Abstract
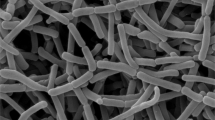
An actinomycete strain RM579T isolated from roots of Pithecellobium dulce in Thailand was studied using the polyphasic taxonomic approach. The results of comparative 16S rRNA gene sequence and phylogenetic analysis showed that strain RM579T belonged to the family Pseudonocardiaceae and it was most closely related to members of the genera Amycolatopsis (≤94.5% sequence similarity) and Haloechinothrix (≤93.4% sequence similarity). A phylogenetic tree constructed by neighbor-joining method indicated that strain RM579T was positioned within the clade of the genus Amycolatopsis and formed a monophyletic cluster with Amycolatopsis taiwanensis 0345M-7T, Amycolatopsis pigmentata TT00-43T and Amycolatopsis helveola TT99-32T. Strain RM579T formed white aerial mycelium and yellowish-brown substrate mycelium that fragments into rod-shaped elements. Morphological features and chemotaxonomic characteristics of strain RM579T were consistent with those of the genus Amycolatopsis. The cell wall of strain RM579T contained meso-diaminopimelic acid. Arabinose, galactose, mannose, and ribose were detected as whole-cell sugars. Mycolic acids were absent. The acyl type of the muramic acid in the cell wall was N-acetyl. Diphosphatidylglycerol, hydroxyl-phosphatidylethanolamine, phosphatidylinositol, and an unidentified phospholipid were detected as polar lipids. The major cellular fatty acids (>10%) were iso-C16:0, iso-C16:0 2-OH, and C16:1 cis9. The resultant data indicated that strain RM579T should be classified as representative of a novel species in the genus Amycolatopsis, for which the name Amycolatopsis pithecelloba sp. nov. is proposed. The type strain is RM579T (=TBRC 1849T =NBRC 106096T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lechevalier MP, Prauser H, Labeda DP, Ruan J-S. Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int J Syst Evol Microbiol. 1986;36:29–37.
Embley MT, Smida J, Stackebrandt E. The phylogeny of mycolate-less wall chemotype IV actinomycetes and description of Pseudonocardiaceae fam. nov. Syst Appl Microbiol. 1988;11:44–52.
Warwick S, Bowen T, McVeigh H, Embley TM. A phylogenetic analysis of the family Pseudonocardiaceae and the genera Actinokineospora and Saccharothrix with 16S rRNA sequences and a proposal to combine the genera Amycolata and Pseudonocardia in an emended genus Pseudonocardia. Int J Syst Bacteriol. 1994;44:293–9.
Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–43.
Lechevalier MP. Identification of aerobic actinomycetes of clinical importance. J Lab Clin Med. 1968;71:934–44.
Lechevalier MP, De Bievre C, Lechevalier HA. Chemotaxonomy of aerobic Actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–60.
Xu X, Han L, Zhao L, Chen X, Miao C, Hu L, et al. Echinosporin antibiotics isolated from Amycolatopsis strain and their antifungal activity against root-rot pathogens of the Panax notoginseng. Folia Microbiol. 2019;64:171–5.
Xu L, Huang H, Wei W, Zhong Y, Tang B, Yuan H, et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genomics. 2014;15:363.
Nigam A, Almabruk KH, Saxena A, Yang J, Mukherjee U, Kaur H, et al. Modification of rifamycin polyketide backbone leads to improved drug activity against rifampicin-resistant Mycobacterium tuberculosis. J Biol Chem. 2014;289:21142–52.
Mingma R, Duangmal K, Trakulnaleamsai S, Thamchaipenet A, Matsumoto A, Takahashi Y. Sphaerisporangium rufum sp. nov., an endophytic actinomycete from roots of Oryza sativa L. Int J Syst Evol Microbiol. 2014;64:1077–82.
Küster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–9.
Tseng M, Yang SF, Li WJ, Jiang CL. Amycolatopsis taiwanensis sp. nov., from soil. Int J Syst Evol Microbiol. 2006;56:1811–5.
Tamura T, Ishida Y, Otoguro M, Suzuki K. Amycolatopsis helveola sp. nov. and Amycolatopsis pigmentata sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2010;60:2629–33.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Jacobson E, Grauville WC, Fogs CE. Color harmony manual, 4th ed. Chicago: Container Corporation of America; 1958.
Gordon RE, Barnett DA, Handerhan JE, Pang CH-N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol. 1974;24:54–63.
Gordon RE, Mihm JM. A comparative study of some strains received as nocardiae. J Bacteriol. 1957;73:15–27.
Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–22.
Uchida K, Aida K. Acyl type of bacterial cell wall: its simple identification by a colorimetric method. J Gen Appl Microbiol. 1977;23:249–60.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–31.
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M. Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol. 1977;27:104–17.
Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–30.
Tomiyasu I. Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J Bacteriol. 1982;151:828–37.
Take A, Matsumoto A, Omura S, Takahashi Y. Streptomyces lactacystinicus sp. nov. and Streptomyces cyslabdanicus sp. nov., producing lactacystin and cyslabdan, respectively. J Antibiot. 2015;68:322–7.
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Fitch WM. Toward defining the course of evolution: minimal change for a specific tree topology. Syst Zool. 1971;20:406–16.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Mingma R, Pathom-aree W, Trakulnaleamsai S, Thamchaipenet A, Duangmal K. Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World J Microbiol Biotechnol. 2014;30:271–80.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5.
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4.
Acknowledgements
This research was supported by Kasetsart University Research and Development Institute (KURDI, Project Code 2559:25.60); Kitasato Institute for Life Sciences, Kitasato University, Japan; Faculty of Science, Kasetsart University, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mingma, R., Inahashi, Y., Matsumoto, A. et al. Amycolatopsis pithecelloba sp. nov., a novel actinomycete isolated from roots of Pithecellobium dulce in Thailand. J Antibiot 73, 230–235 (2020). https://doi.org/10.1038/s41429-019-0271-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0271-z
This article is cited by
-
Amycolatopsis acididurans sp. nov., isolated from peat swamp forest soil in Thailand
The Journal of Antibiotics (2021)