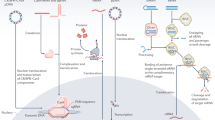
Abstract
Nucleic acid delivery offers tremendous potential for the treatment of acquired and hereditary diseases. Despite limited successes, the use of nucleic acid therapies has been hampered by the lack of safe and efficient delivery approaches. To address this challenge, Epps, Sullivan, and coworkers developed a new nucleic acid delivery framework predicated on a photo-responsive cationic block copolymer (BCP) that enabled tunable nucleic acid binding and precise spatiotemporal control over gene expression. This innovative platform, in which the polymer moieties directly responsible for nucleic acid complexation could be cleaved from the polymer upon photo-stimulation, significantly enhancing nucleic acid release. Furthermore, temporal control over polyplex disassembly facilitated the development of a simple, and potentially universal, kinetic modeling scheme for intracellular small interfering RNA (siRNA), messenger RNA, and protein concentrations, and that model was quantitatively validated using various genes across several animal cell lines and human primary cells. This versatile BCP-based framework easily accommodated: anionic excipients that increased siRNA potency by ~200% (on a per mass basis) over comparable polyplex systems; quantum dots that unlocked theranostic applications without impacting silencing performance; and small-molecule lipid co-formulations that enhanced transfection in human primary cells. Altogether, the system described herein shows great promise for the clinical translation of gene therapeutics.
Similar content being viewed by others
Introduction
Gene therapy involves the transfer of DNA or RNA into cells to alter gene expression and thereby treat hereditary or acquired diseases [1,2,3]. Gene therapy is highly attractive in concept because it addresses common challenges faced by traditional drug delivery methods, such as low specificity, unwanted side effects, high cost, and repetitive administration [4]. Moreover, it offers a versatile route for the in situ production of protein therapeutics and enables sophisticated manipulation of cell phenotype in applications ranging from regenerative medicine to immunotherapies. However, the translation of gene therapies into clinically relevant treatments has not been as rapid or successful as expected [1]. Glybera, the Western world’s first approved gene therapy, was announced in 2012 [5], several decades after the concept of gene therapy was introduced [2, 6]. Due in part to its high price and limited demand, Glybera was discontinued in 2017 [7]. Also in 2017, the US Food and Drug Administration approved its first in vivo gene therapy, Luxturna, for the treatment of an inherited rare retinal disorder. The cost of the treatment was $425,000 per eye [8]. The high prices of these therapies have raised concerns about the sustainability of gene therapy in the long term. Furthermore, the slow and expensive translation of preclinical gene therapy studies into effective clinical protocols has been linked to the lack of suitable and versatile nucleic acid delivery methods [4, 9]. Thus, significant research activity has focused on the development of robust delivery vehicles that can successfully package and transport nucleic acids to target cells, while preserving the gene regulating efficacy of these molecules [4, 10].
Among various delivery approaches, nucleic acid delivery by nonviral vectors, such as cationic polymers, has some advantages over other options (i.e., viral vectors), including reduced immunogenicity and increased synthetic/structural versatility [10, 11]. In most cases, the cationic polymers are used to electrostatically bind nucleic acids to form nanoscale polyplexes exhibiting many favorable properties for gene therapy [12]. For example, the encapsulated nucleic acids exhibit reduced nuclease degradation, while also showing increased cellular endocytosis due to their more favorable interactions with the cell membrane [13]. These cationic polymers also are advantageous because they can be easily linked to “stealthy” macromolecules such as polyethylene glycol (PEG) that are able to reduce serum protein adsorption and increase biocompatibility. However, the main drawback of the above-mentioned cationic polyplexes is their low gene expression efficiency, which is linked to both their inefficient trafficking within cells and their suboptimal in vivo properties.
A key unsolved issue is that polyplexes must stably encapsulate their nucleic acid cargoes in blood/tissues yet effect release of the same nucleic acid cargoes within the cytoplasm of cells. The challenges inherent in designing polymers to meet these contradictory needs have been manifested repeatedly in polymer gene carrier analyses to date [14]. For example, a study by Davis and coworkers demonstrated that small interfering RNA (siRNA)/cyclodextrin-containing polymer polyplexes accumulated and disassembled in the glomerular basement membrane due to interactions with heparan sulfate polyanions [14], demonstrating the critical need for polymer binding interactions with sufficient stability to resist competitive displacement by abundant polyanionic sugars. Approaches to improve nucleic acid binding stability through alteration of polymer architecture have focused primarily on increasing the polymer molecular weight (MW) [15,16,17,18] or hydrophobic modification [19,20,21,22,23]. However, a critical challenge with all of these approaches is the need for intracellular unbinding of siRNA from polyplexes to enable efficient RISC assembly and silencing activity [24,25,26].
To address this challenge, Epps, Sullivan, and coworkers developed novel photo-responsive nucleic acid delivery vehicles comprised of mPEG-b-poly(5-(3-(amino)propoxy)-2-nitrobenzyl methacrylate) (mPEG-b-P(APNBMA)) that served as a versatile platform for the investigation of nucleic acid binding vs. release and provided a new strategy for enhanced nucleic acid delivery [27, 28]. A block copolymer (BCP) architecture was used to ensure nanoscale size control through self-assembly. The PEG-polycation structure led to the formation of stable polyplexes with the nucleic acid payloads binding electrostatically in the cores and the PEG “stealthy” corona to resisting both salt-induced aggregation and protein adsorption [27].
Several key features made this system promising for nucleic acid delivery applications. First, the balance between nucleic acid binding vs. release was optimized by tuning the overall electrostatic interactions via two simple and elegant methods: the use of BCPs of different cationic block lengths (as well as various mixtures of those copolymers) and the incorporation of anionic excipients [29, 30]. A combination of such methods permitted the formulation of polyplexes with maximal nucleic acid delivery and gene regulating efficiency in a range of different cell types. Second, the use of biocompatible, photo-responsive o-nitrobenzyl (o-NB) groups to link the cationic groups of the APNBMA side chains to the polymer backbone facilitated efficient, light-induced side chain cleavage (e.g., in both the free polymer and polyplex, the labile ester bonds exhibited a τ½ of approximately 5 min under mild irradiation conditions of 200 W m−2 [28], typical of o-NB polymers [31, 32]). Polymer cleavage triggered the rapid release of nucleic acid cargoes via light-induced charge reversal of the polymer side chains and also provided a robust method for the spatiotemporal control over gene expression [33]. This control is extremely important in applications such as regenerative medicine and cancer because it allows an extra level of specificity and minimizes unwanted off-target effects [4, 10]. Third, the ability to regulate nucleic acid delivery in both space and time enabled the development of simple kinetic models that accurately predicted the dynamics of nucleic acid-mediated gene silencing in both animal cell lines and human cells [29, 30, 33, 34]. These kinetic models were vital to formulating dosing regimens that sustained therapeutic effects over the course of several days to a week [30, 34]. This model-driven approach is particularly attractive for the successful treatment of chronic diseases in which multiple doses may be required to suppress disease symptoms over prolonged timescales [35]. Accurate dosing regimens also lead to less frequent drug administration, which could further reduce cost and associated risks of side effects.
Versatility and adaptability are other significant characteristics of the mPEG-b-P(APNBMA) system. For example, the incorporation of lipid components into the polyplex formulations enhanced transfection of human primary cells and was easily tailorable to improve specificity and reduce off-target effects for other cell types [34]. Additionally, the facile incorporation of anionic excipients such as quantum dots (QDs) introduced in situ theranostic capabilities, while simultaneously improving nucleic acid release [34, 36]. Finally, the universal principles of electrostatic binding between nucleic acids and cationic blocks permitted the efficient delivery of various nucleic acids without further macromolecular modifications. This characteristic is beneficial for the treatment of more complex genetic diseases in which two or more nucleic acid types must be administered together to achieve a desired therapeutic outcome. Taken together, the flexibility of nucleic acid delivery platforms such as mPEG-b-P(APNBMA) can enhance the efficiency and affordability of gene therapy, bridging the gap between conceptual potential and clinical translation.
This article highlights Epps’ and Sullivan’s development of robust and efficient nucleic acid delivery vehicles that address current drawbacks impeding the success of gene therapeutics in the clinic. The team leveraged combined expertize in chemical synthesis, polymer chemistry, biomaterials, and gene/drug delivery to design nanoscale delivery vehicles that introduced exquisite, rapid, and tunable control over nucleic acid binding and release. The majority of activities focused on siRNA for gene silencing via an RNA interference (RNAi) approach, and the overall system, which is summarized in Fig. 1, is translatable to other nucleic acid types. The presented framework was synthesized by straightforward methods, easily adapted to different cell types, capable of carrying different nucleic cargoes, and exhibited tremendous salt and serum stability along with low toxicity, all toward the goal of improving the efficacy of gene therapy treatments.
Schematic of nucleic acid delivery approaches. Adapted with permission [29]. Copyright © 2017 American Chemical Society
Balancing nucleic acid binding vs. release through formulation design
Many drawbacks of nucleic acid delivery vehicles made from cationic polymers are linked to a lack of control over binding vs. release [1]. Efficient carriers must satisfy the seemingly contradictory requirements of remaining stable extracellularly but disassembling efficiently intracellularly. At one extreme, weak nucleic acid binding often causes premature nucleic acid release in the extracellular environment, which leads to nucleic acid degradation in the presence of nucleases or anionic proteoglycans such as heparin sulfate [14, 37]. Common strategies to enhance binding include increasing the charge density and/or chain length of the complexation-inducing polycations, and/or incorporating hydrophobic groups within the polycations [15, 19, 38, 39]. At the same time, strong binding of nucleic acids can impede release and hinder gene expression [25, 40], and higher cationic charge densities have been linked to greater cytotoxicity [41, 42]. Thus, the balance between nucleic acid binding vs. release is one of the most crucial, and fundamental, design criteria for any nucleic acid delivery vehicle.
In the mPEG-b-P(APNBMA) system, the optimal nucleic acid-cationic polymer binding was probed via two straightforward methods: the use of polyplexes containing BCPs of different cationic block lengths (as well as mixtures of those copolymers) and the incorporation of anionic excipients. These methods eliminated the need to synthesize huge libraries of cationic polymers to probe the impact of changing macromolecular parameters on packaging, uptake, and gene silencing.
Mixed BCPs formulations for screening polyplex binding vs. release
One common strategy to modulate nucleic acid binding is to vary the MW and/or charge density of the cationic components. Generally, increasing the number of cationic groups improves the polymer/nucleic acid binding efficiency and the cellular uptake of the polyplexes; however, increased positive charge has been linked to greater cell death and reduced nucleic acid release [25, 41, 42]. While the use of polymers with intermediate binding forces could offer a suitable balance, systematic approaches to generating such macromolecules often require the synthesis of comprehensive materials libraries, a time-consuming endeavor. A more practical strategy to provide tunable control over siRNA binding affinity is the use of mixed polyplexes made from BCPs of different compositions to rapidly screen the parameter space (Fig. 2a) [30]. The net cationic charge can be modulated by adjusting the molar ratios of as few as two polymers, offering a simple and rapid method to screen for suitable nucleic acid binding [30].
a Schematic of mPEG-b-P(APNBMA)/siRNA polyplexes made from mixed BCPs with different cationic block lengths. b GAPDH protein silencing efficiencies of mixed mPEG-b-P(APNBMA)/siRNA polyplexes. The polyplex composition was defined as the ratio of short/long BCPs, on a molar basis of cationic amine groups. Adapted with permission [30]. Copyright © 2017 Acta Biomaterialia Inc
Work by Greco et al. demonstrated enhanced silencing of the endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene in NIH/3T3 cells using mixed mPEG-b-P(APNBMA)/GAPDH-targeting siRNA polyplexes formulated from two different cationic P(APNBMA) block lengths (Fig. 2a), when compared to “unmixed” polyplexes made from either of the single cationic block lengths (Fig. 2b) [30]. Maximum gene silencing was achieved with the 50/50 formation, showing ~70% GAPDH protein knockdown. It was noteworthy that a BCP was subsequently synthesized with a block length that matched the average length of the 50/50 formulation, and polyplexes from that material exhibited nearly identical silencing to the mixed counterpart, demonstrating the robustness of the screening approach. Furthermore, the silencing efficiency realized with this formulation was very close to the maximum achievable with a single dose of siRNA given the half-lives of the target messenger RNA (mRNA) and protein [43,44,45].
Incorporation of anionic excipients
An alternative approach to modulate nucleic acid release from cationic delivery vehicles is the incorporation of anionic excipients that have strong binding to cationic polymers (e.g., highly negatively charged polymers, peptides, or glycosaminoglycans) [46]. These excipients can improve the stability and biocompatibility of polyplexes, while enabling the selective release of nucleic acid cargoes due to their capacity for competitive displacement of the nucleic acids [40, 47,48,49]. Excipient incorporation was of interest in the mPEG-b-P(APNBMA)/siRNA system because a fraction of the siRNAs remained inaccessible within the polyplexes, even after complete polymer photocleavage [33]. The efficacy of excipients, such as poly(acrylic acid) (PAA), was investigated in the mPEG-b-P(APNBMA)/siRNA system as outlined in Figure 3a [29]. PAA was chosen because of its high-anionic charge density and extensive use in FDA-approved products [50, 51]. Gene silencing efficiency was measured as a function of PAA MW and weight fraction in polyplexes. Several key insights and excipient design rules were gleaned from this effort. First, formulations containing PAA with MWs that were approximately one order of magnitude greater than the MW of a typical siRNA showed the greatest release and most efficient silencing (Fig. 3b) [29]. Second, the mPEG-b-P(APNBMA)/PAA/siRNA formulations were stable for at least one week in serum-supplemented media without any loss in activity [29]. Third, the gene silencing capacity of the excipient-containing polyplexes could be quantitatively predicted using heparin-induced siRNA release assays to generate heat maps that forecast the optimal co-formulation PAA MWs and weight fractions (Fig. 3c) [29]. Finally, the optimized systems resulted in photo-responsive nanocarriers in which the siRNA usage was at least 100% more efficient on a per mass basis than cases where PAA was not used [29].
a Schematic of mPEG-b-P(APNBMA)/siRNA polyplexes co-formulated with PAA and/or QD anionic excipients. b Silencing efficiency fold change of ternary mPEG-b-P(APNBMA)/PAA/siRNA system, as a function of PAA MW and weight fraction in the polyplexes. c Model predictions of silencing efficiency fold change of mPEG-b-P(APNBMA)/PAA/siRNA polyplexes as a function of PAA MW and weight fraction. d Image of an isolated QD-containing polyplex using cryo-TEM. Adapted with permission [29]. Copyright © 2017 American Chemical Society
Given the success of incorporating high-charge density anionic excipients such as PAA to positively impact gene silencing, QDs were explored to introduce self-reporting and imaging capabilities into the mPEG-b-P(APNBMA)-based polyplexes [29]. QDs are an ideal alternative labeling agent for monitoring polyplex trafficking and nucleic acid release because of their high-extinction coefficients, precisely tunable optical properties, and reduced susceptibility to photobleaching [36]. QDs also serve as efficient donors for fluorescence resonance energy transfer (FRET) to provide spatially resolved information about intracellular interactions at the nanometer scale [52]. In relation to gene therapeutics, this type of excipient is attractive for theranostic applications [36], as it does not require the conjugation of the diagnostic moieties to the drugs or nanocarriers, an approach that can negatively impact the activity of therapeutic agents or the intracellular trafficking of the delivery vehicles [52, 53]. The FRET-tracking of the QD-containing system was robust, and additionally, the siRNA release efficiency was improved by 100% as a result of QD loading [29]. Furthermore, polyplex behavior, interrogated using QD-based FRET, demonstrated the stability of the nanocarriers in the absence of the photo-stimulus but clearly indicated polyplex disassembly following several minutes of photo-irradiation. Polyplexes comprised of both excipients (QDs and PAA) displayed the greatest enhancement in disassembly, with the dual excipients effecting an ~200% increase in the per mass efficiency of the siRNA therapeutics [29]. Finally, the contrast afforded by the high-electron density of QDs permitted direct imaging of water-swollen polyplexes via cryogenic-transmission electron microscopy (Fig. 3d) [29], without the need to stain or otherwise fix samples [54]. Overall, these studies demonstrated the versatility of the mPEG-b-P(APNBMA) platform to incorporate additional components for materials optimization (e.g., improve siRNA silencing efficiency, facilitate imaging and diagnostics, self-report nanoscale disassembly).
Spatiotemporal control over gene expression
Nanocarriers with tunable nucleic acid binding vs. release also offer important opportunities for improved spatiotemporal control over gene expression. Broadly, the capacity to control gene expression in both space and time addresses many fundamental challenges faced by drug delivery systems such as low efficacy and off-target effects, while also providing unique capacity to manipulate individual and/or collective cell phenotypes [55]. The predominant strategies for the spatiotemporal alteration of gene expression include inducible promoter and optogenetic regulation systems [55,56,57,58,59,60,61]. Optogenetic regulation systems have demonstrated especially exciting capacity for tunable, light-based control over gene expression [56]. However, significant drawbacks include the inability to regulate endogenous genes, the inherent difficulty in delivering multicomponent optogenetic machinery, and immunogenicity or mutation/oncogene activation [56, 61]. Thus, an alternative approach to regulating spatiotemporal gene silencing is essential.
Stimuli-responsive polymers that regulate nucleic acid binding vs. release offer a particularly appealing strategy to control nucleic acid activity [62,63,64]. This binding/release approach mimics histone-mediated transcriptional regulation in which the charge state of histone proteins is dynamically altered by chromatin-modifying enzymes in order to transition between silenced heterochromatin and active euchromatin structures. Stimuli-responsive delivery also offers more precise control than spatially constrained nucleic acid application, which is limited by diffusion considerations. Photo-stimuli are especially advantageous, as light application can be controlled both spatially and temporally with high resolution [65, 66]; accordingly, light-based tools are increasingly prevalent in clinical practice [67,68,69], with a wide range of devices to facilitate access to remote tissues and provide precise light exposure (e.g., Veritas Medical; LiteCure).
The structure of the mPEG-b-P(APNBMA) BCPs incorporated biocompatible, photocleavable o-NB moieties [70, 71], which linked pendant ammonium cations to the polymer backbone and enabled light-induced hydrolysis and charge reversal in the polymer chain. These BCPs complexed with nucleic acids to form highly stable polyplexes that did not disassemble or lose activity, even after a week-long incubation at 37 °C in serum/salt solutions [27, 33]. Moreover, in the absence of light, no siRNA activity was noted even after a multiday exposure to cells, demonstrating the fully responsive nature of the polyplexes. However, upon exposure to light, o-NB cleaved to introduce negatively charged carboxylate ions in the polymer side chains [28, 72, 73], and this charge reversal led to the release of siRNA cargoes from polyplexes, with the manifestation of gene silencing activity that was directly correlated with light dose [33, 74]. The light exposure was shown to induce rapid and tunable intracellular release of siRNA, as demonstrated by fluorescence correlation spectroscopy (FCS) and FRET analyses of polyplex disassembly and siRNA release. FCS and FRET analyses also demonstrated the fully intact nature of the polyplexes in the absence of light [29]. Furthermore, the ability to rapidly transition from the complexed to the free siRNA states was manifested in the precise (cellular length scale) on/off spatial control over exogenous green fluorescent protein (GFP) activity (Fig. 4) [33]. NIH/3T3 cells that were irradiated with a biocompatible dosage of light [75] displayed tunable reductions in GFP expression up to the point when GFP expression was no longer detectable [33]. Conversely, NIH/3T3 cells that were protected from UV irradiation by a photomask exhibited robust GFP expression, with a sharp interface at the edge of the photomask [33].
On/off spatiotemporal control over nanocarrier disassembly to modulate the expression of an exogenous gene. Zoomed-in view of line-patterning of GFP expression with cell-to-cell resolution. NIH/3T3 cells were cotransfected with GFP DNA (in Lipofectamine polyplexes) and GFP-targeted siRNA (in mPEG-b-P(APNBMA) polyplexes), and half of the plate was covered with a photomask prior to light irradiation. The dashed red line represents the edge of the photomask, and the scale bars represent 250 μm. Adapted with permission [33]. Copyright © 2016 American Chemical Society
In addition to the control of exogenous genes, the efficiency and tunability of gene silencing also were shown using GAPDH [33]. Regulating the light exposure time in NIH/3T3 cells transfected with mPEG-b-P(APNBMA)/GAPDH-targeting siRNA polyplexes allowed control of GAPDH silencing over a range of 0–86% when adjusting the duration of UV irradiation between 0 and 20 min [33]. Transitioning from the exogenous (GFP) to endogenous (GAPDH) targets proceeded through simple substitution of siRNA sequences and required no adjustments in the composition or assembly protocols for the polyplexes. The above results demonstrated that silencing of a range of gene products can be controlled with ease and high accuracy using the versatile photo-responsive mPEG-b-P(APNBMA) polyplexes. These advantages are especially important given the increasing relevance of multi-gene silencing designs and personalized therapeutic approaches, as our optimized polymer assemblies can be directly formulated to contain a wide range of target molecules [44, 76, 77].
Dosing regimen design using kinetic modeling
The design of accurate siRNA dosing regimens is an important challenge underlying the clinical translation of RNAi therapies. Control over the extent and duration of gene silencing is essential for efficacy in cancer therapeutics, regenerative medicine, and other RNAi applications, and often, such control cannot be realized with a single bolus dose of siRNA. For example, in diseases that are characterized by rapid cell division such as cancer, frequent dosing typically is necessary to counteract the rapid dilution of siRNA that occurs during cell division [43]. Meanwhile, the treatment of chronic diseases can require administration of multiple siRNA doses to provide sustained alleviation of disease-causing discomfort and pain [35, 78, 79]. The ability to connect the quantity of delivered siRNA to its silencing outcome also is necessary to predict/minimize off-target effects and minimize cost. However, a majority of reported dosing schedules are chosen through trial and error, requiring expensive and time-consuming analyses that provide limited design insight into alternative dosing schedules [30, 80]. Moreover, even when quantitative data exist on siRNA biodistribution, total siRNA in tissues typically does not correlate well with silencing efficacy [81, 82]. These observations highlight the need for improved approaches to control and predict siRNA outcomes within cells.
The kinetics of gene silencing depends upon a series of biologically defined rates, such as cell doubling time, as well as protein and mRNA half-lives. Additionally, gene silencing depends upon the efficiency with which siRNA enters cells, escapes into the cytoplasm, and unbinds from its nanocarrier [43, 83]. Mathematical modeling has been employed in a few instances to provide more holistic and quantitative insights into the kinetics of the RNAi process, with promising benefits for dosing design [43, 80, 83, 84]. However, while prior modeling approaches accurately captured the effects of biological processes such as cell dilution, modeling was most often applied to commercial gene delivery systems lacking the ability to precisely control the amount of free intracellular siRNA [43, 52, 85]. Delivery vehicles with efficient intracellular trafficking and well-defined siRNA release would thereby enable more accurate prediction of dose-response behavior.
The mPEG-b-P(APNBMA)/siRNA polyplex design capitalized upon stimuli-responsive and tunable siRNA release to provide precise control over the amount of intracellular siRNA, and thereby facilitated the development of a noncompartmentalized cellular reaction kinetics model [30, 74]. The model was implemented as a simplified system of ordinary differential equations (Eqs. 1–3), which were designed to capture changes in protein and mRNA levels due to siRNA-mediated silencing or proteolytic/nucleolytic turnover, as well as dilution through cell division. The model considered only the most important, rate-determining processes, using target-specific protein and mRNA half-lives reported in the literature [43,44,45], and free siRNA concentrations that were determined experimentally [30, 74].
Model predictions of GAPDH gene silencing efficiency in NIH/3T3 cells corresponded well with experimental measurements, confirming that the key cellular factors limiting the efficiency of the knockdown response were the long relative half-lives governing the degradation of GAPDH protein and mRNA, and the short cell doubling time (Fig. 5a, b) [30]. Additionally, the ability to predict dynamic changes in mRNA and protein following siRNA treatment enabled the design of a two-dose model that sustained silencing over several days (Fig. 5c, d) [30], with protein expression reduced two-fold below the minimum level achievable with a single dose. These results showcase the versatility and power of combining simple kinetic models with stimuli-responsive nanocarriers, an approach that offered new mechanistic insights and accurate a priori predictions of gene silencing relevant to dosing design in a wide range of applications.
a–d Dosing regimens development for GAPDH silencing process using kinetic modeling with comparisons to experimental gene silencing data. a, c Kinetic model prediction of the dynamic nature of the GAPDH silencing process with a single dose (a) and double dose (c) of siRNA. b, d GAPDH mRNA and protein expression levels determined through qPCR (quantitative PCR) and western blot experiments, respectively, following either a single dose (b) or double dose (d) of siRNA. Model predictions of mRNA (green) and protein (orange) expression levels at the end points of 48 and 75 h are presented as solid bars; experimental values are presented as diagonally striped bars. Adapted with permission [30]. Copyright © 2017, Acta Biomaterialia Inc
Gene silencing in human primary cells: reducing maladaptive responses in aortic adventitial fibroblasts (AoAFs)
Recent work on mPEG-b-P(APNBMA) has showcased the versatility of the photo-responsive platform to address disease targets in human primary cells, which are more refractory to gene modulation than immortalized cell lines [86]. This ability to effectively transfect human primary cells is a critical requirement for gene delivery methods in tissue engineering and regenerative medicine [87]. More specifically, a gene silencing approach was investigated for the treatment of maladaptive adventitial remodeling that occurs at anastomostic sites following cardiovascular bypass graft surgery. These maladaptive responses are largely driven by injury-induced adventitial fibroblast (AF) proliferation and differentiation, which control the gradual progression of intimal hyperplasia and fibrosis [88, 89]; thus, methods to provide sustained suppression of key proliferative effectors and phenotypic modulators in AFs would provide a potential route to improve healing outcomes [90, 91].
To facilitate the transfection of human primary AoAFs with the BCP-based polyplexes, a hybrid nanocomplex design was necessary. Nanocomplexes were formulated using a combination of Lipofectamine RNAiMAX and the BCPs, enabling photo-controlled, spatiotemporal release of siRNA and efficient gene silencing (Fig. 6a) [34]. These hybrid nanocomplexes were generated in a sequential fashion in which siRNA was complexed with Lipofectamine, PAA was added to change the surface charge, and finally, the photo-responsive BCP was added to complete the multicomponent nanocarrier. This lipid/polymer combination successfully married the on/off responsiveness of the BCP system with the efficient silencing of the lipid-only system (Fig. 6b) [34].
a Schematic depicting the formulation of the hybrid nanocomplexes. b Photo-controlled IL1β protein silencing with mPEG-b-P(APNBMA) polyplexes, Lipofectamine RNAiMAX lipoplexes, and hybrid nanocomplexes. c Representative immunocytochemistry micrographs of AoAFs 4 days post-transfection following a single dose of siRNA. AoAFs were stained for F-actin (magenta, [indicative of fibroblasts]) and αSMA (green, (indicative of myofibroblasts)). Scale bars = 50 µm. d, e Dynamics of IL1β and CDH11 protein silencing following a single dose (d) or double dose (e) of siRNA as predicted by kinetics modeling (dashed lines) and measured experimentally (solid symbols). Adapted with permission [34]. Copyright © 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim
The above-mentioned hybrid system was used for the co-delivery of interleukin 1 beta (IL1β)-targeting and cadherin 11 (CDH11)-targeting siRNAs to investigate their cooperative roles in mitigating maladaptive responses in AoAFs, a key hallmark of which is the suppression of fibroblast proliferation and fibroblast-to-myofibroblast differentiation [91]. Cellular proliferation was reduced through the down regulation of IL1β expression [34]. Furthermore, significant attenuation of differentiation was achieved upon knockdown of IL1β-only (Fig. 6c), as well as with knockdown of both IL1β and CDH11 [34]. These aggregate results have major implications in the development of spatially targeted clinical approaches to reduce and/or eliminate fibrosis at the junction (e.g., anastomosic) sites, i.e., the typical failure locations of many vascular bypass grafts.
Dosing regimens for the silencing of IL1β and CDH11 were determined via kinetic modeling of gene silencing dynamics, using the framework mentioned in the previous section. It is noteworthy that, after obtaining the appropriate mRNA and protein half-lives, the same set of ordinary differential equations that were used for the murine cell analyses could be employed for the human primary cell modeling with quantitative results (Fig. 6d) [34]. For example, the model predicted that an initial transfection at 0 h, followed by a second transfection at 72 h, would enable the knockdown of both genes to the desired threshold of 20% relative to untreated controls, with sustained suppression over a one-week duration (Fig. 6e) [34]. This timescale was chosen because it matches the duration of gene suppression necessary to control adventitial remodeling and fibroblast differentiation following surgically induced injury [92]. Most importantly, the model prediction was in excellent agreement with experimental gene silencing results over the same one week timescale (Fig. 6e) [34, 93]. In vivo studies of these hybrid polyplexes are on-going, and the current data regarding the extent and duration of silencing foreshadow potential promising outcomes for vascular bypass surgeries, along with other regenerative medicine applications.
Conclusions
The cationic and photo-responsive BCP platform, based on mPEG-b-P(APNBMA), has demonstrated enormous utility and potential in the development of robust approaches to gene therapy and regenerative medicine. Epps, Sullivan, and coworkers have leveraged combined expertize in macromolecular synthesis, nanoscale soft materials assembly, nanocarrier trafficking in cellular environments, and nucleic acid delivery to create a modular system that can overcome numerous barriers to clinical translation in nucleic acid nanocarriers. In particular, the mPEG-b-P(APNBMA) framework is an innovative platform that facilitated the precise spatiotemporal control over gene expression in a tunable, versatile, and cell-compatible package. Key aspects of the photo-responsive polyplexes include: (1) a unique macromolecular architecture in which the cationic moieties were attached to the polymer backbone via photocleavage groups that also underwent charge reversal to accelerate nucleic acid release; (2) a modular self-assembly approach that incorporated various nucleic acids and polymer MWs into a single polyplex to tune siRNA release; (3) a precise on/off release nature that enabled the development of a quantitative, and possibly universal, gene silencing model that could be adapted across numerous genes and cell types; (4) a multicomponent design that benefitted from the inclusion of anionic excipients to increase the potency of siRNA by over 200% on a per mass basis relative to non-excipient-containing systems; (5) a stable and robust framework that was augmented with QDs and other imaging agents to unlock theranostic applications without negatively impacting therapeutic efficacy; and (6) a hybrid formulation, containing a mixture of polymer and lipids, that optimized photo-responsive triggering and transfection in human primary cells for attenuating maladaptive responses following surgical injury. In short, the efforts highlighted herein demonstrate the enormous opportunities provided by this photo-responsive system to effect gene silencing in multiple cell types, with the potential to positively impact the treatment of various diseases. Furthermore, the inherent adaptability of the framework also can accommodate the incorporation of other additives moving forward, such as upconverting nanoparticles, to unlock deep tissue-based applications [94]. Overall, the described photo-responsive BCP design is a significant advance in the quest for safer, more efficient, more reliable, and lower-cost gene therapeutics.
Equations 1–3
Ordinary differential equations to model time-dependent changes in mRNA, protein, and siRNA concentration. kmRNA, ksiRNA, and kprot are the rate constants for the production of mRNA, siRNA, and protein, respectively. km,deg, ks,deg, and kp,deg are the rate constants for the degradation of mRNA, siRNA, and protein, respectively. Adapted with permission [33]. Copyright © 2016, American Chemical Society.
References
Draghici B, Ilies MA. Synthetic nucleic acid delivery systems: present and perspectives. J Med Chem. 2015;58(10):4091–130.
Wolff JA, Lederberg J. An early history of gene transfer and therapy. Hum Gene Ther. 1994;5(4):469–80.
Zhang Y, Satterlee A, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol Ther. 2012;20(7):1298–304.
Vile RG. Gene therapy. Curr Biol. 1998;8(3):R73–5.
Mullin E. The world’s most expensive medicine is being pulled from the market, April 21, 2017. https://www.technologyreview.com/s/604252/the-worlds-most-expensive-medicine-is-being-pulled-from-the-market/.
Gallagher J. Gene therapy - Glybera approved by European Commission, November 2, 2012. http://www.bbc.com/news/health-20179561.
Sagonowsky E. With its launch fizzling out, UniQure gives up on $1M + gene therapy Glybera, April 20, 2017. https://www.fiercepharma.com/pharma/uniqure-gives-up-1m-gene-therapy-glybera.
Scutti B. Gene therapy for retinal disorder to cost $425,000 per eye, January 3, 2018. https://www.cnn.com/2018/01/03/health/luxturna-price-blindness-drug-bn/index.html.
Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59(2–3):75–86.
de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6(6):443–53.
Zhou J, Shum KT, Burnett JC, Rossi JJ. Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals. 2013;6(1):85–107.
Haussecker D. The Business of RNAi Therapeutics. Human Gene Ther. 2008;19(5):451–62.
Kelley EG, Albert JNL, Sullivan MO, Epps TH III. Stimuli-responsive copolymer solution and surface assemblies for biomedical applications. Chem Soc Rev. 2013;42(17):7057–71.
Zuckerman JE, Choi CH, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci USA. 2012;109(8):3137–42.
Liu X, Howard KA, Dong M, Andersen MØ, Rahbek UL, Johnsen MG, Hansen OC, Besenbacher F, Kjems J. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28(6):1280–8.
Johnson RN, Chu DSH, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: Optimization of peptide length and polymer molecular weight. J Control Release. 2011;155(2):303–11.
Kang HC, Kang H-J, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32(4):1193–203.
Mann A, Thakur G, Shukla V, Singh AK, Khanduri R, Naik R, Jiang Y, Kalra N, Dwarakanath BS, Langel U, Ganguli M. Differences in DNA condensation and release by lysine and arginine homopeptides govern their DNA delivery efficiencies. Mol Pharm. 2011;8(5):1729–41.
Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7(10):8870–80.
Kizzire K, Khargharia S, Rice KG. High-affinity PEGylated polyacridine peptide polyplexes mediate potent in vivo gene expression. Gene Ther. 2012;20:407.
Philipp A, Zhao X, Tarcha P, Wagner E, Zintchenko A. Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA Delivery. Bioconjugate Chem. 2009;20(11):2055–61.
Nguyen HK, Lemieux P, Vinogradov SV, Gebhart CL, Guérin N, Paradis G, Bronich TK, Alakhov VY, Kabanov AV. Evaluation of polyether-polyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 2000;7:126.
Werfel TA, Jackson MA, Kavanaugh TE, Kirkbride KC, Miteva M, Giorgio TD, Duvall C. Combinatorial optimization of PEG architecture and hydrophobic content improves ternary siRNA polyplex stability, pharmacokinetics, and potency in vivo. J Control Release. 2017;255:12–26.
Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130(1):57–63.
Shim MS, Chang S-S, Kwon YJ. Stimuli-responsive siRNA carriers for efficient gene silencing in tumors via systemic delivery. Biomater Sci. 2014;2(1):35–40.
Shim MS, Kwon YJ. Acid-responsive linear polyethylenimine for efficient, specific, and biocompatible siRNA delivery. Bioconjug Chem. 2009;20(3):488–99.
Foster AA, Greco CT, Green MD, Epps TH III, Sullivan MO. Light-mediated activation of siRNA release in diblock copolymer assemblies for controlled gene silencing. Adv Healthc Mater. 2015;4(5):760–70.
Green MD, Foster AA, Greco CT, Roy R, Lehr RM, Epps TH III, Sullivan MO. Catch and release: photocleavable cationic diblock copolymers as a potential platform for nucleic acid delivery. Polym Chem. 2013;5(19):5535–41.
Greco CT, Andrechak JC, Epps TH III, Sullivan MO. Anionic polymer and quantum dot excipients to facilitate siRNA release and self-reporting of disassembly in stimuli-responsive nanocarrier formulations. Biomacromolecules. 2017;18(6):1814–24.
Greco CT, Muir VG, Epps TH III, Sullivan MO. Efficient tuning of siRNA dose response by combining mixed polymer nanocarriers with simple kinetic modeling. Acta Biomater. 2017;50:407–16.
Il’ichev YV, Schwörer MA, Wirz J. Photochemical reaction mechanisms of 2-nitrobenzyl compounds: methyl ethers and caged ATP. J Am Chem Soc. 2004;126(14):4581–95.
Zhao H, Sterner ES, Coughlin EB, Theato P. o-nitrobenzyl alcohol derivatives: opportunities in polymer and materials science. Macromolecules. 2012;45(4):1723–36.
Greco CT, Epps TH III, Sullivan MO. Mechanistic design of polymer nanocarriers to spatiotemporally control gene silencing. ACS Biomater Sci Eng. 2016;2(9):1582–94.
Greco CT, Akins RE, Epps TH III, Sullivan MO. Attenuation of maladaptive responses in aortic adventitial fibroblasts through stimuli-triggered siRNA release from lipid-polymer nanocomplexes. Adv Biosyst. 2017;1(8):1700099.
Alliegro M, Ferla R, Nusco E, De Leonibus C, Settembre C, Auricchio A. Low-dose gene therapy reduces the frequency of enzyme replacement therapy in a mouse model of lysosomal storage disease. Mol Ther. 2016;24(12):2054–63.
Kelkar SS, Reineke TM. Theranostics: combining imaging and therapy. Bioconjug Chem. 2011;22(10):1879–903.
Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. Monitoring the disassembly of siRNA polyplexes in serum is crucial for predicting their biological efficacy. J Control Release. 2010;141(1):38–41.
Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93.
Thibault M, Astolfi M, Tran-Khanh N, Lavertu M, Darras V, Merzouki A, Buschmann MD. Excess polycation mediates efficient chitosan-based gene transfer by promoting lysosomal release of the polyplexes. Biomaterials. 2011;32(20):4639–46.
Han L, Tang C, Yin C. Effect of binding affinity for siRNA on the in vivo antitumor efficacy of polyplexes. Biomaterials. 2013;34(21):5317–27.
Grandinetti G, Ingle NP, Reineke TM. Interaction of poly(ethylenimine)-DNA polyplexes with mitochondria: implications for a mechanism of cytotoxicity. Mol Pharm. 2011;8(5):1709–19.
Reineke TM, Davis M. Structural effects of carbohydrate-containing polycations on gene delivery. 1. Carbohydrate size and Its distance from charge centers. Bioconjug Chem. 2003;14:247–54.
Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34(1):322–33.
Dani C, Piechaczyk M, Audigier Y, Sabouty S, Cathala G, Marty L, Fort P, Blanchard J-M, Jeanteur P. Characterization of the transcription products of glyceraldehyde 3-phosphate-dehydrogenase gene in HeLa cells. Eur J Biochem. 1984;145(299–304):299–304.
Franch HA, Sooparb S, Du J, Brown NS. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem. 2001;276(22):19126–31.
Kurosaki T, Kitahara T, Fumoto S, Nishida K, Nakamura J, Niidome T, Kodama Y, Nakagawa H, To H, Sasaki H. Ternary complexes of pDNA, polyethylenimine, and γ-polyglutamic acid for gene delivery systems. Biomaterials. 2009;30(14):2846–53.
Duceppe N, Tabrizian M. Factors influencing the transfection efficiency of ultra low molecular weight chitosan/hyaluronic acid nanoparticles. Biomaterials. 2009;30(13):2625–31.
Lee J, Ryoo NK, Han H, Hong HK, Park JY, Park SJ, Kim YK, Sim C, Kim K, Woo SJ, Park KH, Kim H. Anti-VEGF polysiRNA polyplex for the treatment of choroidal neovascularization. Mol Pharm. 2016;13(6):1988–95.
Tseng S-J, Zeng Y-F, Deng Y-F, Yang P-C, Liu J-R, Kempson IM. Switchable delivery of small interfering RNA using a negatively charged pH-responsive polyethylenimine-based polyelectrolyte complex. Chem Commun. 2013;49(26):2670–2.
Hoffman AS. Stimuli-responsive polymers: biomedical applications and challenges for clinical translation. Adv Drug Deliv Rev. 2013;65(1):10–16.
Schlegel A, Largeau C, Bigey P, Bessodes M, Lebozec K, Scherman D, Escriou V. Anionic polymers for decreased toxicity and enhanced in vivo delivery of siRNA complexed with cationic liposomes. J Control Release. 2011;152(3):393–401.
Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 2006;116(1):83–9.
Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjug Chem. 2007;18:1391–6.
Kuntsche J, Horst JC, Bunjes H. Cryogenic transmission electron microscopy (cryo-TEM) for studying the morphology of colloidal drug delivery systems. Int J Pharm. 2011;417(1–2):120–37.
Aytar BS, Muller JP, Kondo Y, Abbott NL, Lynn DM. Spatial control of cell transfection using soluble or solid-phase redox agents and a redox-active ferrocenyl lipid. ACS Appl Mater Interfaces. 2013;5(17):8283–8.
Muller K, Naumann S, Weber W, Zurbriggen MD. Optogenetics for gene expression in mammalian cells. Biol Chem. 2015;396(2):145–52.
Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134(40):16480–3.
Sauers DJ, Temburni MK, Biggins JB, Ceo LM, Galileo DS, Koh JT. Light-activated gene expression directs segregation of co-cultured cells in vitro. ACS Chem Biol. 2010;5(3):313–20.
Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20(10):1041–4.
Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2011;13(1):21–35.
Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–55.
Chertok B, Langer R, Anderson DG. Spatial control of gene expression by nanocarriers using heparin masking and ultrasound-targeted microbubble destruction. ACS Nano. 2016;10(8):7267–78.
Kami D, Takeda S, Itakura Y, Gojo S, Watanabe M, Toyoda M. Application of magnetic nanoparticles to gene delivery. Int J Mol Sci. 2011;12(6):3705–22.
Wu B, Qiao Q, Han X, Jing H, Zhang H, Liang H, Cheng W. Targeted nanobubbles in low-frequency ultrasound-mediated gene transfection and growth inhibition of hepatocellular carcinoma cells. Tumour Biol. 2016;37(9):12113–21.
Lin DT, Liebert CA, Esquivel MM, Tran J, Lau JN, Greco RS, Mueller CM, Salles A. Prevalence and predictors of depression among general surgery residents. Am J Surg. 2017;213(2):313–7.
Liu YC, Ny ALML, Schmidt J, Talmon Y, Chmelka BF, Lee CT. Photo-assisted gene delivery using light-responsive catanionic vesicles. Langmuir. 2009;25(10):5713–24.
Hashmi JT, Huang Y-Y, Osmani BZ, Sharma SK, Naeser MA, Hamblin MR. Role of low-level laser therapy inneurorehabilitation. PM R. 2010;2(12Suppl 2):S292–S305.
Holanda VM, Chavantes MC, Wu X Fau - Anders JJ, Anders JJ. The mechanistic basis for photobiomodulation therapy of neuropathic pain by near infrared laser light. Lasers Surg Med. 2017;49(5):516–24.
Migliorati C, Hewson I, Lalla RV, Antunes HS, Estilo CL, Hodgson B, Lopes NNF, Schubert MM, Bowen J, Elad S. Systematic review of laser and other light therapy for the management of oral mucositis in cancer patients. Support Care Cancer. 2013;21(1):333–41.
Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324(5923):59–63.
Tomatsu I, Peng K, Kros A. Photoresponsive hydrogels for biomedical applications. Adv Drug Deliv Rev. 2011;63(14-15):1257–66.
Aujard I, Benbrahim C, Gouget M, Ruel O, Baudin JB, Neveu P, Jullien L. O-nitrobenzyl photolabile protecting groups with red-shifted absorption: syntheses and uncaging cross-sections for one- and two-photon excitation. Chemistry. 2006;12(26):6865–79.
Holmes CP. Model studies for new o-nitrobenzyl photolabile linkers - substituent effects on the rates of photochemical cleavage. J Org Chem. 1997;62:2370–80.
Greco CT, Epps TH III, Sullivan MO. Predicting gene silencing through the spatiotemporal control of siRNA release from photo-responsive polymeric nanocarriers. J Vis Exp. 2017;125:55803.
Tibbitt MW, Kloxin AM, Dyamenahalli KU, Anseth KS. Controlled two-photon photodegradation of PEG hydrogels to study and manipulate subcellular interactions on soft materials. Soft Matter. 2010;6(20):5100–8.
Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23(8):1868–76.
Qian J, Berkland C. pH-sensitive triblock copolymers for efficient siRNA encapsulation and delivery. Polym Chem. 2015;6(18):3472–9.
Adams D, Suhr OB, Dyck PJ, Litchy WJ, Leahy RG, Chen J, Gollob J, Coelho T. Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017;17(1):181.
Gambacorti-Passerini C, Antolini L, Mahon FX, Guilhot F, Deininger M, Fava C, Nagler A, Della Casa CM, Morra E, Abruzzese E, D’Emilio A, Stagno F, Le Coutre P, Hurtado-Monroy R, Santini V, Martino B, Pane F, Piccin A, Giraldo P, Assouline S, Durosinmi MA, Leeksma O, Pogliani EM, Puttini M, Jang E, Reiffers J, Valsecchi MG, Kim DW. Multicenter independent assessment of outcomes in chronic myeloid leukemia patients treated with imatinib. J Natl Cancer Inst. 2011;103(7):553–61.
Bartlett DW, Davis ME. Impact of tumor-specific targeting and dosing schedule on tumor growth inhibition after intravenous administration of siRNA-containing nanoparticles. Biotechnol Bioeng. 2008;99(4):975–85.
Abrams MT, Koser ML, Seitzer J, Williams SC, Dipietro MA, Wang W, Shaw AW, Mao X, Jadhav V, Davide JP, Burke PA, Sachs AB, Stirdivant SM, Sepp-Lorenzino L. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol Ther. 2010;18(1):171–80.
Pei Y, Hancock PJ, Zhang H, Cherrin C, Innocent N, Burke PA, Sachs AB, Sepp-Lorenzino L, Barnett SF. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010;16(12):2553–63.
Raab RM, Stephanopoulos G. Dynamics of gene silencing by RNA interference. Biotechnol Bioeng. 2004;88(1):121–32.
Cuccato G, Polynikis A, Siciliano V, Graziano M, di Bernardo M, di Bernardo D. Modeling RNA interference in mammalian cells. BMC Syst Biol. 2011;5:1–12.
Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4(5):438–46.
Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Lett. 2007;7(4):874–9.
Hamm A, Krott N, Breibach I, Blindt R, Bosserhoff AK. Efficient transfection method for primary cells. Tissue Eng. 2002;8(2):235–45.
Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol. 2013;65:108–19.
Weman SM, Salminen Us Fau - Penttila A, Penttila A Fau-Mannikko A, Mannikko A Fau - Karhunen PJ, Karhunen PJ. Post-mortem cast angiography in the diagnostics of graft complications in patients with fatal outcome following coronary artery bypass grafting (CABG). Int J Legal Med. 1999;112(2):107–14.
Nie T, Akins RE, Kiick KL. Production of heparin-containing hydrogels for modulating cell responses. Acta Biomater. 2009;5(3):865–75.
Robinson KG, Nie T, Baldwin AD, Yang EC, Kiick KL, Akins RE. Differential effects of substrate modulus on human vascular endothelial, smooth muscle, and fibroblastic cells. J Biomed Mater Res Part A. 2012;100(5):1356–67.
Shi Y, O’Brien JE, Fard A, Mannion JD, Wang D, Zalewski A. Adventitial myofibroblasts contribute to neointimal formation in injured porcine coronary arteries. Circulation. 1996;94(7):1655–64.
Robinson KG, Scott RA, Hesek AM, Woodford EJ, Amir W, Planchon TA, Kiick KL, Akins RE. Reduced arterial elasticity due to surgical skeletonization is ameliorated by abluminal PEG hydrogel. Bioeng Transl Med. 2017;2(2):222–32.
Yan B, Boyer JC, Branda NR, Zhao Y. Near-infrared light-triggered dissociation of block copolymer micelles using upconverting nanoparticles. J Am Chem Soc. 2011;133(49):19714–7.
Acknowledgements
The authors thank the National Institute of General Medical Sciences of the National Institutes of Health (NIH) for financial support through an Institutional Development Award (IDeA) under grant number P20GM103541, along with grant number P20GM10344615 during the writing of this manuscript. The statements herein do not reflect the views of the NIH. T.H.E. also thanks Thomas & Kipp Gutshall Professorship for financial support. The authors also thank Joy Smoker for assistance with the graphical abstract image.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Epps, T.H., Vi, T. & Sullivan, M.O. Design and development of a robust photo-responsive block copolymer framework for tunable nucleic acid delivery and efficient gene silencing. Polym J 50, 711–723 (2018). https://doi.org/10.1038/s41428-018-0077-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-018-0077-z