Abstract
Mitochondrial myopathies are a heterogeneous group of disorders associated with a wide range of clinical phenotypes. We present a 16-year-old girl with a history of exercise intolerance since childhood. Acylcarnitine species suggestive of multiple acyl-CoA dehydrogenase deficiency were found in serum, however genetic analysis did not reveal variants in genes associated with this disorder. Biochemical analyses of skeletal muscle mitochondria revealed an isolated and extremely low activity of cytochrome c oxidase (COX). This finding was confirmed by enzyme histochemistry, which demonstrated an almost complete absence of fibers with normal COX activity. Whole-exome sequencing revealed a single base-pair deletion (m.8088delT) in MT-CO2, which encodes subunit 2 of COX, resulting in a premature stop codon. Restriction fragment length polymorphism-analysis confirmed mtDNA heteroplasmy with high mutant load in skeletal muscle, the only clinically affected tissue, but low levels in other investigated tissues. Single muscle fiber analysis showed segregation of the mutant genotype with respiratory chain dysfunction. Immuno-histochemical studies indicated that the truncating variant in COX2 has an inhibitory effect on the assembly of the COX holoenzyme.
Similar content being viewed by others
Introduction
Mitochondrial disorders constitute a large, heterogeneous group of disorders with molecular defects affecting the mitochondrial respiratory chain. Complex IV (cytochrome c oxidase, COX) is composed of 13 subunits, of which three form the catalytic core of the enzyme and are encoded by the mitochondrial genome. Disease-associated variants have been identified in all three of the mtDNA-encoded subunits presenting with highly variable clinical phenotypes, whereas only a few disease-associated variants in the ten nuclear genes have been identified [1,2,3,4].
To date, about ten disease-associated variants in MT-CO2 have been reported, associated with a variety of more or less severe phenotypes, such as MELAS [5], Alpers-Huttenlocher-like disease [6], myopathy [7], myalgia and myoglobinuria [8, 9], non-syndromic mitochondrial encephalomyopathy [10,11,12], and severe lactic acidosis [13].
We present clinical, histological, biochemical, and genetic findings in a patient carrying a novel heteroplasmic variant in MT-CO2, leading to truncation of COX2, and affecting the assembly of the COX holoenzyme.
Subject
This 16-year-old girl with normal early development has experienced exercise intolerance since childhood. She was first referred to the pediatric cardiologist at the age of 14 because of exercise intolerance. Initial cardiac evaluation was unremarkable. Due to persistent symptoms of exercise intolerance and fatigue, another cardiac evaluation was performed 1.5 years later, including an exercise stress test. The stress test was terminated after 3 min due to exercise intolerance. The metabolic work-up directly after the exercise stress test showed hyperlactatemia, slightly increased transaminases and mild anemia. Further laboratory investigations showed slightly increased resting P-lactate and normal resting creatine kinase in serum (S-CK). Urine and serum creatinine, cerebrospinal fluid/plasma glucose and albumin ratios were also normal. Organic acids in urine showed an increased excretion of lactate and 2-hydroxyglutarate. Urine and serum myoglobin were not measured.
Neuromuscular evaluation disclosed muscle weakness predominantly of the proximal muscles and exercise intolerance characterized by breathlessness and tachycardia. The results of the 6-min walk test were below the 3rd percentile, with exercise intolerance observed already after the first minute.
The patient was then referred for mitochondrial investigation, In the meanwhile, initial metabolic work-up revealed a serum acylcarnitine profile suggestive of multiple acyl-CoA dehydrogenase deficiency (Supplementary Figure 1). The patient was treated with riboflavin 300 mg daily for ~1 month without effect.
Methods
Standard morphological and enzyme histochemical analyses of diagnostic fresh frozen skeletal muscle biopsy were performed according to established protocols [14]. Immunohistochemical analysis of Complex IV was performed with antibodies directed against the three mtDNA encoded subunits (COX1-3) and the nuclear-encoded COX4 (Supplementary Table 1). An antibody to VDAC1 served as a mitochondrial marker. Isolation of skeletal muscle mitochondria, oximetric measurements of fresh mitochondria and spectrophotometric analyses were performed essentially as described previously [15]. Single COX-deficient and normal muscle fibers were dissected to analyze the mutant load using a tungsten needle as described previously [16].
Whole-exome sequencing was performed on total DNA extracted from skeletal muscle tissue using the Sure SelectXT Human All Exon kit version 6 (Agilent Technologies, Santa Clara, CA, USA) and sequenced on the HiSeq2500 platform (Illumina, San Diego, CA, USA) as paired-end reads. Base calling was performed with the Illumina pipeline. Sequence reads were aligned to the reference genome (hg19) and variant calling was performed using the CLC Biomedical Genomics workbench (Qiagen). Filtering of called variants was performed using Ingenuity Variant Analysis (www.ingenuity.com/products/variant-analysis). Variants predicted to affect normal protein function in nuclear genes potentially associated with mitochondrial disease and the entire mtDNA were analyzed. Variants of interest were further evaluated using in silico prediction tools: PhyloP, SIFT, PolyPhen-2, and MutationTaster.
Results
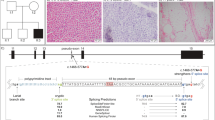
The muscle biopsy demonstrated profound COX deficiency with only a few scattered fibers with positive COX staining and multiple fibers with subsarcolemmal accumulation of mitochondria (Fig. 1a–d). Lipid accumulation was seen in many fibers (Fig. 1f). Ultrastructural analysis revealed mitochondrial accumulation and mitochondria of slightly abnormal size and shape (Fig. 1g, h).
Morphological features of the vastus lateralis muscle from the patient. a Muscle architecture is relatively normal (Hematoxylin & Eosin). b Cytochrome c oxidase (COX) enzyme histochemistry shows profound deficiency in the majority of the fibers. c COX/succinate dehydrogenase (SDH)-double staining shows only scattered muscle fibers with normal COX activity. d SDH enzyme histochemistry demonstrates mitochondrial proliferation. e Several fibers with mitochondrial proliferation may be classified as ragged-red fibers (Gomori trichrome). f Vacuoles containing lipid deposits are observed in many fibers (Sudan black). g and h On electron microscopy, the accumulation of mitochondria is clearly seen and they show slightly abnormal size and shape. Scale bar a–f, 20 μm
Biochemical analyses of the respiratory chain enzyme complexes revealed an isolated and extremely low activity of cytochrome-c oxidase (Supplementary Table 2).
Only COX-positive cells expressed normal immunoreactivity for COX subunits 1–4. Expression of COX4 was preserved in COX-negative cells, albeit to a lesser extent than in COX-positive cells. There was no difference in the protein expression of VDAC1 between normal and COX-deficient cells (Fig. 2).
Serial sections of the patient’s skeletal muscle. COX/SDH enzyme histochemistry with fibers lacking COX activity in blue. Immunohistochemistry of the three mtDNA encoded subunits and the nuclear encoded subunit 4 of COX demonstrate that only COX-positive cells express normal immunoreactivity for COX subunits. Expression of COX4 is present in COX-negative cells, albeit to a lesser extent. There is no difference in the protein expression of VDAC1, serving as a marker for mitochondrial mass, between normal and COX-deficient cells. Asterisks marking identical myofibers. Insets show staining of an age-matched control. Scale bar, 50 μm
Whole-exome sequencing revealed a near homoplasmic single base pair deletion (m.8088delT) in MT-CO2 (ClinVar Accession nr SCV000678293, NC_012920.1, https://www.ncbi.nlm.nih.gov/clinvar/variation/488349/), resulting in a premature stop codon, p.(Leu168*). This was confirmed by Sanger sequencing (Fig. 3a).
a Sanger sequencing demonstrating high levels of a single base pair deletion (m.8088delT) in the MT-CO2 gene, which encodes subunit 2 of COX, in muscle homogenate from the patient, resulting in a premature stop codon. b The proportion of mutated DNA as determined by PCR and restriction fragment length polymorphism in single muscle fibers using a mismatch reverse primer to introduce a recognition site for the restriction endonuclease MslI. Primer sequences are available upon request. The median mutation load in COX-negative muscle fibers (100%, n = 14) is significantly higher than in COX-positive cells (40%, n = 14, P < 0.05, Mann–Whitney U-test)
The proportion of mutated DNA as determined by PCR-RFLP was 96% in muscle homogenate, and between 2 and 15% in other examined tissues (hair follicles 2%, blood 5%, fibroblasts 6%, buccal mucosa 12%, and urinary epithelial cells 15%). The median mutational load in COX-deficient muscle fibers was significantly higher than in COX-positive cells (Fig. 3b). The variant was not detected in the mother’s blood, urinary epithelial cells, buccal mucosa, or hair roots, suggesting that the variant had arisen de novo.
Discussion
We describe a new variant in MT-CO2, m.8088delT, changing a leucine at position 168 to a premature stop codon, in a patient with exercise intolerance and proximal muscle weakness. The variant was absent in the asymptomatic mother, suggesting that it had appeared de novo.
The pathogenicity of the m.8088delT variant is supported by several lines of evidence. First, the variant is heteroplasmic and present at high levels in skeletal muscle, the only clinically affected tissue. The mutant load was below a level needed to cause symptoms in other examined tissues [17]. Second, the variant is predicted to lead to a premature stop codon in the gene encoding subunit 2 of COX and therefore affect COX activity, which was indeed demonstrated by biochemical and enzyme histochemical investigations. Third, single fiber analysis showed that almost all COX-negative fibers only contained mutant DNA, while the median mutant load was 40% in biochemically normal fibers, demonstrating segregation of the m.8088delT variant with respiratory chain dysfunction. Fourth, immunohistochemistry demonstrated loss of the COX2 protein.
Studies on assembly of the COX holoenzyme have identified COX1 as the scaffold around which the other subunits are sequentially assembled. Several intermediates have been identified, the first of which contains COX1 and COX4. COX2 and COX3 are then added in an ordered manner, followed by the other nuclear-encoded subunits to complete the formation of the holoenzyme complex [18]. Immunohistochemical analysis in our patient shows reduced immunoreactivity for COX2, as well as for other COX subunits in COX-deficient fibers, suggesting that the truncating variant has an inhibitory effect on the assembly of the COX holoenzyme. In a previously described patient, a missense variant in MT-CO2 resulted in a similar clinical phenotype and biochemical defect as our patient [7]. This m.7671T>A variant caused a severe reduction in COX2 and COX3, and a milder reduction in COX1 and COX4. A similar pattern of COX subunit destabilization has also been reported in a patient with MELAS caused by a m.7630delT variant in MT-CO2 [5].
In disorders of fatty acid oxidation, acylcarnitine species accumulate and are released into the circulation. The pattern of these species can be diagnostic for specific disorders. Respiratory chain disorders and fatty acid oxidation disorders may share phenotypic features. At the same time fatty acid oxidation disorders also present with secondary mitochondrial dysfunction due to disrupted energy and redox mitochondrial homeostasis, making the differential diagnosis even more challenging [19]. Our patient exhibited an acylcarnitine profile with increased levels of multiple acylcarnitine species, which was suggestive of multiple acyl-CoA dehydrogenase deficiency, but lacked disease-associated variants in any of the three disease-causing genes (ETFA, ETFB, and ETFDH). A similar pattern of a deranged acylcarnitine profile has previously been described in an adult patient with complex IV deficiency, myalgia and exercise intolerance associated with a heteroplasmic single-base duplication in the MT-CO2 gene [9].
In conclusion, COX deficiency can present with a plethora of phenotypes, with relatively poor genotype–phenotype correlation. We present a patient with an isolated and extremely low COX activity harboring a MT-CO2-variant causing premature termination of translation of COX2 and affecting assembly of the COX holoenzyme. Despite this, the patient’s phenotype was relatively mild, with the variant being present at high levels only in muscle, supporting the concept that the mutant load in different tissues is predictive for the phenotype severity. Furthermore, our findings demonstrate that COX2 deficiency may cause a perturbed acylcarnitine profile mimicking multiple acyl-CoA dehydrogenase deficiency. The mechanism by which COX2 deficiency causes increased acylcarnitine species remains unknown and warrants further studies.
References
Abdulhag UN, Soiferman D, Schueler-Furman O, et al. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur J Hum Genet. 2015;23:159–64.
Rak M, Benit P, Chretien D, et al. Mitochondrial cytochrome c oxidase deficiency. Clin Sci. 2016;130:393–407.
Hallmann K, Kudin AP, Zsurka G, et al. Loss of the smallest subunit of cytochrome c oxidase, COX8A, causes Leigh-like syndrome and epilepsy. Brain. 2016;139:338–45.
Shteyer E, Saada A, Shaag A, et al. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am J Hum Genet. 2009;84:412–7.
Rossmanith W, Freilinger M, Roka J, et al. Isolated cytochrome c oxidase deficiency as a cause of MELAS. J Med Genet. 2008;45:117–21.
Uusimaa J, Finnila S, Vainionpaa L, et al. A mutation in mitochondrial DNA-encoded cytochrome c oxidase II gene in a child with Alpers-Huttenlocher-like disease. Pediatrics. 2003;111:e262–268.
Rahman S, Taanman JW, Cooper JM, et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am J Hum Genet. 1999;65:1030–9.
McFarland R, Taylor RW, Chinnery PF, Howell N, Turnbull DM. A novel sporadic mutation in cytochrome c oxidase subunit II as a cause of rhabdomyolysis. Neuromuscul Disord. 2004;14:162–6.
Vissing CR, Duno M, Olesen JH, et al. Recurrent myoglobinuria and deranged acylcarnitines due to a mutation in the mtDNA MT-CO2 gene. Neurology. 2013;80:1908–10.
Horvath R, Schoser BG, Muller-Hocker J, Volpel M, Jaksch M, Lochmuller H. Mutations in mtDNA-encoded cytochrome c oxidase subunit genes causing isolated myopathy or severe encephalomyopathy. Neuromuscul Disord. 2005;15:851–7.
Clark KM, Taylor RW, Johnson MA, et al. An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am J Hum Genet. 1999;64:1330–9.
Campos Y, Garcia-Redondo A, Fernandez-Moreno MA, et al. Early-onset multisystem mitochondrial disorder caused by a nonsense mutation in the mitochondrial DNA cytochrome C oxidase II gene. Ann Neurol. 2001;50:409–13.
Wong LJ, Dai P, Tan D, et al. Severe lactic acidosis caused by a novel frame-shift mutation in mitochondrial-encoded cytochrome c oxidase subunit II. Am J Med Genet. 2001;102:95–99.
Dubowitz V, Sewry CA, Oldfors A. Muscle Biopsy: A Practical Approach. Elsevier; Philadelphia, PA: 2013:1–592.
Tulinius MH, Holme E, Kristiansson B, Larsson NG, Oldfors A. Mitochondrial encephalomyopathies in childhood. I. Biochemical and morphologic investigations. J Pediatr. 1991;119:242–50.
Roos S, Lindgren U, Ehrstedt C, Moslemi AR, Oldfors A. Mitochondrial DNA depletion in single fibers in a patient with novel TK2 mutations. Neuromuscul Disord. 2014;24:713–20.
Hellebrekers DM, Wolfe R, Hendrickx AT, et al. PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Hum Reprod Update. 2012;18:341–9.
Shoubridge EA. Cytochrome c oxidase deficiency. Am J Med Genet. 2001;106:46–52.
Wajner M, Amaral AU. Mitochondrial dysfunction in fatty acid oxidation disorders: insights from human and animal studies. Biosci Rep. 2015;36:e00281.
Acknowledgements
The authors gratefully acknowledge the expert technical assistance of Brith Leidvik and Armita Mandegari. This work was supported by the Swedish Research Council (Proj No 2012–2014).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Roos, S., Sofou, K., Hedberg-Oldfors, C. et al. Mitochondrial complex IV deficiency caused by a novel frameshift variant in MT-CO2 associated with myopathy and perturbed acylcarnitine profile. Eur J Hum Genet 27, 331–335 (2019). https://doi.org/10.1038/s41431-018-0286-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0286-0