, and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the ethics committee of the Dijon University Hospital. Informed consents were provided for the study, with separate consent obtained for the use of photographs.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Garret, P., Chevarin, M., Vitobello, A. et al. A second look at exome sequencing data: detecting mobile elements insertion in a rare disease cohort. Eur J Hum Genet 31, 761–768 (2023). https://doi.org/10.1038/s41431-022-01250-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01250-3
This article is cited by
-
GPAD: a natural language processing-based application to extract the gene-disease association discovery information from OMIM
BMC Bioinformatics (2024)
-
2023 in the European Journal of Human Genetics
European Journal of Human Genetics (2024)
-
Unusual genomic variants require unusual analyses
European Journal of Human Genetics (2023)