Abstract
Northern Pakistan is home to many diverse ethnicities and languages. The region acted as a prime corridor for ancient invasions and population migrations between Western Eurasia and South Asia. Kho, one of the major ethnic groups living in this region, resides in the remote and isolated mountainous region in the Chitral Valley of the Hindu Kush Mountain range. They are culturally and linguistically distinct from the rest of the Pakistani population groups and their genetic ancestry is still unknown. In this study, we generated genome-wide genotype data of ~1 M loci (Illumina WeGene array) for 116 unrelated Kho individuals and carried out comprehensive analyses in the context of worldwide extant and ancient anatomically modern human populations across Eurasia. The results inferred that the Kho can trace a large proportion of their ancestry to the population who migrated south from the Southern Siberian steppes during the second millennium BCE ~110 generations ago. An additional wave of gene flow from a population carrying East Asian ancestry was also identified in the Kho that occurred ~60 generations ago and may possibly be linked to the expansion of the Tibetan Empire during 7th to 9th centuries CE (current era) in the northwestern regions of the Indian sub-continent. We identified several candidate regions suggestive of positive selection in the Kho, that included genes mainly involved in pigmentation, immune responses, muscular development, DNA repair, and tumor suppression.
Similar content being viewed by others
Introduction
South Asia saw the development of one of the earliest urban societies and currently encompasses at least one-fourth of humanity. South Asian populations harbor the highest genetic diversity in Eurasia with large effective population sizes and a complex history [1]. Ancient invasions and migrations resulted in the admixture of different populations that eventually led to the complex linguistic and genetic patterns found across South Asia today [2]. Present-day north-western Pakistan served as a prime corridor for the influx of invaders and immigrants to South Asia from the northwest. The area currently houses diverse tribes and ethnicities with excessive linguistic diversity, where different Indo-European and Tibeto-Burman languages are spoken [3]. Previous genetic studies reported significant West Eurasian genetic contributions, assumed to be derived from Neolithic Iranians and Middle-Late Bronze Age steppe, among populations from northwestern Pakistan and neighboring north India [4, 5].
Several Pakistani populations have been studied over the past two decades as part of worldwide population genetic projects, including the Human Genome Diversity Project [6], the 1000 Genomes Project, Human Genome Organisation (HUGO) Pan-Asian SNP Consortium, and GenomeAsia 100 K Project [7]. However, genetic characterization of some of the smaller isolated population groups is still lacking and represents a significant gap in the understanding of Pakistani as well as South Asian population genetic history. One such population includes the Kho who reside in the Chitral Valley in the Hindukush Mountain range close to the Kalash, an enigmatic isolated ethnic group from Pakistan [8]. Kho speak the Indo-European Khowar language, which is the predominant language of the region. Khowar is phylogenetically and typologically related to the Kalasha language and according to Morgenstierne’s assumption both Khowar and Kalasha belong to the first wave of Indo-Aryan immigrants from the North [9]. Furthermore, a number of phonological, grammatical, and lexical features of Kalasha were reported to infer a close historical relationship to neighboring Indo-Aryan Khowar [10]. The approximate population size of Kho is about 0.24 million [3].
The Kho population originating in the context of pre-historical and extant South and Central Asian groups has so far only been studied using a dental morphometric approach [11]. This study inferred that the Kho exhibit distant affinities to prehistorical Central Asian and present-day North Indian groups, and concluded that the Kho represented either highly isolated or peripheral population of the rough Hindu Kush highlands [11]. A previous report using a limited number of mtDNA markers also showed the presence of major western Eurasian haplogroups in the Kho with few low frequency South Asian specific haplogroups, but offered no consolidated evidence about Kho origins [12].
In the current study, we produced genome-wide data for the Kho population residing in the mountainous Chitral Valley of the Hindu Kush Mountains in northwestern Pakistan, near the border with Afghanistan, and carried out a population genetic analysis aimed at unveiling their demographic history and identifying genomic regions characteristic of adaptation. We also examined any possible genetic affinity between the Kho and Kalash groups and compared them with modern and ancient human populations.
Materials and methods
DNA sampling
This project protocol was approved by the Ethical Review Committee of the Abdul Wali Khan University Mardan (AWKUM), Pakistan (AWKUM/Biochem/ERC/2018/574). A total of 116 unrelated Kho individuals were enrolled in the current study. The self-reported Kho ethnicity of these individuals was confirmed up to five generations ago. Written informed consent was provided by all participants. The whole blood specimens of the participants were collected on Whatman 903 cards and dried.
Genotyping assay
Genomic DNA was extracted from dried blood spots using DP-318 Kit (Tiangen Biotechnology, Beijing). Genotyping was performed on the Illumina WeGene V2 Array (1,194,791 SNPs) by Illumina iScan System at the WeGene genotyping center, Shenzhen, China. A minimal genotyping call of 98.5% was required for a valid sample.
Data processing
Genetic marker quality control
Indels, heterosomal loci, and loci with more than two allelic states were removed from the genotyping data. For each sample, SNP markers were filtered with PLINK V1.9 [13] with parameters “--maf 0.001 --geno 0.05”. Only the intersection of the two arrays with identical allelic states was retained.
Genotype phasing
Eagle V2.3.5 [14] was employed for a reference panel-free genotype phasing via default parameters for the WeGene and 1000 Genomes Project phase 3 dataset.
Reference datasets
We downloaded the v42.4 1240 K (1,233,013 sites) and 1240 K_HO (597,573 sites) datasets from David Reich Lab website: (https://reich.hms.harvard.edu). We additionally included modern individuals from [15] available at the same source, in the 1240K_HO dataset. We used the 1240K datasets for the analysis on both modern and ancient individuals, since it has a higher number of sites. However, for comparison with modern population samples, we also used the 1240K_HO dataset because it includes a higher number of individuals. We converted the data to a PLINK format using ADMIXTOOLS convertf [16] and then merged it with the Kho genotype data using PLINK 1.9 [13], keeping only overlapping autosomal SNPs and excluding triallelic sites. We pruned the data for linkage disequilibrium using plink --indep-pairwise command with parameters 50 10 0.1 and --maf 0.05, thus retaining 66,503 and 168,874 SNPs for the 1240K_HO and 1240K datasets, respectively. For the ADMIXTURE analysis, we used as a reference panel a subset of the data presented in Yelmen et al. (2019) [1] pruned for LD as described above.
Principal component analysis
We performed the PCA using LASER-2.04 [17]. The genotype data was converted to vcf using PLINK and PCA analysis was performed with parameters -pca 1 and -k 100 retaining 3192 individuals from 126 different populations (Table S1).
ADMIXTURE, f-statistics, and ALDER analysis
We ran ADMIXTURE [18] on modern genomes in unsupervised mode with K ranging from 3 to 16. After inspecting cross-validation errors (CV) and finding no obvious best K, we chose to focus on K = 10 and K = 11 as the ones where the CV lower plateau seemed to start (Fig. S16). Additionally, we performed a supervised analysis with K = 7 using ancient genomes as sources. We converted the PLINK files to EIGENSTRAT format using Admixtools convertf [16], then performed f3 with default parameters. Standard errors were computed using a block jackknife with a size of 0.050 cM. We inferred admixture dates using ALDER [19]. We used a dataset not pruned for LD to carry out these analyses.
Selection scans
To identify putatively selected regions, we assembled a new reference dataset comprising of the Punjabi (PJL) and Yoruba (YRI) populations from the 1000 Genomes phase 3 dataset [20]. We removed indels and joined biallelic sites from both 1KG and Kho data before merging, keeping only autosomal sites with <10% missingness. A total of ~1 M SNPs were retained. We used the scikit-allel package to compute PBSn1 [21] score for each available position with the allele.pbs function with window size = 1, window step = 1 and normed = True.
Plots and figures
All graphs were plotted using R (version 4.0.5 - “Shake and Throw”) and the GUI RStudio (version 1.4.1103 - “Wax Begonia”). Circular admixture graphs were plotted using the Ancestry Painter software [22].
Results
Population demography
Principal component analysis
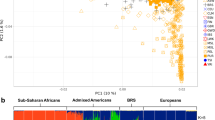
We performed a PCA to visualize the relationship between Kho and other Eurasian populations (Fig. 1, Supplementary Figs. S1, S2, and Table S1). The plot partially resembles the geographic distribution of these populations with the first principal component separating Europeans and South Asians from East Asians. All populations from Pakistan, with the exception of the Hazara, are located between Iranians and South Asians. The Kho individuals are all clustered together and are close to other populations from Pakistan. However, they are slightly shifted in the direction of East Asians, a feature that they share with the Burusho who reside in the neighboring Karakoram Mountain ranges close to Kho homeland in northwest Pakistan.
ADMIXTURE
To infer Kho ancestry we also conducted an unsupervised ADMIXTURE [18] analysis in the context of global populations with K clusters ranging from 3 to 16 to infer the Kho ancestry composition (Fig. 2; Supplementary Figs. S4–17). The Kho shows genetic affinity with populations from West Eurasia, South Asia, Central Asia, and East Asia. At K = 5 to K = 7 the West Eurasian populations, i.e., French, Basque, and Sardinian showed their own component sharing majorly with the Kho population as well (Supplementary Figs. S6–S8). At K = 8 the Paniya, i.e., a South Asian population makes its own component that is also present in Kho (Supplementary Figs. S9–S15). The Kho inferred no significant genetic differentiation from other surrounding Pakistani populations, i.e., Sindhi, Brahui, and Burusho from K = 5–10 and show ancestry components similar to Kalash at K = 10 (Fig. 2A). However, they exhibit an additional three extra components, one that is high in East Asian populations, one high in South Asian (Paniya), and a third component common in Europeans (Sardinian). The Burusho population from northern Pakistan show a similar profile, but their “East Asian” component was found to be higher. At K = 11 (Fig. 2B), the Kalash population stands out, while the Kho retain their three additional components.
Additionally, we performed a supervised analysis using the following seven as source populations: Yamnaya, Iran Neolithic, Anatolia Neolithic, Han, Irula, Serbia Mesolithic, and Yoruba (Supplementary Fig. S17, Table S4). The results show that Kho possess genetic ancestry components associated with European Neolithic farmers (Anatolia Neolithic + Mesolithic European Hunter Gatherers) and Yamnaya; these three components taken together are characteristic of Middle Bronze Age populations from the steppe region, while Early Bronze Age steppe populations (Yamnaya) lack the Anatolian Neolithic and European Hunter Gatherer components [4].
Comparisons with modern and ancient populations
To follow up the exploratory results we obtained with Admixture, we computed f3-outgroup [16] statistics in the form f3out (Kho, X, Mbuti) to identify the populations sharing the highest amount of drift with Kho (Supplementary Fig. S18, Table S2). We found populations from Eastern Europe and Caucasus regions, but also Kalash and Burusho from Pakistan at the higher positions. To test whether the Kho can be described as an admixture of Kalash with another Eurasian population, we performed an f3-admixture analysis in the form of f3adm (Kalash, X, Kho), while X being every Eurasian population present in our dataset. However, we did not obtain any significant results (Z < −3). In contrast, f3adm (Kalash, X, Burusho) was significantly negative for many East Asian populations, confirming East Asian admixture in the Burusho (Table S3). Since f3-admixture test is not suitable for negative tests, we further tested whether Kho show a higher proportion of East Asian ancestry compared to Kalash using a D-statistic test, which yielded a significant result (Z-score = 6) for the test Dstat (Kho, Kalash, Han, Mbuti). Such a contribution is found at a higher proportion in the Burusho population, as shown by both ADMIXTURE and f4-ratio test: f4 (Kho, Kalash, Han, Mbuti)/f4 (Japanese, Kalash, Han, Mbuti) = 0.05 while f4 (Burusho, Kalash, Han, Mbuti)/f4 (Japanese, Kalash, Han, Mbuti) = 0.11.
We subsequently compare the Kho with ancient DNA data and selected ~100 Middle-Late Bronze Age samples from the steppe region (Table S5). The f3adm (Steppe_MLBA, Han, Kho) result was highly significant with Z = −21. We used ALDER [19] to compute weighted linkage disequilibrium and date the admixture event identified with the previous analysis. Using Steppe_MLBA + Han (as a proxy for East Asia) inferred an admixture date of 62.37 ± 2.55 generations ago. The two 1-ref decay rates, which aim to describe the studied admixture event from the perspective of either source populations, suggest that the populations related to Steppe_MLBA and Han contributed to the ancestry of Kho at different times. For this reason, we performed an additional run with Steppe_MLBA and Turkmenistan_C_Geoksyur (a group of samples preceding the Bronze Age relatively close to Pakistan) [5] as sources. This analysis resulted in an inferred admixture date of 110.45 ± 10.63, compatible with the 1-ref result for Steppe_MLBA in the previously described iteration of ALDER (96.82 ± 11.83).
Natural selection signatures in Kho population
We computed the normalized version [21] of Population Branch Statistic—PBS [25] to identify putative signs of selection in the Kho, using Punjabi and Yoruba samples from 1000 Genomes Project as reference. We averaged the PBS score over 50-Kb windows and annotated the top results with PBS score based on the confidence percentile cut-off threshold ≥ 99.9% and 99.5%—shown as red and blue dash lines in Manhattan plot (Fig. 3, Table S6).
The top 53 windows (n = 53) were ranked according to the ≥99.9% percentile (PBS ≥ 0.165601) threshold and based on the ≥99.5% percentile (PBS ≥ 0.080014) a total of 265 windows were highlighted. The windows shortlisted based on these cut-off thresholds were annotated along with nearby (i.e., ~50 kb upstream and ~50 kb downstream) regions. The genes within and nearby the 50-kb windows acquired from top threshold, i.e., ≥99.9% percentile were annotated into five biological categories (Table 1).
Furthermore, a SNP-based enrichment analysis was performed for the top SNPs prioritized based on 99.9% percentile (PBS ≥ 0.366879, n = 973) and 99.5% percentile (PBS ≥ 0.134140, n = 4862) cut-off thresholds (Supplementary Fig. S3). GWAS catalog, Ensembl, and gnomAD repositories were used for SNP annotation and unveiled more than 500 genes underline selection in Kho population based on the confidence percentile 99.9% PBS score criteria (Table S7).
A number of genes associated with pigmentation and immune response to pathogens were highlighted by the PBS analysis (Table 1). GTF2IRD2 stood out within the top 50-kb window prioritized based on the 99.9% percentile with the highest PBS score of ≅ 0.89344. GTF2IRD2 is involved in the neurodevelopmental disorder Williams–Beuren syndrome and its functioning is important in cognitive phenotype and neuropsychological implications. The GTF2IRD2 positive selection feature has formerly been reported in Human [26].
Besides, the genes related to the innate immune system, i.e., DDB1, and VAT1 [27, 28] was also identified within the 99.9% percentile cut-off threshold. Both of the genes have an important role in the epidermis repair mechanism. The other mucosal membrane genes like IGHA2 and IGHA1 play critical immune function role in the recognition phase (i.e., mucous membranes) of the humoral immunity on exposure to pathogens. In addition to mucus recognition phase genes, other associated genes like IGHE, IGHG1, IGHEP1, and pulmonary inflammation, psoriasis linked gene, i.e., FBXL19, exhibited selection signals, possibly drive against pathogens [29, 30]. Another top-ranked window, having a high average PBS score of ≅ 0.8066, harbors TP53TG3D that is involved in the Wolf Wolf-Hirschhorn Syndrome.
The POTE family of genes, involved in several cancers’ diseases, also map within the positive selection regions. Previously, the functional SNPs of such p53-target genes have been reported to undergo positive selection, influencing the p53 mediated transcription regulation and hence affect cancer susceptibility [31]. Besides, several other tumor suppressor genes, including the GOLGA8N, NBR2, BRCA1, BRCA2, ARHGAP11A, ULK1, anticancer mature miRNA (microRNA) encoding gene MIR1270-2, MORF4L1, and RUNDC1 with p53/TP53 inhibitor are actively involved in tumor proliferation inhibition, suppression, and migration control [32] were also among the top PBS selection windows ranked on the 99.9% percentile confidence threshold. Besides, several important genes are shortlisted based on 99.9% percentile confidence (Table 1).
In addition, we scanned the COSMIC database [33] to check for possible correlation with cancer-related genes. Out of 20,539 protein-coding genes in the Ensembl GRCh37 Release 104, total 705 are classified as cancer-related genes in the COSMIC database. We identified 464 protein-coding genes in or nearby ~50 kb upstream and ~50 kb downstream the PBS top-scoring windows (99.5% percentile cut-off threshold). Among these, the 21 were identified as cancer-related genes (Table 1). The ratio of cancer-related genes in the positively selected genes was found higher than in the entire genes list (0.045 vs 0.034), but this was statistically not significant (chi-squared = 1.6734, df = 1, p value = 0.1958).
The PBS and FST based selection scan analysis of Kalash population is formerly reported [8]. However, we found no shared genes that possibly underlie selection in both the Kho and Kalash groups.
Discussion
Our results show that, while being included within the broader West-South Asian genetic cline, Kho display unique features which are telling of their peculiar demographic past. Similar to the many present-day South Asian populations residing in the north and western part of the Indian sub-continent, the Kho genetic ancestry has been heavily influenced by the immigration of Bronze Age populations from the steppe region of Southern Siberia during the second millennium BCE. This event has been well characterized archeologically, linguistically, and genetically [4, 5] and fits very well with the oldest of the admixture events being identified in the current study (i.e., Steppe_MLBA – Turkmenistan_C_Geoksyur 110 ± 10 generations ago) that resulted in an ancestry component known as Ancestral North Indian [34]. Other population sharing a similar history up to this point are the Kalash, another ethnic minority residing in nearby valleys in the Hindu Kush Mountain ranges. While the Kalash remained isolated and experienced intense drift [8], the ancestors of Kho received gene flow from a population carrying East Asian ancestry. We date this event to ~60 generations ago during the first centuries of the Common Era. The Kho share this feature with the nearby Burusho population from northern Pakistan that shows a comparatively higher proportion of East Asian ancestry, as shown by f4 ratio results. Such an admixture, estimated to be ∼26% proportion, has also been observed in the neighboring Balti population that resides in the Karakoram Mountain valleys and is dated to around 21–39 generations ago. This admixture event may be linked with the expansion of the Tibetan Empire in 869–1391 CE in this area [35] as well as with many other events that linked the Eastern and Western portion of the broader Eurasian continent [36]. The first millennium CE hosted several population movements in the area which may have brought West and East Eurasian components on the Steppe-like background shared by several neighboring populations, and which we here date to around 100 generations ago in the area, in agreement with genetics, archeological and linguistic studies that suggest a Late Bronze Age chronology for the arrival of steppe ancestry and Indoeuropean languages in South Asia [37, 38].
From the natural selection perspective, we observed a number of genomic regions which specifically differentiated Kho from the nearby Punjabi population that constitutes the majority of the Pakistani population, and which could be considered as plausible targets of adaptation to the local environment. Several regions depicted evidence of natural selection in Kho possibly with respect of immune responses to pathogens. DEFB130, an antimicrobial beta-defensin family protein, is located within a window having an average PBS score of ≅ 0.4. Up regulation of DEFB130 within macrophages has been reported to have a possible role in malarial parasite response [39]. The Chromosome 17: 43551389-43601388 window with PBS score ≅ 0.38 annotated for PLEKHM1, i.e., involved in autophagosomes maturation. The PLEKHM1 is reported to be targeted by Salmonella enterica effector protein, i.e., SifA, and hence the pathogen possibly hijacks the host endosomal system. Therefore, the PLEKHM1 acts like an interface between the host endolysosome and microbial infection [40]. The positive selection signatures exhibited by the PLEKHM1 might be mediated in response to such microbial infection. ERV3-1 located within ≈1 Mb region of a selection scan window (chromosome 7: 64543748-64593747) has an average PBS score ≅ 0.36. It is a retrovirus group 3 member protein which mediates the receptor recognition during early infection. The ERV3 locus is conserved in the primate genomes possibly due to its important evolutionary role [41]. Likewise, the MRC1L1 gene exhibits selection signatures and is also involved in microbial infection and acts as a major target and receptor of dengue virus and other pathogens including bacteria [42]. This MRC1L1 loci may also undergo selection in the context of immune response mechanisms against pathogens. In addition to replicating several well-characterized loci involved in skin pigmentation and immune responses to pathogens several of the loci contained genes reported to be involved in cancer pathogenesis and could reflect the response to increased exposure to ultraviolet radiation at high altitudes (i.e., average 1500 m) and may play an important role in cancer development in the Kho population. Besides, the toxic elements are considered as potential carcinogenic risk. Studies have reported about high cancerous risk hazardous in water and soil samples from district Chitral, where the Kho individuals reside [43]. Several important carcinoma-associated genes exhibited significant positive selection features in current investigation. Among these, the RALGAPA and RAB6C are involved in breast cancer [44], while ZAR1L and NBR2 are associated with breast-ovarian cancer pathways [45]. Moreover, several other general cancer-linked genes, including PSG1, CDK2AP2, CEACAM1 [30], carcinoembryonic antigen, STAG3L2, and LIMS1 involved in colorectal cancer, endocervical adenocarcinoma, lung cancer susceptibility, and exocervical carcinoma [46] were also annotated within the top positive selection regions.
The selection scan window, ranked based on 99.5% percentile threshold on chromosome17: 39701389-39751388 mapped close to the cluster of keratins encoding genes, including KRT9, and KRT15. KRT9 plays a role in keratin filament assembly and affects the footpad morphology and structure of the palmoplantar epidermis [47]. During ecological adaptation, KRTAPs genes encoding the major structural hair shaft proteins, evolve rapidly in response to intense selective pressure like heat, ultraviolet radiation, water loss, and mechanical force. This evolution allows successful ecological adaptation by modifying and diversifying the hair keratin [48]. A significant selection signature based on 99.5% percentile threshold was also detected on chromosome 17: 10,301,389-10,351,388 across the myosin heavy chain (MYH) gene cluster in the Kho. MYH genes are expressed in different developmental stages of muscle fiber [49]. The signatures in the keratin and muscle fibers encoding gene clusters may underline selection in Kho as an adaptation to their lifestyle in the Hindu Kush Mountain valleys. Among the major psychotic disorders, the schizophrenia-associated genes, i.e., ASPHD1, TMEM219, INO80E, and HIRIP3 [50] were also annotated among the top 27 selection scan windows. From an evolutionary perspective, schizophrenia is considered an adaptation phenomenon influenced by leaving the familiar and safe home to a stressful environment and building new social networks during migration [51]. Male schizophrenia pathogenesis has a reduced reproduction rate compared to the non-affected individuals [52]. This might be the possible cause of significant selection signals in Kho individuals across the few male fertility spermatogenesis-associated genes, such as PMCH involved in spermatocyte differentiation, and DHX32, ASTL, and CYP3A5 involved in testosterone biosynthesis [53].
Moreover, regarding the migratory events across the region/continents, changing language trajectories cause language disorder in children after ages 5–6 years [54]. Consequently, detecting selection signature at expressive language disorder associated gene, i.e., SRCAP [55], strengthens the evidence of cross-continent migration events in the Kho population. The GTF2IRD2 and POTE family of genes were identified as top selection candidates. However, these genes are reported to be recently evolved in primates via duplication events [26] and therefore may not reflect any Kho demography-specific adaptation.
Conclusion
With our work, we elucidated putative genetic origins of the Kho ethnic minority living in remote Chitral Valley of north-western Pakistan. The Kho exhibit cross-continental admixture signal of steppe immigration of Southern Siberia to South Asian region. Together with the Burusho and Balti they also share a unique additional wave of East Asian ancestry admixture possibly during expansion of the Tibetan Empire during the first millennium CE. The Kho share the Middle-Late Bronze Age ancestry with the neighboring Kalash, an enigmatic isolated population of South Asia. We highlighted several genes as candidates of natural selection in the Kho population that may implicate in diseases etiology and adaptation to the local environment.
References
Yelmen B, Mondal M, Marnetto D, Pathak AK, Montinaro F, Gallego Romero I, et al. Ancestry-specific analyses reveal differential demographic histories and opposite selective pressures in modern South Asian populations. Mol Biol Evol. 2019;36:1628–42.
Majumder PP. The human genetic history of South Asia. Curr Biol. 2010;20:R184–R187.
Liljegren H, & Akhunzada F. Linguistic diversity, vitality and maintenance: a case study on the language situation in northern Pakistan. Multiethnica. Meddelande från Centrum för multietnisk forskning, Uppsala universitet, 2017; 61–79.
de Barros Damgaard P, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374.
Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, et al. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487.
Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012.
GenomeAsia100K Consortium. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019;576:106.
Ayub Q, Mezzavilla M, Pagani L, Haber M, Mohyuddin A, Khaliq S, et al. The Kalash genetic isolate: ancient divergence, drift, and selection. Am J Hum Genet. 2015;96:775–83.
Heegard Petersen J. Kalasha texts–With introductory grammar. Acta Linguistica Hafniensia. 2015;47:1–275. sup1
Morch IE. How fast will a langauge die when it is officially no longer spoken? Odense Working Papers Language Communications. 2000;2:125–34.
Hemphill BE, Ali I, Blaylock S, Willits N. Are the Kho an indigenous population of the Hindu Kush?: A dental morphometric approach. In: Tosi M, Frenez D (eds.) South Asian Archaeology 2007. Oxford: Archaeopress-BAR; 2013.
Aziz S, Nawaz M, Afridi SG, Khan A. Genetic structure of Kho population from north-western Pakistan based on mtDNA control region sequences. Genetica. 2019;147:177–83.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443.
Wang, CC, Yeh HY, Popov AN, Zhang HQ, Matsumura H, Sirak K, et al. Genomic insights into the formation of human populations in East Asia. Nature. 2021;591:413–9.
Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93.
Wang C, Zhan X, Bragg-Gresham J, Kang HM, Stambolian D, Chew EY, et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet. 2014;46:409–15.
Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64.
Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–54.
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68.
Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, et al. A genomic history of Aboriginal Australia. Nature. 2016;538:207–14.
Feng Q, Lu D, Xu S. AncestryPainter: a graphic program for displaying ancestry composition of populations and individuals. Genom Proteom Bioinform. 2018;16:382–5.
Song S, Tian D, Li C, Tang B, Dong L, Xiao J, et al. Genome Variation Map: a data repository of genome variations in BIG Data Center. Nucleic Acids Res. 2018;46:D944–D949.
BIG Data Center Members. Database resources of the BIG data center in 2019. Nucleic Acids Res. 2019;47:D8. Database issue
Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78.
Nowick K, Stubbs L. Lineage-specific transcription factors and the evolution of gene regulatory networks. Brief Funct Genom. 2010;9:65–78.
Alekseev S, Luijsterburg MS, Pines A, Geverts B, Mari PO, Giglia-Mari G, et al. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol Cell Biol. 2008;28:7402–13.
Shan X, Wang K, Tong X, Wang Z, Wu F, Liu X, et al. High expression of VAT1 is a prognostic biomarker and predicts malignancy in glioblastoma. Oncol Rep. 2019;42:1422–30.
Aristizábal, B, & González, Á. Innate immune system. In: Autoimmunity: from bench to bedside [Internet]. El Rosario University Press, 2013.
Weng CY, Hu XY, Wang YJ. Integrated analysis of gene expression, alteration and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 1 in cancer. 3 Biotech. 2020;10:1–19.
Zeron-Medina J, Wang X, Repapi E, Campbell MR, Su D, Castro-Giner F, et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013;155:410–22.
Gao N, Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 2020;20:1–12.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2018;47:D941–D947.
Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–94.
Yang XY, Rakha A, Chen W, Hou J, Qi XB, Shen QK, et al. Tracing the genetic legacy of the tibetan empire in the Balti. Mol Biol Evol. 2021;38:1529–36.
Vincent AS. The early history of India, Vol. II - from the sixth century B.C. to the Mohammedan conquest, including the invasion of Alexander the Great. The Gupta Empire and the White Huns. Chapter 12. Oxford: Clarendon Press; 1906. p. Page 275–92.
de Barros Damgaard P, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, et al. The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 2018;360:eaar7711.
Kroonen G, Barjamovic G, Peyrot M. Linguistic supplement to Damgaard et al. 2018: Early Indo-European languages, Anatolian, Tocharian and Indo-Iranian. Zenodo. 2018. https://doi.org/10.5281/zenodo.1240524.
Terkawi MA, Takano R, Furukawa A, Murakoshi F, Kato K. Involvement of β-defensin 130 (DEFB130) in the macrophage microbicidal mechanisms for killing Plasmodium falciparum. Sci Rep. 2017;7:1–11.
McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, et al. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17:58–71.
Fei C, Atterby C, Edqvist PH, Pontén F, Zhang WW, Larsson E, et al. Detection of the human endogenous retrovirus ERV3-encoded Env-protein in human tissues using antibody-based proteomics. J R Soc Med. 2014;107:22–29.
Miller JL, Dewet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, et al. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17.
ur Rehman I, Ishaq M, Ali L, Khan S, Ahmad I, Din IU, et al. Enrichment, spatial distribution of potential ecological and human health risk assessment via toxic metals in soil and surface water ingestion in the vicinity of Sewakht mines, district Chitral, Northern Pakistan. Ecotoxicol Environ Saf. 2018;154:127–36.
Fohlin H, Bekkhus T, Sandström J, Fornander T, Nordenskjöld B, Carstensen J, et al. Low RAB6C expression is a predictor of tamoxifen benefit in estrogen receptor positive/progesterone receptor negative breast cancer. Mol Clin Oncol. 2020;12:415–20.
Coppa A, Nicolussi A, D’Inzeo S, Capalbo C, Belardinilli F, Colicchia V, et al. Optimizing the identification of risk‐relevant mutations by multigene panel testing in selected hereditary breast/ovarian cancer families. Cancer Med. 2018;7:46–55.
Huang C, Li Y, Li Z, Xu Y, Li N, Ge Y, et al. LIMS1 promotes pancreatic cancer cell survival under oxygen-glucose deprivation conditions by enhancing HIF1A protein translation. Clin Cancer Res. 2019;25:4091–103. https://doi.org/10.1158/1078-0432.CCR-18-353
Fu DJ, Thomson C, Lunny DP, Dopping-Hepenstal PJ, McGrath JA, Smith FJ, et al. Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J Investig Dermatol. 2014;134:754–63.
Khan I, Maldonado E, Vasconcelos V, O’Brien SJ, Johnson WE, Antunes A. Mammalian keratin associated proteins (KRTAPs) subgenomes: disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genom. 2014;15:1–19.
Yoon SJ, Seiler SH, Kucherlapati R, Leinwand L. Organization of the human skeletal myosin heavy chain gene cluster. Proc Natl Acad Sci USA. 1992;89:12078–82.
Greenwood TA, Lazzeroni LC, Maihofer AX, Swerdlow NR, Calkins ME, Freedman R, et al. Genome-wide association of endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia (COGS) study. JAMA Psychiatry. 2019;76:1274–84.
Scheepers FE, De Mul J, Boer F, Hoogendijk WJ. Psychosis as an evolutionary adaptive mechanism to changing environments. Front Psychiatry. 2018;9:237.
MacCabe JH, Koupil I, Leon DA. Lifetime reproductive output over two generations in patients with psychosis and their unaffected siblings: the Uppsala 1915-1929 birth cohort multigenerational study. Psychol Med. 2009;39:1667–76.
Vaarala MH, Mattila H, Ohtonen P, Tammela TL, Paavonen TK, et al. The interaction of CYP3A5 polymorphisms along the androgen metabolism pathway in prostate cancer. Int J Cancer Res. 2008;122:2511–6.
Norbury CF, Vamvakas G, Gooch D, Baird G, Charman T, Simonoff E, et al. Language growth in children with heterogeneous language disorders: a population study. J Child Psychol Psychiatry. 2017;58:1092–105.
Nogueira E, Garma C, Lobo C, Del Olmo B, Arroyo JM, et al. Severe developmental expressive language disorder due to a frameshift mutation in exon 18 of SRCAP gene, far away from the mutational hotspot in exons 33 and 34 associated to the Floating-Harbor syndrome. Neurol Sci. 2021;42:4349–52.
Acknowledgements
The authors acknowledge the Kho people from Chitral, Pakistan for their active participation in the study and blood samples donation.
Funding
This work was supported by the Scientific and Technology Committee of Shanghai Municipality (18490750300), Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and the 111 Project (B13016). AK acknowledges the Higher Education Commission (HEC), Pakistan for grant support (NRPU-7438).
Author information
Authors and Affiliations
Contributions
AK and S-QW conceived the research idea. SA and AK collected the samples. AK, LV, HK, KZ, and KN performed data analyses. L-XW and S-QW contributed in array based genotype data generation. QA and LP supervised the data analyses. AK, LP, QA, and SA wrote the manuscript basic draft. S-QW supervised the study and contributed in manuscript revisions..
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study has been approved by the Ethical Review Committee of the Abdul Wali Khan University Mardan, Pakistan under the AWKUM/Biochem/ERC/2018/574.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Khan, A., Vallini, L., Aziz, S. et al. Cross-continental admixture in the Kho population from northwest Pakistan. Eur J Hum Genet 30, 740–746 (2022). https://doi.org/10.1038/s41431-022-01057-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01057-2
This article is cited by
-
Whatâs new in genetics in June 2022?
European Journal of Human Genetics (2022)