Abstract
Myotonic dystrophy type 1 (DM1) is an autosomal dominant muscular dystrophy that results from a CTG expansion (50–4000 copies) in the 3′ UTR of the DMPK gene. The disease is classified into four or five somewhat overlapping forms, which incompletely correlate with expansion size in somatic cells of patients. With rare exception, it is affected mothers who transmit the congenital (CDM1) and most severe form of the disease. Why CDM1 is hardly ever transmitted by fathers remains unknown. One model to explain the almost exclusive transmission of CDM1 by affected mothers suggests a selection against hypermethylated large expansions in the germline of male patients. By assessing DNA methylation upstream to the CTG expansion in motile sperm cells of four DM1 patients, together with availability of human embryonic stem cell (hESCs) lines with paternally inherited hypermethylated expansions, we exclude the possibility that DMPK hypermethylation leads to selection against viable sperm cells (as indicated by motility) in DM1 patients.
Similar content being viewed by others
Introduction
Myotonic dystrophy type 1 [DM1, (OMIM 160900)] is an autosomal dominant muscular dystrophy that results from a trinucleotide CTG repeat expansion (50–4000 copies) in the 3′ UTR of the dystrophia myotonica protein kinase gene (DMPK) [1, 2]. When the CTG tract expands beyond 300 repeats, they lead to abnormal CpG methylation next to the repeats in a way that is age and tissue dependent [3, 4]. The disease is classified into four [5] or five [6] somewhat overlapping forms, which incompletely correlate with expansion size in somatic cells of patients. With rare exception, it is affected mothers who transmit the congenital (CDM1 >1000 CTGs) and most severe form of the disease [7,8,9].
Why CDM1 is hardly ever transmitted by fathers remains unresolved. One potential explanation would be frequent oligo/azoospermia among male patients with the adult form of the disease leading to reduced fecundity [10]. This may be in addition to behavioral changes that often result in male patients less frequently acquiring partners in particular populations [11]. However, both factors are not sufficient to explain the acute tendency toward maternal CDM1 transmissions. A different explanation for the underrepresentation of paternal CDM1 cases may be the absence of very large expansions in the sperm of DM1 males. Indeed, while exceedingly small mutations are prone to further expand in the male germline, larger mutations (>80 repeats) often contract in a way that negatively correlates with expansion size [12]. Hence, although the role of male transmission is to trigger repeat instability at low mutation range (as exemplified by excess of transmitting grandfathers among children with the congenital form of the disease [13]), for larger mutations it most likely limits allele length by yet to be identified factors/mechanism(s).
Previously, Barbe et al. reported rigorous data concerning a strong correlation between aberrant methylation immediately upstream to the CTG repeats [CTCF1 site], and maternal transmission of CDM1 by the analysis of a large cohort of DM1 patients [14]. In light of their findings, a model that relies on the systematic elimination of particularly large mutations by hypermethylation upstream to the repeats in the male germline was proposed to explain the unresolved question of why CDM1 is almost always transmitted by affected mothers [14]. This model is especially attractive since it parallels the observed involvement of a similar mechanism in Fragile X syndrome [4, 15,16,17].
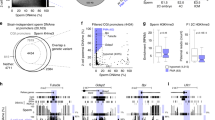
To test this hypothesis and determine the timing of DMPK hypermethylation in DM1, we tested sperm from four male patients undergoing preimplantation genetic diagnosis (PGD) procedures for DM1 (Table 1, IRB 110/11 followed by IRB 90/17) (for methods see Supplementary Material). Sperm isolation was carried out by a two-layer gradient system which is designed to guarantee that the sample is predominately composed of motile sperm with normal morphology. Methylation analysis was carried out on residual sperm samples by DNA bisulfite massive parallel sequencing (pyrosequencing). In this set of experiments, CTG repeat allele size could not be determined simultaneously with methylation status because residual sperm cells were available to us only in small quantities. Nevertheless, we assayed methylation levels in a restricted region located 650 bp upstream to the CTG repeats (13 CpG sites) (see Fig. 1A) which was formerly identified as a disease associated DMR (Differentially Methylated Region) that becomes hypermethylated in a way that best correlates with the inherited size of the mutation (progenitor allele length) and putatively acts as a regulatory element for SIX5 transcription [4]. In addition, because methylation systematically and uninterruptedly spreads from the upstream flanking region toward the repeats, methylation status 650 bp upstream to the CTGs (referred to as region E in ref. [4]) could be directly inferred from, and thus compared to, alleles that are heavily methylated at the CTCF1 binding site (the region analyzed by Barbe et al.). Hence, we aimed our methylation analysis at region E and found abnormal methylation levels of 13% and 30% (corresponding to 26% and 60% on the background of the expanded allele) in viable sperm cells of patients A and I, respectively (see Table 1 and Fig. 1B). Taking advantage of an informative SNP which resides within the DMR sequence (rs635299), we further validated the results in patient A by bisulfite allele-specific colony DNA sequencing, demonstrating methylation levels of nearly 30% exclusive to the mutant allele (Fig. 1C, altogether 26 CpG sites, bisulfite conversion rate nearly 100%). Of note, methylation levels among these patients did not correlate with sperm quality (Table 1). In addition, it should be mentioned that all four patients are now fathers to at least two healthy children following PGD procedures.
A Graphic illustration of the human DM1 locus, including DMWD, DMPK, and SIX5 genes. Indicated in detail the differentially methylated region (DMR) upstream to the CTG repeats. Dashed line in orange designates the amplicon that was used for the methylation analysis in this study covering SNP rs635299 (G/T), while dashed line in gray designates the amplicon that was used for the methylation analysis in Barbe et al. [14]. CTCF1 binding site is designated by green ellipse. The numbering represents chromosome 19 positioning according to the hg19 human genome assembly. B Absolute methylation levels in region E (488–777 bp upstream of the CTGs), as determined by bisulfite pyrosequencing. The 13 CpG sites tested are indicated by shaded squares according to methylation levels (see colored legend). Mean absolute methylation levels across all tested CpG positions are indicated to the right in each line. C Allele-specific bisulfite sequencing of the expanded allele (CTGEXP) within the differentially methylated region of DMPK (region E in [4]), overlapping SNP2 (according to [4]), in DNA extracted from sperm sample of patient A. Full circles correspond to methylated CpGs, whereas empty circles represent unmethylated CpGs. Methylation levels (%) were determined by the analysis of 20 molecules by bisulfite colony sequencing. Methylation levels (29%) are indicated at the bottom.
Although we analyzed a different region from Barbe et al. a region located a bit further upstream to the CTG repeats, our findings are consistent with upstream methylation not affecting spermatogonial stem cell death and/or the production of nonviable sperm cells, thus narrowing the timing of selection, if at all, to not before fertilization. On the contrary, it is apparent that paternal hypermethylated mutations are compatible with embryo viability, at least until the embryo implantation stage. This stems from the availability of a considerable number of hESC lines which we established from affected embryos with a paternally inherited mutation (some derived from patient A in Table 1) that are methylated along the entire region upstream to the repeats, including the region that is designated as a CTCF binding site (referred to CTCF1 in ref. [14] or region F in ref. [4]). Importantly, in DM1 hESCs, methylation levels and spreading are totally dependent on expansion size and not parental origin of the mutation, provided that the number of the repeats is >300 [4]. The larger the mutation, the farther methylation spreads from intron 13 of DMPK toward the CTGs. Yet, considering that all of the hESC lines were established from intracytoplasmic sperm injection (ICSI)-derived embryos (as is regularly carried out in all PGD cycles), we may have bypassed a natural barrier while producing these embryos by assisted reproduction techniques (ART) i.e.,; the failure of mutant hypermethylated mature sperm cells to fertilize an oocyte or to trigger embryonic genome activation after penetrating the egg [18].
Therefore, all things considered, the data presented here suggest that the stem/mature sperm cell selection model is one of the following: (i) incorrect, (ii) leaky, and (iii) correct except that haploid sperm containing methylated expansions are fertilization incompetent or are unable to trigger embryonic genome activation because of methylation, expansion size or some other yet identified paternal factor. Although the dynamics of hypermethylation in the early embryo may be a bit different so that hypermethylation is first erased and then reestablished (as opposed to directly inherited from the sperm), this should not conflict with the claim that the suggested model for sperm cell selection by upstream methylation is most likely incorrect. Alternative models for maternal transmission of CDM1 by sperm cell selection should be sought such as, but not limited to, differences in the activity level of mismatch proteins [19] or other repeat length controlling modifiers among male and female germlines.
For further investigation, we suggest to extend this study, which was limited to a very small number of individuals, to a much larger cohort of patients. In addition, progenitor mutations in DMPK should preferably be classified by size, age and purity of the repeats to better substantiate our findings. Moreover, it would be informative to follow the dynamics of hypermethylation along with expansion size during other stages of spermatogenesis. Furthermore, assessment of the competence of sperm cells with methylated alleles to fertilize an egg without the involvement of ART is warranted in order to gain more insight into the timing and mechanisms underlying these biological events.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385.
Aslanidis C, Jansen G, Amemiya C, Shutler G, Mahadevan M, Tsilfidis C, et al. Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature. 1992;355:548–51.
López Castel A, Nakamori M, Tomé S, Chitayat D, Gourdon G, Thornton CA, et al. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet. 2011;20:1–15.
Yanovsky-Dagan S, Avitzour M, Altarescu G, Renbaum P, Eldar-Geva T, Schonberger O, et al. Uncovering the role of hypermethylation by CTG expansion in myotonic dystrophy Type 1 using mutant human embryonic stem cells. Stem Cell Rep. 2015;5:221–31.
Hecht BK, Donnelly A, Gedeon AK, Byard RW, Haan EA, Mulley JC. Direct molecular diagnosis of myotonic dystrophy. Clin Genet. 1993;43:276–85.
De Antonio M, Dogan C, Hamroun D, Mati M, Zerrouki S, Eymard B, et al. Unravelling the myotonic dystrophy type 1 clinical spectrum: a systematic registry-based study with implications for disease classification. Rev Neurol. 2016;172:572–80.
Lavedan C, Hofmann-Radvanyi H, Rabes JP, Roume J, Junien C. Different sex-dependent constraints in CTG length variation as explanation for congenital myotonic dystrophy. Lancet. 1993;341:237.
Harley HG, Rundle SA, MacMillan JC, Myring J, Brook JD, Crow S, et al. Size of the unstable CTG repeat sequence in relation to phenotype and parental transmission in myotonic dystrophy. Am J Hum Genet. 1993;52:1164–74.
Harper PS, Dyken PR. Early-onset dystrophia myotonica. Evidence supporting a maternal environmental factor. Lancet. 1972;2:53–5.
Puy V, Mayeur A, Levy A, Hesters L, Raad J, Monnot S, et al. CTG Expansion in the DMPK gene: semen quality assessment and outcome of preimplantation genetic diagnosis. J Clin Endocrinol Metab. 2020;105:1137–44.
Veillette S, Perron M, Mathieu J. Myotonic dystrophy: II. Marital status, fertility and gene transmission. Can J Neurol Sci. 1989;16:114–8.
Martorell L, Gámez J, Cayuela ML, Gould FK, McAbney JP, Ashizawa T, et al. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology. 2004;62:269–74.
López de Munain A, Cobo AM, Poza JJ, Navarrete D, Martorell L, Palau F, et al. Influence of the sex of the transmitting grandparent in congenital myotonic dystrophy. J Med Genet. 1995;32:689–91.
Barbé L, Lanni S, López-Castel A, Franck S, Spits C, Keymolen K, et al. CpG methylation, a parent-of-origin effect for maternal-biased transmission of congenital myotonic dystrophy. Am J Hum Genet. 2017;100:488–505.
Sun YJ, Baumer A. Nonrandom X inactivation and selection of fragile X full mutation in fetal fibroblasts. Am J Med Genet. 1999;86:162–4.
Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, et al. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet. 1997;15:165–9.
Salat U, Bardoni B, Wöhrle D, Steinbach P. Increase of FMRP expression, raised levels of FMR1 mRNA, and clonal selection in proliferating cells with unmethylated fragile X repeat expansions: a clue to the sex bias in the transmission of full mutations? J Med Genet. 2000;37:842–50.
Schulz KN, Harrison MM. Mechanisms regulating zygotic genome activation. Nat Rev Genet. 2019;20:221–34.
Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol Cell Biol. 2004;24:629–37.
Mangin A, de Pontual L, Tsai YC, Monteil L, Nizon M, Boisseau P, et al. Robust detection of somatic mosaicism and repeat interruptions by long-read targeted sequencing in myotonic dystrophy Type 1. Int J Mol Sci. 2021;22:2616.
Acknowledgements
We would like to thank Dr. David Zeevi for the critical reading of the paper and Prof. Genevieve Gourdon for assisting with determining mutation size by PCR in patient D.
Funding
This work was partly supported by the Ministry of Science and Technology (MOST) Israel (3-17372, RE) and the kind donation of Rabbi David Fuld.
Author information
Authors and Affiliations
Contributions
SYD and SEL contributed to the conception and design of the study, the collection and assembly of the data, data analysis and interpretation, and paper writing. EC and PM contributed to the assembly of the data. OS, TEG, and GA contributed to the collection of data. RE contributed to the conception and design of the study, financial support, data analysis and interpretation, and paper writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethics approval for this study was provided by the Shaare Zedek Medical Center Ethics Committee (IRB 110/11 followed by IRB 90/17) under the strict guidelines and regulations outlined by the Israeli National Ethics Committee. A written informed consent was obtained for the donation of residual sperm cells after ICSI procedure from all patients.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yanovsky-Dagan, S., Cohen, E., Megalli, P. et al. DMPK hypermethylation in sperm cells of myotonic dystrophy type 1 patients. Eur J Hum Genet 30, 980–983 (2022). https://doi.org/10.1038/s41431-021-00999-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00999-3
This article is cited by
-
Exome sequencing—one test to rule them all?
European Journal of Human Genetics (2022)