Abstract
DNA origami technology enables the precise assembly of well-defined two-dimensional and three-dimensional nanostructures with DNA, an inherently biocompatible material. Given their modularity and addressability, DNA origami objects can be used as scaffolds to fabricate larger higher-order structures with other functional biomolecules and engineer these molecules with nanometer precision. Over the past decade, these higher-order functional structures have shown potential as powerful tools to study the function of various bio-objects, revealing the corresponding biological processes, from the single-molecule level to the cell level. To inspire more creative and fantastic research, herein, we highlight seminal works in four emerging areas of bioapplications of higher-order DNA origami structures: (1) assisting in single-molecule studies, including protein structural analysis, biomolecule interaction analysis, and protein functional analysis, (2) manipulating lipid membranes, (3) directing cell behaviors, and (4) delivering drugs as smart nanocarriers. Finally, current challenges and opportunities in the fabrication and application of DNA origami-based functional structures are discussed.
Similar content being viewed by others
Introduction
The hierarchical self-assembly of simple units, such as proteins and nucleic acids, into sophisticated objects, such as organelles and cells, is the basic characteristic of life. Biological processes at different scales jointly determine the fate of living organisms. For example, nucleosome remodeling can influence gene expression; motor proteins are responsible for transporting various cargos; and cells communicate with each other to exchange materials or work together. Significant efforts have been devoted to developing methods to study these processes at multiple scales. However, they are more or less limited by the low precision and harsh experimental conditions. DNA nanotechnology, especially DNA origami technology, which is known for its high programmability and nanoscale precision, provides a powerful tool to study the function of various objects and reveal the corresponding biological processes.
DNA, as a self-assembly unit, is known for its unique properties: predictable and specific interactions, well-studied structural characteristics, convenient synthesis, and customizable functional modification. These properties make DNA the most potent material for nanofabrication. The history of DNA nanotechnology can be traced back to the proposal of “immobile junctions” by Ned Seeman in 19821. Subsequently, various DNA nanostructures were created and assembled into larger structures, including one-dimensional (1D) ribbons, two-dimensional (2D) arrays, and three-dimensional (3D) crystals2. Another milestone of DNA nanotechnology was the development of DNA origami, proposed by Rothemund in 20063, which enables the fabrication of almost arbitrarily shaped 2D and 3D DNA nanostructures4. These DNA origami nanostructures can be assembled into higher-order structures with other functional biomolecules, in which the DNA origami objects act as robust scaffolds to control the guest molecules at nanoscale precision5. Over the past decade, an increasing number of higher-order functional DNA origami structures have been created to probe the functions of molecules and cells during biological processes.
In this review, we intend to summarize recent advances in this field and inspire more fantastic research and applications. After a brief introduction to the development of DNA origami nanotechnology, we highlight design strategies for the fabrication of higher-order DNA origami structures, including the hierarchical assembly of DNA origami structures and the construction of hybrid DNA origami structures. Then, we focus on recent advances in using higher-order DNA origami structures as powerful tools to probe biological processes at multiple scales, from the single-molecule level to the cell level (Scheme 1). We illustrate how these functional structures have been used to facilitate protein structural analysis, detect interactions between biomolecules, study protein functions, regulate lipid membranes, and direct cell behavior. Design strategies to obtain DNA origami-based smart drug delivery systems for therapy are also discussed. Finally, we outline questions that remain outstanding and put forward our thoughts on future applications.
DNA origami nanotechnology and higher-order assembly of DNA origami nanostructures
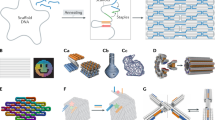
DNA origami is a bottom-up nanofabrication approach in which long single-stranded DNA (called scaffold DNA) is folded into well-defined shapes with the help of a set of short “staple strands” designed to be complementary to different parts of the scaffold DNA (Fig. 1a). In the original work, Rothemund created several single-layered planar structures, such as the rectangle, the triangle, the five-pointed star, and the representative smiley face (Fig. 1b)3. Subsequently, a variety of intricate 2D shapes were constructed, including a map of China and a dolphin6,7. To construct 3D DNA origami objects, different strategies have been developed8,9,10,11,12,13,14. Douglas and colleagues created various solid 3D objects by stacking pleated layers of helices into a honeycomb lattice in which each DNA helix is connected to three neighboring helices by crossovers (Fig. 1c)8. Later, Ke et al. proposed a square lattice to arrange DNA helices at a higher density (Fig. 1d)9. In their initial design, the double-helical twist density was higher than the preferred 10.5 bp per turn, which led to a compensatory global right-handed twist of the entire structure. By deleting some bases periodically, the twist can be diminished. In contrast, by rational design of the base density gradient, Dietz and coworkers created 3D DNA origami nanostructures with programmable twists and curvatures (Fig. 1e)11. In addition to the rigid lattice model, Han et al. constructed intricate DNA objects with curved surfaces, such as a DNA nanoflask (Fig. 1f), by manipulating the position and pattern of crossovers among a series of concentric rings12. Another vital strategy for DNA origami design was wireframe DNA origami. In 2013, Han et al. developed this strategy and used four-arm junctions to construct gridiron-like DNA nanostructures13. Subsequently, Zhang et al. used multiarmed junctions with tunable angles to construct a series of intricate finite-size wireframe DNA origami nanostructures, such as a flower-and-bird image (Fig. 1g)15. Compared to conventional DNA origami objects, whose helices are densely packed, the relatively loose wireframe origami structure requires fewer cations to shield the electrostatic repulsion between the helices, which may facilitate its bioapplications under the physiological environment.
a Schematic illustration of the assembly of DNA origami structures. A long single-stranded scaffold DNA (blue) is folded by a set of short staples (colored) into a prescribed shape. b DNA origami smiley face. (Reproduced with permission3, Copyright 2006, Macmillan Publishing Ltd.). c Square nut constructed from a honeycomb DNA lattice. (Reproduced with permission8, Copyright 2009, Macmillan Publishing Ltd.). d Solid block constructed from a square lattice. (Reproduced with permission9, Copyright 2009, American Chemical Society). e A curved 6-helix bundle. (Reproduced with permission11, Copyright 2009, AAAS). f A DNA nanoflask constructed from concentric rings. (Reproduced with permission12, Copyright 2011, AAAS). g A flower-and-bird image constructed from multiarmed DNA junctions. (Reproduced with permission15, Copyright 2015, Macmillan Publishing Ltd.
DNA origami structures with larger sizes and more complex conformations are desired to accomplish sophisticated tasks. Extending the length of the scaffold is a direct way to build larger DNA origami objects16,17,18. Alexandria and colleagues produced an ultralong 51-knt ssDNA scaffold using a λ/M13 hybrid virus and experimentally demonstrated the efficient assembly of the scaffold into asymmetric origami sheets18. However, as the length of the scaffold increases, many more staple strands are needed to fold it, which increases the cost. The hierarchical assembly of DNA origami nanostructures is a more feasible approach. Taking individual DNA origami objects as building blocks, we are able to fabricate various higher-order DNA origami structures, both finite and infinite19,20,21. Sticky-end hybridization and shape complementarity are two basic strategies for the hierarchical assembly of DNA origami objects. Iinuma et al. employed a self-limiting design strategy for the one-step self-assembly of wireframe DNA polyhedra via sticky ends (Fig. 2a)22. In this work, they tested two connector designs for the assembly of polyhedra and demonstrated that the dynamic connector design could increase assembly yields. Various wireframe DNA polyhedra were constructed using a stiff three-arm-junction DNA origami building block with controlled angles and arm lengths22. A DNA origami lattice was also constructed via sticky-end hybridization. Ke and coworkers designed a family of hexagonal DNA origami tiles and successfully constructed micrometer-scale 2D honeycomb lattices and tubes (Fig. 2b)23. It should be noted that the two interfaces responsible for connection should match each other, and nonspecific interactions between two interfaces would cause the unexpected aggregation of structures3,24. Considering DNA origami tiles as patch particles and adding flexible spacer segments between the long sticky ends and the main structure can avoid the impact of complex interfaces but sacrifices structural integrity25. However, the flexibility brought by the spacer segments could facilitate the assembly of DNA origami crystals26,27. The shape-complementarity strategy depends on base stacking, which is considered the primary force stabilizing DNA duplex structures28. In 2011, Rothemund et al. used base stacking to establish orthogonal stacking bonds, in the form of binary codes or shape complementarity, for the hierarchical assembly of 2D DNA origami tiles29. Subsequently, the Dietz group further expanded this method to establish specific stacking bonds at both the edges and the surfaces of 3D DNA origami nanostructures. Using this strategy, they created a variety of higher-order DNA origami structures with sizes approaching those of viruses and cellular organelles, such as 3D polyhedral assemblies and DNA origami lattices (Fig. 2c, d)30,31. In addition, they constructed many reversibly reconfigurable multistate DNA devices, e.g., the DNA origami robot31, based on the responsiveness of stacking bonds to environmental parameters, including temperature and cation concentrations. Other ingenious assemblies were constructed through special strategies. For example, a fractal assembly process was used to create micrometer-scale DNA origami arrays with arbitrary patterns, such as Mona Lisa (Fig. 2e)19. In addition, “meta-base pairs” were established to fold DNA origami six-helix bundles into meta DNA structures32. Recently, by integrating computer-aided engineering with a versatile computer-aided design approach, Huang et al. developed an iterative design pipeline and were able to design large, multicomponent DNA origami superstructures within several minutes33.
a A hexagonal prism assembled from 12 DNA origami tripods. (Reproduced with permission22, Copyright 2014, AAAS). b A 2D honeycomb lattice assembled from DNA origami hexagonal tiles. (Reproduced with permission23, Copyright 2016, American Chemical Society). c A 3D dodecahedron structure assembled from a DNA origami vertex. (Reproduced with permission30, Copyright 2017, Macmillan Publishing Ltd.). d A two-stranded filament assembled from a DNA origami hexagonal prism. (Reproduced with permission31, Copyright 2015 AAAS). e A 2D DNA origami array with a pattern of Mona Lisa. (Reproduced with permission19, Copyright 2017, Macmillan Publishing Ltd.). f Assembly of the 2D streptavidin protein array on the biotin-tagged DNA origami plate. (Reproduced with permission53, Copyright 2010, Macmillan Publishing Ltd.). g Spatially assembled thrombin nanoarray on the aptamer-tagged DNA origami plate. (Reproduced with permission54, Copyright 2007, American Chemical Society). h In situ assembly of tobacco mosaic virus coat protein on the DNA origami bond directed by RNA templates. (Reproduced with permission55, Copyright 2018, American Chemical Society). i Hierarchical assembly of DNA origami cuboid and coiled-coil peptide into 1D nanofibers. (Reproduced with permission59, Copyright 2020, American Chemical Society). j Hierarchical assembly of DNA origami plates and collagen-mimetic peptides into face-to-face stacking nanostructures via electrostatic attraction. (Reproduced with permission61, Copyright 2017, American Chemical Society).
Because of the modularity and addressability of DNA origami building blocks, other functional materials can also participate in the hierarchical assembly of DNA origami objects into higher-order structures, such as inorganic nanoparticles, polymers, and biomolecules34,35,36. Protein is the most important biological machine, and great effort has been devoted to the construction of DNA‒protein hybrid complexes37,38. A straightforward method is the use of a bifunctional crosslinking reagent to covalently conjugate an oligonucleotide tag to the reactive residues (e.g., lysine and cysteine) on the protein surface, which can hybridize to the complementary strand anchored on the DNA origami scaffold (Table 1). However, the abundant homogenous residues on the protein surface could result in poor control of the number and position of oligonucleotide tags, which could cause loss of activity of the protein and uncontrolled orientation of protein on the scaffold. Many efforts have been devoted to developing controllable protein modification strategies, such as reactive tag fusion and noncanonical amino acid mutation (Table 1)39,40,41,42,43,44,45,46,47,48. The histidine tag is widely used in the purification of proteins because of its high binding affinity for a nitrilotriacetic acid-Ni2+ (NTA-Ni2+) substrate through metal ion-mediated chelation. Goodman et al. designed a simple modification strategy that allows DNA functionalized with three NTA groups to be linked to the His6-tag of the recombinant proteins49. This noncovalent linkage is reversible and site specific, and has a high affinity (Kd∼6 nM). Shen et al. used NTA-DNA handles to pattern histidine-tagged enhanced green fluorescent proteins (EGFR) onto a 2D DNA origami nanosheet50. In addition to building connections directly, the concept of DNA-templated protein conjugation (DPTC) was proposed based on reversible His-tag-Ni2+-NTA-DNA conjugation51. A complementary DNA strand carrying a reactive group, e.g., N-hydroxysuccinimide (NHS), hybridizes with the tris(NTA)-DNA template, which carries the NHS group to a target area on the protein and facilitates the formation of a covalent crosslink of NHS to a lysine ε-amine. Due to the templating effect, the reaction is relatively rapid and stoichiometrically controllable. Natural histidine clusters on the protein surface could also be used to direct covalent conjugation51,52. Using this strategy, Ouyang and coworkers docked antibodies into the cavities of a DNA origami structure. Two tris(NTA)-DNA strands are fixed in the cavity to form NTA-Ni2+ complexes with the histidine clusters on the Fc domain. Subsequently, activated NHS esters form covalent linkages to the antibody. As a result, antibodies are immobilized on the DNA origami substrate, and the cavity allows fine control over the orientation of the antibody. Other noncovalent receptor‒ligand interactions could also be utilized to immobilize proteins without chemical modification. The advantages of such strategies are the predictable number of linkages and binding sites, which can facilitate the precise control of protein molecules and prevent the adverse effects of chemical modification on protein function. Voigt et al. utilized biotinylated DNA handles to assemble a 2D streptavidin (STV) array on a DNA origami plate (Fig. 2f)53. By selectively cleaving or coupling the biotin linkers, they also demonstrated single-molecule chemical reactions on the DNA origami. This strategy is attractive because of the high affinity of STV-biotin binding (Kd ≈ 10−14 M). However, due to the large size of STV, many proteins may not be appropriate for STV fusion due to the potential disruption of native protein structures38.
Aptamer-protein interaction is another attractive strategy to attach proteins onto the DNA origami scaffold since aptamers can readily hybridize to the DNA origami templates and bind to a specific site on the target protein as the natural antibody does. As a proof of concept, thrombin aptamers were placed on the DNA origami surface to pattern thrombin proteins into defined shapes (Fig. 2g), and the specificity of the aptamer-protein interaction also allows the precise patterning of different proteins on the same template54. In addition, RNA templates can be used to assemble proteins, such as virus coat protein, and DNA origami tiles into higher-order structures. Zhou and colleagues used tobacco mosaic virus (TMV) genome-mimicking RNA strands to guide the assembly of TMV coat proteins onto DNA origami scaffolds (Fig. 2h)55. They also showed the dynamic assembly of TMV coat proteins via controlled release of the RNA template from the DNA origami scaffold56. Similar to RNA templates, we can utilize specific binding between DNA and protein to fabricate hybrid structures as well. An exciting design was presented by Praetorius et al. in which transcription activator-like (TAL) effector protein dimers can be used as staple proteins to fold double-stranded DNA templates into user-defined shapes57. Every staple protein is built by connecting the C-terminus of one TAL to the N-terminus of another TAL and can recognize two distinct sites of the dsDNA scaffold. By carefully arranging the binding sites, the dsDNA scaffold could coassemble with TAL staple proteins into user-defined geometric shapes, such as the circle, the square, and the four-helix bundle. In another case, Mayo et al. used a computational approach to construct a DNA‒protein hybrid structure. They fused a three-helix protein, Drosophila engrailed homeodomain (ENH), which contains a native DNA-binding domain, with a two-helix homodimerization protein. The authors constructed irregular bulk nanoparticles or nanowires with single-molecule widths by tuning the number and position of protein-binding sites on the DNA strand58. Although the structures in these works are not strictly DNA origami structures, these cleaving strategies could be readily applied to the construction of other higher-order DNA origami structures.
The conjugation of oligonucleotide tags with other biomolecules, such as peptides and lipids, is relatively easy. Buchberger et al. hybridized two DNA-modified peptides to the opposite surfaces of a cubic DNA origami block (Fig. 2i)59. The two peptides can comprise a coiled-coil heterodimer pair and assemble the DNA origami building blocks into micrometer-long 1D bundles. A similar structure was assembled to quantify the interaction between two peptides60. In addition, peptides can control the assembly of DNA origami objects by mimicking collagen. Jiang et al. designed two peptides, CP++ and sCP++, with a sequence comprising a central neutral block and two positively charged domains at the N- and C-termini. The collagen-mimetic peptides can bind to the negatively charged DNA origami objects and direct face-to-face stacking of the 2D DNA origami nanosheets to form 1D nanowires (Fig. 2j)61. Additionally, Udomprasert and coworkers created a DNA origami platform to guide the assembly of amyloid fibrils62. They positioned an amyloid fibril peptide-DNA conjugate inside the DNA origami nanotube, which could serve as a nucleation site and organize amyloid fibrils. The DNA nanotube-fibril complex could be further assembled with a DNA origami platform, thereby controlling the orientation and location of the amyloid fibril. Lipid molecules could be covalently crosslinked with functionalized DNA handles and precisely anchored on the surface of DNA origami. These lipid molecules could anchor DNA origami tiles onto the lipid membrane surface and assist the hierarchical assembly of DNA origami tiles63. They can also act as nucleation sites to recruit other free lipid molecules to direct the assembly of lipid molecules under the control of DNA origami scaffolds64. We will discuss some interesting examples in Section “Regulating lipid membranes”.
Elucidation of biological processes with higher-order DNA origami structures
With the development of DNA origami technology, the focus of attention in this field has turned from the construction of sophisticated and beautiful structures to the fabrication of higher-order functional structures as powerful tools to explore problems in other fields, e.g., physics, biophysics, biology, materials science, and computer science65,66,67,68. Over the past decade, DNA origami nanotechnology has shown potential in various bioapplications. At the heart of all these applications lies the precise control of biomolecules by DNA origami scaffolds in these higher-order structures. There are three key parameters. First, the number of biomolecules can be well controlled by the DNA origami nanostructure. The most direct method is to attach a defined number of handles that can catch biomolecules to the DNA origami template. By controlling the number of different handles, two or more biomolecules can be combined in a specific stoichiometric ratio. As mentioned above, two proteins can be patterned on the same DNA origami plate without interfering with each other by controlling the number and position of two different aptamers54. The precise control of protein number is of great significance for exploring a series of biological processes, including molecule transportation in vivo and the catalytic mechanism of enzymes.
Second, the position of biomolecules can be regulated via protruding handles at different sites on the DNA origami object. Most DNA origami structures are based on B-form DNA, which is a right-handed double helix with a helical pitch of 10.5 base pairs or 3.4–3.6 nm and a diameter of 2 nm. Along the direction of the DNA helix, the orientation of the staple strand changes periodically, and any outward point can be set as an anchor point to immobilize guest molecules. It should be noted that not all outward points can be utilized at the same time. The resolution is generally limited to the minimal distance between handles attached to staple strands, typically 3–5 nm. In 2020, Wickham et al. designed a 3D DNA-barrel pegboard that provides a rhombic-lattice canvas of a thousand pixels with a pitch of ~8 nm69. When two or more molecules are placed on the DNA origami object, the spacing can also be precisely tuned by adjusting the position of each molecule. With this ingenious design, the average distance between fluorescent molecules could be adjusted from 1.5 to 9 nm. The smallest displacement step possible is 0.04 nm, which is slightly less than the Bohr radius70.
Third, the orientation of each biomolecule can be controlled as well. Early studies on the control over the orientation of biomolecules were mainly carried out on small DNA nanostructures. In 2006, Erben et al. constructed a rigid tetrahedral DNA cage and used the intrinsic helicity of DNA to control the orientation of cytochrome c71. The protein was covalently conjugated to the 5’ end of oligonucleotide S1, which combined with other DNA strands to form tetrahedra. By altering the sequence of S1, the orientation of cytochrome c was shifted from the inside to the outside of the cage. In DNA origami systems, the linker used to immobilize biomolecules to DNA origami objects is usually soft and does not participate in the assembly of the main structure, resulting in relatively poor control over the biomolecule. Selecting the appropriate fixed point according to the shape of the DNA origami structure can help control the orientation of the protein molecule72. In an interesting case, Ouyang et al. designed a DNA origami cube with a cavity to control the orientation of a Y-shaped antibody. The Fc domain of the antibody was docked into the cavity, and the wall of the cavity oriented the Fab domain outward52. Recently, Zhou et al. demonstrated that the relative orientation of TMV rods could be controlled with DNA origami. By tuning the position and length of the TMV genome mimic RNA strands on the DNA origami template, the orientation of TMV rods was precisely controlled55. This kind of control is mainly derived from the steric hindrance of the DNA origami structure. Additionally, precise control of protein orientation can be achieved by utilizing specific interactions between protein and DNA-binding sites. Martin et al. designed a hollow 3D DNA origami structure with a binding site for transcription factor p53 lying in the center. Thus, the orientation of the p53 protein can be tuned by changing the position of the binding site73. In addition, using a rigid connector could be helpful in controlling the orientation of the protein74.
We emphasize that all the biological applications of higher-order DNA origami structures discussed in this section are based on precise control of the number, position and orientation of biomolecules by the DNA origami scaffolds. Therefore, in the following parts of this section, we focus on utilizing higher-order functional structures to reveal biological processes at different scales, from manipulating biomolecules to regulating lipid membranes to directing cell behaviors.
Manipulating biomolecules
Assisting in structural analysis of protein
Proteins are the basic building blocks of living issues and the most important players in almost all biological processes. How a protein works and what it does is determined by its 3D shape. Solving the structure of proteins is crucial to elucidating their functional mechanisms. DNA origami structures have been applied to assist in the structural analysis of proteins. Indeed, the birth of nanotechnology originated from the idea of Professor Seeman to use oligonucleotides as a structural material to create self-assembled 3D crystals and then to use these addressable frameworks to immobilize proteins in a periodic 3D array75. Significant efforts have been devoted to constructing protein‒DNA hybrid crystals while obtaining a good crystal and solving the structure of the protein in the crystal remain challenging76,77.
The development of cryo-electron microscopy technology dramatically facilitates the determination of protein structure, while it remains challenging to obtain a high resolution of the target protein, especially for those that are small and prone to aggregation78,79. Higher-order DNA origami structures could assist in the structural analysis of proteins in many ways. First is image acquisition: by combination with large DNA origami signposts, small proteins, which have a low contrast against the background, can be more easily discerned in images. In addition, DNA origami can protect the protein from the detrimental effects of cryo-EM sample preparation, such as adsorbing to the air‒water interface, which also facilitates the acquisition of high-quality images. Second, the position and orientation of the target protein can be tuned precisely in the higher-order DNA origami structure, and the robust and highly anisotropic structure of DNA origami can function as a marker to facilitate the class averaging of images of the target protein. In 2016, Martin et al. designed a hollow 3D DNA origami structure with a tunable binding site for transcription factor p53 lying in the center (Fig. 3a)73. The helix containing the binding site could be used to adjust the relative orientation of p53 complexes to provide more structural information. They reconstructed the structure to a final resolution of ~15 Å and settled an ongoing debate about the symmetry of the p53 tetramer bound to DNA. In 2020, Aksel and coworkers constructed another DNA origami goniometer to solve the structure of a DNA binding protein, BurrH, to a resolution of 6.5 Å. The DNA origami goniometers had 14 different stage configurations to orient the protein, and each of them had a distinct barcode pattern, which facilitated particle classification80. Last but not least, higher-order DNA origami structures can provide a favorable environment for characterizing the structure of membrane proteins by attaching lipids in the cavity of a DNA origami object81. Dong et al. designed a barrel-like DNA origami object with an inner hydrophobic lipid channel to accommodate a membrane protein82. The membrane proteins docked into the cavity could maintain the monodispersed and native-like state. Using this method, they successfully characterized the 3D structure of α-hemolysin at a resolution of approximately 30 Å. Compared to lipid nanodiscs, the DNA origami scaffold can be readily tuned to match membrane proteins with different sizes. In early experiments, the use of DNA origami structures to assist in the structural analysis of proteins did not meet expectations. Many factors could hinder the application of DNA origami objects in this area, such as the relative displacement between the DNA origami support and protein and the superposition of the signal from the support structure and the target protein. Developing a method capable of establishing rigid and well-defined connections between proteins and DNA origami scaffolds is key to facilitating this application. Although the development of structural biology methods has made it easier to solve protein structures, DNA origami can still help in several ways. For example, DNA origami templates can be used to fix the conformation of a protein. By controlling the distance between two interacting proteins, such as antibodies and antigens, the protein structure under a specific situation could be determined. More structural details could be obtained in such a way.
a A DNA origami support with a docking site for the DNA-binding P53 protein for cryo-EM structure analysis. The relative angle of the P53 protein is controlled by rotating the docking site in the DNA origami object, and the P53 protein is protected in the hollow support structure from the detrimental effects of sample preparation. (Reproduced with permission73, Copyright 2016, Proceedings of the National Academy of Sciences). b A DNA origami clip consists of two beams and a torsional spring for determining the interaction between two nucleosomes. The distance and relative orientation of two nucleosomes can be adjusted, and the interaction between two nucleosomes will shift the angle distribution of the clips, which reflects the energy landscape of nucleosome-nucleosome stacking. (Reproduced with permission94, Copyright 2016, AAAS). c A DNA origami rod for exploring the mechanisms of the coordinate motor ensemble. Dynein and kinesin can be immobilized on the DNA origami rod in different combinations, and the fluorescent signal is used to characterize the walking of the complex structure on the microtubule track. (Reproduced with permission103, Copyright 2012, AAAS). d The GOx/HRP enzyme pair arranged at different distances on a DNA origami plate for interenzyme substrate diffusion investigation. (Reproduced with permission114, Copyright 2012, American Chemical Society). e A DNA origami-protein hybrid Nano Trap for studying the selective filtering properties of intrinsically disordered Phe-Gly-rich nucleoporins. (Reproduced with permission130, Copyright 2021, American Chemical Society).
Assisting in the analysis of interactions between biomolecules
Identifying interactions between biomolecules, such as protein−protein interactions (PPIs), is of pivotal importance in revealing many biological processes because most proteins do not function alone83,84. Previous studies proved that abnormal PPIs account for many diseases, including cancer, neurodegenerative diseases, and autoimmune diseases85,86. As a result, determining the mode and strength of PPIs is of great clinical significance and could facilitate drug discovery, such as inhibitors and oncology drugs87,88. Many methods have been developed to detect PPIs, e.g., coimmunoprecipitation technology, yeast two-hybrid technology, surface plasmon resonance technology, and protein microarray technology89. However, conventional detection techniques are usually experimentally time-consuming and lack single-molecule manipulation precision.
The programmable flexibility and addressability of DNA origami objects enable the fabrication of nanodevices that show great potential for detecting interactions between proteins. Proteins of interest can be placed at designed spacing and relative orientation on the DNA origami scaffold at nanoscale resolution, which permits the analysis of the interaction between two biomolecules in a given mode. More specifically, DNA origami scaffolds can engineer two molecules in an unusual status, which could be associated with the protein status in disease.
Ke et al. designed a rhombus-shaped DNA‒protein hybrid “nanoactuator” that can serve as a general platform for studying weak molecular interactions at the single-molecule level90. Two split eGFP fragments were anchored on two arms of the “nanoactuator”, the conformations of which could be controlled by adding different struct-locking strands. Then, they used fluorescence to monitor the conformational change of the hybrid “nanoactuator” and thus reveal the interaction between the two split eGFP fragments. In this study, the eGFP fragments were hybridized to extending handles on the arms, although the flexibility of the handles may disadvantage the control of the orientation of the protein fragment. In the design created by Shaw and colleagues, the oligonucleotide handles conjugated to the protein were involved in forming the main structure of the DNA origami plate rather than bonded to the flexible handles to achieve more precise control of the spacing between antigens. Using this system, they found the tolerance of IgG antibodies to be between 3 and 17 nm and demonstrated considerable differences in spatial tolerance between IgM and IgG and between low- and high-affinity antibodies91. The structure and dynamics of nucleosomes regulate the interaction of genomic DNA with transcription, replication, and DNA repair. To investigate the structural dynamics of nucleosomes, Le et al. designed a DNA-based nanocaliper and attached the two DNA ends of the nucleosome to the ends of the two arms. Thus, the angle of the nanocaliper can be used as a direct readout of structural changes in nucleosomes. With this device, the authors detected the process of transcription factor (TF) binding to its target site and demonstrated that TF binding would partially unwrap the nucleosome and increase the probability of TF occupancy92. At almost the same time, Funke and colleagues constructed a high-resolution DNA origami spectrometer to analyze the interactions within nucleosome complexes93,94. They first studied the salt-induced disassembly of nucleosome core particles93. The unwrapping of the nucleosomal template DNA was coupled to the conformation of the DNA spectrometer and could be directly measured by TEM and Förster resonance energy transfer (FRET). They found that nucleosome dissociation was constant at different ionic strengths and was not affected by the integration of nucleosomes. The same group also employed the DNA origami positioning spectrometer to reveal the energy landscape for nucleosome association (Fig. 3b)94. In their device, single DNA strands, which hybridize with the complementary strands on the beams of the DNA origami scaffold, are added to the nucleosome template DNA to immobilize nucleosomes at selected positions on the two beams. Then, the positioning device placed the two nucleosomes close to each other in a defined relative orientation, and a tunable DNA spring at the hinge was used to counteract the attraction interactions between the nucleosomes. With this device, Funke et al. were able to measure nucleosome-nucleosome distance frequencies via single-particle electron imaging and derived the energy landscape for nucleosome pair interactions. As a result, they found a shallow but long-range (~6 nm) attractive nucleosome pair potential with a minimum of −1.6 kcal/mol close to direct contact distances. Furthermore, they also found that modification of histones, such as H4 acetylation and removal of the histone tail, dramatically decreased the interaction strength.
In addition, DNA origami nanostructures could be used to analyze DNA‒DNA interactions and DNA‒protein interactions. Nickels et al. created a DNA origami nanoscopic force clamp that can work as an array, enabling high-throughput study of interactions95. The single-stranded DNA springs exerted tunable tension in the low piconewton range on a molecular system whose conformational changes could be monitored by measuring single-molecule FRET between two dyes on the DNA origami framework. Using this force clamp, they observed the TATA-binding protein-induced bending of a DNA duplex under tension and further proved the mechanosensitivity of gene regulation. In 2020, Kramm used the same device to analyze the effect of DNA strain on the formation of the initiation complex96. Increasing the number of binding partners is a routine approach to obtain robust binding of the target molecule. At the same time, the arrangement of the binding partners in space also plays an important role. Rafat and coworkers constructed a 2D array of DNA origami-based nanocavities in which aptamers could be placed with known mechanical properties at defined distances and orientations, and each type of nanocavity was labeled with specific barcodes for distinction97. The DNA origami array allowed analysis of the interaction between aptamers and the target protein in parallel under the same conditions and in high throughput. They concluded that the binding geometry and mechanical properties of aptamers have a dramatic effect on the binding strength. Such higher-order DNA origami structures could be extended to probe interactions between protein ligands and receptors or between small molecules and protein receptors after appropriate modification. By arranging single DNA origami structures into arrays, interactions between larger objects could also be detected. Dutta et al. designed the first DNA origami tension probe (DOTP), which reflects the changes in tension on the probe via changes in fluorescence intensity. They used the DOTP to measure tension forces applied by human blood platelets during initial adhesion and activation98.
Due to their customizability and excellent mechanical properties, DNA origami objects can also be used as components to advance other molecular mechanical technologies. Optical tweezers are a method to exert forces or torques on individual molecules or to directly measure the forces or torques generated in their biochemical reactions99. Molecules of interest are linked to the probes of optical tweezer via a molecular linker. However, the flexibility of the linker is disadvantageous for measurement with a low force since it could result in a low signal-to-noise ratio. DNA origami objects can function as a stiff linker to enhance the signal-to-noise ratio and arrange the biomolecules in a defined conformation. To demonstrate this concept, Pfitzner et al. used a stiff DNA helix bundle as the molecular linker to probe the interaction of the DNA hairpins100. In such a way, they improved the resolution of single-molecule force spectroscopy in the low-force regime and provided a multivalent platform for conjugating other target molecules. Similarly, Kilchherr and colleagues designed a stiff beam-like DNA origami nanostructure as the linker to measure the forces and lifetimes of DNA base-pairing and base-stacking interactions with dual-beam optical tweezers101. The stiff DNA origami beams not only suppressed noise but also enabled the repeated detection of unbinding and rebinding of stacking contacts through a flexible polymer tethering two beams. They tested all 16 sequence combinations of blunt ends and found that the strength of the base stacking was sequence dependent: the strongest stacking force (−3.42 kcal/mol) occurred between a CG:CG pair, and the weakest stacking was between a TA:CG pair (−0.81 kcal/mol)101. In these studies, in addition to serving as a rigid linker to suppress background noise, DNA origami objects also provided a template to precisely control the number and position of the molecules to be detected. This approach has the potential to detect some complex molecular interactions, such as the interaction between bispecific antibodies and antigens. With some ingenious design, it allows the repeated probing of a given pair of molecules, which is critical in the analysis of biomolecule interactions101.
Assisting in functional studies of proteins
In organisms, the realization of a function often requires the cooperation of multiple biomolecules. By controlling how they cooperate, we can regulate their function and thus dissect the underlying mechanism. Relying on the precise controllability of DNA nanotechnology, a wide variety of higher-order DNA origami structures were fabricated to study the function of biomolecules, especially proteins. In this section, we discuss the construction of DNA‒protein hybrid structures to probe motor protein movement, enzyme catalysis, and the function of nuclear pore proteins as typical examples.
Cytoplasmic dynein and kinesin are microtubule-based motors that transport cargos in opposite directions and are involved in various essential biological processes102. In 2012, Derr and colleagues designed a DNA origami-based system to investigate the mechanisms that coordinate motor ensemble behavior (Fig. 3c)103. Motor proteins were anchored at the surface of the DNA origami bundle in tunable types, numbers, spacings, and orientations. By monitoring the movement of the DNA origami-motor protein complex on the microtubule (MT) at the single-molecule level using a fluorescence signal, they demonstrated that the motor number had a minimal effect on directional velocity. When opposite-polarity motor proteins were placed on the same DNA origami, they engaged in a tug-of-war, and the motor with higher affinity won the game. Recently, Abdellatef et al. fabricated a model system to study the oscillatory movement of dynein104. Two kinesin-modified DNA origami bundles were used to crosslink two MTs to restrict their relative movements caused by dynein. They observed obvious oscillatory movement and bending motions of the dynein-MT-DNA-origami complexes. Myosin, which interacts with actin to generate force and movement, is the motor protein responsible for muscle function. Hariadi and coworkers assembled groups of myosin V and VI motors onto a 2D DNA origami scaffold to investigate the collective motion in myosins105. They found that single myosin V and VI dimers displayed similar skewed trajectories on the keratocyte actin network. However, compared to myosin V with a flexible arm, the trajectories of myosin VI, whose arm is relatively rigid, progressively straightened as the number of myosin VI molecules increased. The results of Hariadi et al. suggested that structural features of myosin may confer selective advantages in cellular functions. The same group also applied a DNA origami tube to reveal the motion of larger ensembles of motors106. Myosins were anchored on the DNA origami tubes at various spacing, and DNA origami objects were hierarchically assembled into long crystalline 1D tracks with several micrometers. In this way, this higher-order structure could be used to mimic the interactions on the length scale of a muscle sarcomere. Interestingly, similar to cytoplasmic dynein and kinesin, neither myosin density nor spacing had a significant effect on the gliding speed of actin filaments. In the above examples, myosin proteins are directly anchored to the DNA origami structure by DNA hybridization. However, in native thick filament and thin filament complexes, myosin is subject to geometric restriction in force generation. To further explore this process, Fujita and colleagues constructed a DNA origami-based thick filament in which myosin proteins were attached to the DNA bundle backbone in a restricted geometry to mimic the native thick filament74. Using high-speed atomic force microscopy to monitor the movement of the artificial complex, they found that the free myosin head interacted weakly with actin filaments and bound firmly to the forward region. Upon binding, the two-step lever-arm swing stopped mainly at the first step and occasionally reversed direction. In another interesting case, Iwaki and coworkers constructed a DNA origami nanospring to investigate the effect of mechanical forces on the stepping mode of myosin VI proteins107. The authors demonstrated that a hand-over-hand mechanism was used for the transport of cargos at low load, and an inchworm-like mechanism was used at high loads. Recently, Rai et al. used DNA origami nanotubes to explore the multimodal regulation of myosin VI activity through the cargo adaptor GAIP-interacting protein108.
Enzymes, as unique catalysts in organisms, have always attracted great attention. Almost all metabolic processes in cells are inseparable from enzymes. Enzymes can greatly accelerate the rate of chemical reactions in these processes so that the substances and energy produced by metabolism can meet the needs of organisms. DNA origami structures are used to study the function of enzymes in two main approaches The first one is to act as an artificial nanocompartment to regulate the protein and substrate reactions. Indeed, one of the first 3D DNA origami structures was envisioned to encapsulate enzymes109. Grossi et al. constructed a DNA origami nanoscale vault and loaded a bovine alpha-chymotrypsin in its cavity110. The DNA vault could be reversibly opened/closed via a multilock mechanism and thus control the access of substrates to the enzyme in the cavity. Similarly, Ijäs et al. designed a DNA origami nanocapsule to encapsulate horseradish peroxidase111. The DNA nanocapsule was locked by a DNA triplex that is sensitive to the pH of the surrounding solution. Therefore, the nanocapsule could be opened and closed by changing the pH to control the catalysis reaction. To achieve controlled release, other “locks” were also used, including protein recognition and light irradiation, which will be discussed in Section “DNA origami-based smart drug delivery systems”. In another case, a DNA origami plate containing a T7 RNA polymerase and multiple target‒gene substrates was created to function as a gene logic chip112. By controlling the intermolecular distances between the enzyme and the target genes, gene expression could be regulated.
The second approach is to study the enzyme cascades. It should be noted that early explorations of enzymatic cascades were performed on nanostructures assembled by DNA tiles113. With the development of DNA origami technology, its structural rigidity and addressability enable more precise control over the number, position and orientation of enzymes, making it an ideal platform for exploring enzyme cascade reactions114,115,116,117,118. Fu et al. were the first to demonstrate this concept by organizing discrete glucose oxidase-horseradish peroxidase (Gox-HRP) enzyme pairs on a 2D DNA origami plate with tunable spacing (from 10 to 20, 45 and 60 nm) to investigate the distance-dependent catalysis activity (Fig. 3d)114. Strongly enhanced activity was observed when enzyme pairs were closely spaced, while the activity dropped dramatically for enzyme pairs separated by 20 nm or more. The authors proposed that Brownian diffusion of intermediates (H2O2 to this enzyme pair) in solution governed the variations in activity at long distances, while the dimensionally limited diffusion of intermediates contributed to the enhancement in activity at shorter distances. They also found that a noncatalytic protein bridge connecting the hydration shells of the enzyme pair could enhance the activity. Subsequently, Morii et al. used a DNA origami template to study two different enzymes, xylose reductase and xylitol dehydrogenase, at defined distances115. In this study, the authors demonstrated that the intermediate, NADH, diffused to the second enzyme by Brownian motion and contributed to the increased yield when two enzymes were placed in close proximity, which is consistent with the work of Fu et al. Instead of controlling the distance between two paired enzymes, the Yan group developed another exciting strategy to regulate the activity of the enzyme cascade119. In their design, an NAD+ molecule (the intermediate of glucose-6-phosphate dehydrogenase (G6pDH) and malic dehydrogenase (MDH)) was directly organized between the enzymes by a DNA handle to mimic the natural swinging arm, facilitating the direct transfer of substrate between the G6pDH-MDH enzyme pair. Using this device, they systematically studied the effects of interenzyme distances, enzyme orientations, and stoichiometry on enzymatic activity. This swinging arm also afforded high specificity in a complex environment by selectively establishing a substrate channel between two defined enzymes. Subsequently, the same group applied this strategy to study the selective enzyme cascade reaction on a DNA origami scaffold120. The NAD+ swinging arm was anchored among three enzymes (G6pDH, MDH, and lactate dehydrogenase (LDH)) and fixed with two anchor strands between the enzymes. By reconfiguring the swinging arm between different enzyme pathways, they could control substrate channeling and thus regulate the ON-OFF states of different enzyme pairs120. In 2018, Yang et al. assembled an artificial 2D enzyme network of G6pDH-LDH on a wireframe DNA origami scaffold. They found a higher reaction efficiency than that of single enzyme pairs due to the enhanced transfer121.
The strategies mentioned above are mainly based on substrate channeling. Recent work, however, offered different viewpoints. Zhang et al. have shown that the activity of enzymes benefits from the lower pH environment near the protein surface, caused by the negatively charged DNA origami structure, compared to that in the bulk solution, which provides a better reaction environment for the enzyme122. Zhao et al. assembled the Gox-HRP enzyme pair in a DNA origami nanocage to study the impact of both encapsulation and proximal polyanionic surfaces. They found that the distal polyanionic surfaces of the nanocage enhance the stability of active enzyme conformations through a strongly bound hydration layer123. In 2019, Klein et al. evaluated the kinetics of seven different three-enzyme configurations and suggested that the reason for activity enhancement is the increased enzyme stability on the DNA origami scaffold and a localized DNA surface affinity or hydration layer effect instead of the enzyme-to-enzyme channeling mechanism124. Recently, Kahn et al. used an open, 3D DNA wireframe octahedron to create a library of spatial organizations of glucose oxidase and horseradish peroxidase for systematic exploration of the contribution of enzyme distribution to cascade activity72. They demonstrated that the colocalization of cascaded enzymes to the DNA origami structure dominated the increase in cascade activity compared to other spatial parameters, and the DNA origami scaffold could serve as a bridge connecting the enzyme environments at larger length scales. The debate on the reasons for the enhanced catalytic activity of the enzymes in the DNA origami-enzyme complexes is clearly far from over, and more work, both experimental and theoretical, is needed to explore these systems. This point also suggests that when we use DNA origami scaffolds to regulate protein functions, in addition to the influence of the spatial arrangement of proteins, the influence of the DNA origami structure itself should not be ignored.
Apart from enzyme cascades, DNA origami scaffolds have also been used to study the oligomerization process of proteins. Rosier et al. used a DNA origami plate to assemble caspase-9 monomers (both wild-type and inactive protein) into different multiprotein complexes. They demonstrated that enzymatic activity is induced by proximity-driven dimerization with half-of-site reactivity and revealed a multivalent activity enhancement in oligomers of three and four enzymes125. Kurokawa and coworkers designed a perforated DNA origami platform to reconstruct Kir3 K+ channels in vitro126. Local changes in protein structure can also be amplified by DNA origami structures. In an interesting example, Kosuri et al. designed a DNA-enzyme complex to track the DNA rotations that result from unwinding by the helicase and transcription by RNA polymerase127. The DNA origami blades were attached to the enzyme via a double-stranded DNA segment, which also serves as the substrate of the enzymes, and the blades not only amplified the motion of the DNA but also minimized the obscuring effect of Brownian fluctuations. They characterized a series of events during the unwinding of DNA and revealed some mechanisms of helicase and RNA polymerase.
In addition, DNA origami rings that mimic the conformation of natural nuclear pores were used to organize intrinsically disordered nucleoporins into artificial nuclear pore complexes, which acted as model systems to study the function of intrinsically disordered proteins and thus reveal the gatekeeper mechanism of the nuclear pore complex. Fisher et al. constructed a biomimetic DNA origami nanopore with different FG-Nups (nucleoporins rich in Phe-Gly amino acid residues) anchored at the inner wall. They demonstrated that the overall morphology of the assemblies was determined by the type and density of FG repeats128. Using a similar DNA origami ring, Ketter and colleagues compared the assembly behaviors of two different NSP1 Nups (NSP1, a model yeast FG-Nup, and NSP1-S, a hydrophilic mutant) and demonstrated that NSP1 formed a denser pore than NSP1-S129. Derived from the DNA nanoring, a DNA origami nanotrap was constructed to study the selective filtering capability of the nuclear pore complex. Molecular bites were preanchored at the bottom of the trap, and FG-Nups formed a selective barrier above them. By detecting the molecules bound to the bites, they demonstrated the critical parameters of an effective diffusion barrier and how the nuclear transport receptor Ntf2 can selectively transport model cargo through nanotraps (Fig. 3e)130.
Regulating lipid membranes
Lipid membranes are essential elements for all life that serve as a boundary structure to isolate the cytoplasm from the outer environment and maintain the stability of cells. Many biological processes are related to it. It participates in budding, tubulation, fission and fusion, characteristics essential for cell division, biological reproduction, and intracellular membrane trafficking131. Manipulating the shape of lipid membranes in an artificial system to mimic the changes they undergo under physiological conditions can help us understand these biological processes. However, it takes work to achieve this. With the development of chemical conjugation methods, it is easy to conjugate oligonucleotide tags to lipid molecules or hydrophobic moieties. As a result, these molecules can be anchored onto DNA origami structures at designed positions via hybridization to act as handles, enabling the DNA origami objects to interact with lipid membranes and shape them.
In 2014, inspired by the “cytoskeleton–membrane protein–lipid bilayer” principle of the cell membrane, an exciting strategy termed “frame-guided assembly (FGA)” was proposed by the Liu group to direct the assembly of amphiphiles64. In their strategy, leading hydrophobic groups (LHGs) were first anchored at defined positions of the frame, where they functioned as nucleation seeds to guide the amphiphiles to grow and form customized 2D or 3D membrane structures. The group first demonstrated this method on AuNPs132. Poly (aryl ether) dendron was chosen as a model LHG and conjugated to a DNA strand and an oligo (ethylene glycol) (OEG) on two ends to form the DDOEG. Then, DDOEGs were anchored onto the surface of AuNPs, and subsequently, the LHGs could guide amphiphilic molecules to assemble into heterovesicles. By choosing different frames and small amphiphilic molecules, they were able to prepare monodisperse vesicles of various shapes and sizes133. Wang et al. used a transmembrane peptide as the LHG to guide the assembly of lipid molecules on the AuNP frame134. Using pH-sensitive peptides as LHGs, the assembly process of liposomes could be controlled by changing the pH of the solution135. Compared with inorganic nanoparticles, DNA origami structures have richer structural diversity. Moreover, due to the addressability of the DNA origami structure, LHGs can be placed at defined positions on the frame, achieving more precise control over the assembled structure. Zhou et al. applied the FGA strategy to the 2D assembly of amphiphilic molecules based on 2D DNA origami frames. They demonstrated that the shapes of the assemblies were precisely controlled by the LHGs preanchored on the template (Fig. 4a)136. Dong and coworkers further applied this strategy to 3D DNA origami frames137. Subsequently, Yang and coworkers constructed four different circular DNA origami rings as outer frames to synthesize size-tunable liposomes138. DNA strands modified with the hydrophobic group DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine N-[4-(p-maleimidophenyl) butyramide]) were hybridized to the inner handles and pointed toward the center of the ring. These hydrophobic groups acted as nucleation seeds to recruit free lipids for the formation of liposomes (Fig. 4b). Compared to the extrusion method and ultrasound method, the liposomes synthesized in this approach were more uniform due to the limitation of DNA origami frames.
a Frame-guided assembly of amphiphilic molecules on DNA origami nanosheets. (Reproduced with permission136, Copyright 2016, Wiley-VCH). b Synthesis of size-controlled liposomes with DNA origami templates. c Vesicle tubulation controlled by the DNA origami nanosprings. DNA origami monomers are absorbed onto the vesicle with the help of amphipathic peptides and polymerized into nanosprings via linker strands, which can shape vesicles into tubules. (Reproduced with permission143, Copyright 2018, Wiley-VCH). d A synthetic lipid membrane channel created with a DNA origami structure inserted into a lipid membrane via cholesterol moieties. (Reproduced with permission145, Copyright 2012, AAAS). e A synthetic DNA origami channel equipped with a size-selective gating system for the transport of macromolecules. (Reproduced with permission150, Copyright 2019, Macmillan Publishing Ltd.). f A DNA origami-SNARE hybrid platform for exploring the membrane-fusion process. (Reproduced with permission153, Copyright 2016, American Chemical Society).
Hierarchical assembly of DNA origami building blocks could form large templates that can manipulate the lipid membrane at a larger scale. Based on their previous work, Zhang and colleagues utilized modular DNA origami frames to remodel liposomes with designer geometries: tubular liposomes, toroidal liposomes, helical liposomes and lipid tubule arrays139. They also demonstrated the dynamic control of liposome shape by DNA template reconfiguration. In natural biological processes, the remodeling of the cell membrane is constructed by specific proteins. Thus, many works were inspired by natural membrane-sculpting proteins. Czogalla et al. designed an amphipathic DNA origami structure as a membrane-scaffolding tool to mimic the biological activity of coat-forming proteins such as the I-/F-BAR family140. The DNA origami scaffold has a flat membrane-binding interface and can form a 2D array via overhangs at the side facets to induce the deformation of giant vesicles. To further elucidate whether the “banana-like” shape of the BAR protein can be uncoupled from other functional features, Franquelim et al. constructed a variety of BAR protein-mimicking DNA origami structures and characterized their ability to participate in membrane binding and transformation141. They demonstrated that the BAR protein-mimicked DNA origami scaffold could induce membrane deformation dependent on curvature, membrane affinity and surface density and reproduce the activity of membrane-sculpting proteins. In addition, a clathrin-inspired three-armed DNA origami nanostructure was designed by Journot et al. to induce submicrometer deformation of a lipid monolayer142. In addition to the deformation of vesicles mentioned above, the tubular shape is another important membrane structure in cellular processes. Inspired by the ESCRT machine, Grome and colleagues created DNA origami curls to draw tubules from vesicles. After adding linker strands, DNA origami curls preabsorbed to the vesicles would polymerize into “nanosprings” and transform the spherical vesicles into membrane protrusions and tubule vesicles with the help of amphipathic peptides (Fig. 4c)143. Furthermore, the same group designed various DNA origami curls with stiffness and amphipathic peptide density to analyze the effect of structural stiffness and affinity on the remodeling of lipid membranes144. These studies not only reconstructed the lipid membrane shape with a higher-order DNA origami structure but also provided important insights into how the lipid membrane is regulated by natural proteins to regulate cellular behavior.
Membrane channels are essential for allowing cells to exchange materials with the outer environment, through which cells can concentrate valuable chemicals and output metabolic waste, maintaining the internal environment. The construction of artificial membrane channels that mimic natural ones could provide a powerful platform to probe these processes. DNA origami structures show great potential in this field due to their controlled pore size. In 2012, Langecker et al. designed the first DNA origami transmembrane channel145. The DNA origami object consisted of a stem that penetrated and spanned a lipid membrane and a barrel-shaped cap that adhered to the membrane via cholesterol moieties (Fig. 4d). Similar to natural ion channels, this artificial channel also displayed current gating behavior, which was considered to be caused by thermal fluctuations. Langecker et al. also used this artificial channel to study DNA hairpin unzipping and guanine quadruplex unfolding and thus discriminate analyte molecules that pass through145. Subsequently, Krishnan and colleagues synthesized a DNA membrane channel, termed the “T pore”, with a larger central pore (~4 nm)146. After being attached to the membrane, this channel allowed the translocation of single-stranded and double-stranded DNA analytes. Thereafter, DNA origami channels with larger pores were constructed to allow larger molecular cargos to pass through147,148,149. However, as the size of the pore increases, both small and large molecules can pass, resulting in a decrease in the selectivity of the channel. To solve this problem, Thomsen and coworkers designed a 9 nm wide DNA nanopore equipped with a size-selective gating system150. In particular, they used a PEG plug to narrow the nanopore. Thus, only small molecules can pass through. After the removal of the plug, larger molecules can pour out from the vesicles (Fig. 4e). Although size-selective gating could be achieved by the controlled unplugging system, the order is limited from small to large substances, and a concentration gradient of the substances is essential. Recently, Dey and colleagues designed a reversibly gated DNA origami channel, the lid of which could be closed and reopened via a lock-and-key mechanism, allowing the controlled transport of functional proteins through membranes151. It is of great value to design artificial channels that have the ability to transport with specific selectivity or against concentration gradients such as natural channels. In addition, DNA origami objects can assemble into higher-order structures with larger pores or form multiple parallel channels for a closer look at the transport process of substances.
In addition to artificial membrane channels, higher-order DNA origami structures are used to explore other material exchange processes between lipid membranes. Bian and colleagues created a tunable DNA origami platform to study lipid transfer by the synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain of extended synaptotagmin 1152. In this model, the SMP domain was anchored to donor liposomes, and the distances between the donor and acceptor liposomes were controlled via the DNA pillars. They analyzed the lipid transfer via a FRET-based assay and demonstrated that the SMP domain could transfer lipids at a distance that exceeds the length of an SMP dimer152. Membrane fusion is another critical process that drives inter- and intracellular trafficking and communication. Studies have revealed that soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complexes are the engines for membrane fusion, while how SNAREs cooperate remains unclear. Xu et al. designed a DNA origami ring and placed different numbers of SNAREs on the ring. Liposomes were formed in the DNA origami ring, and the DNA‒protein-liposome complexes were anchored to the target membrane via DNA handle hybridization, which enabled bypass of the docking step and allowed direct observation of individual membrane-fusion events. By monitoring the membrane fusion process, they demonstrated that one to two pairs of SNAREs were sufficient to drive fast lipid mixing at a short distance (Fig. 4f)153.
Directing cell behaviors
At a larger scale, where cells become participants in the life process, higher-order functional DNA origami structures could display their power as well. The behavior of the cell is regulated by a set of nanoscale molecular events, e.g., the interaction of ligands and receptors. The binding of ligands to corresponding receptors activates downstream signaling pathways and then directs the behavior of the cell. In the immune system, the continued binding of antigens to B-cell receptors can cause receptors to form clusters and activate B cells154. However, the exact antigen-receptor binding mode, especially the effect of the spatial distribution of antigen on B-cell activation, remains poorly understood. DNA origami nanostructures can form defined higher-order structures with antigens and control the spatial distribution of antigens with nanoscale precision. Previous studies have shown that DNA origami can be used to explore the relationship between the binding affinity of antigen-antibody and the distance between antigens91. Therefore, it is practical to construct “synthetic cells” with DNA origami scaffolds, on which the type, number, and spacing of antigens could be tailored, for studying the process of immune cell activation. Veneziano and coworkers displayed the clinical vaccine immunogen eOD-GT8 on different DNA origami scaffolds (icosahedral and rod-shaped) to systematically investigate the impact of the spatial distribution of eOD-GT8 antigen on B-cell activation in vitro155. By monitoring the activation of B cells with different synthetic antigen-DNA origami particles, they demonstrated that a valency of five or more antigens and nearest-neighbor spacings of approximately 25–30 nm would maximally activate B cells (Fig. 5a). This study also provided a potential platform to investigate the spatial relationships with immunogens of other viral pathogens. DNA origami scaffolds were also used to explore the T-cell activation process. Whether preclustered pMHC contributes to the formation of TCR clusters and activates T cells has long been doubted. Hellmeier et al. designed a robust 2D DNA origami plate and placed antigens at defined positions156. The DNA plate was anchored to fluid-phase planar supported lipid bilayers to allow lateral movement, which enabled nanoscale control over ligand distances without interfering with the dynamics of receptor clustering. They demonstrated that, unlike the high-affinity ligands that activate T-cells only when they are in close proximity, monomeric agonist pMHCs stimulate T-cells via serial, rapid, and short-lived engagement (Fig. 5b)156. Recently, Wang et al. used a DNA origami plate to pattern anti-CD3 into different spacing and geometric arrangements to evaluate the spatial effect of TCR on T-cell signaling157. They suggested that early activation of T cells favored high-density ligand geometric arrangement, while the signal of late T-cell activation was linearly correlated with anti-CD3 valence. Dong et al. designed a DNA origami pegboard functionalized with an engineered receptor system to investigate the influence of the spatial configuration of clusters on the MAPK signaling response in T cells, including the threshold and duration of the MAPK signal158. In addition, a similar DNA origami plate was used to study the phagocytosis process of macrophages driven by Fcγ receptors159. Kern and coworkers constructed an artificial DNA-CARγ system and used the DNA origami plate to control the size and spatial arrangement of ligand clusters while maintaining the total number of ligands on the surface of beads. They found that reducing ligand spacing could potently enhance engulfment, and increasing the number of single ligand‒receptor complexes was insufficient to recapitulate this effect, which suggested the key role of the receptor cluster in phagocytosis. Recently, Zhang and colleagues designed a DNA nanoball ~74 nm in diameter to display SARS-CoV-2 RBDs, forming a virus-like particle (VLP) to enhance the immune response160. They found that, compared to evenly distributed RBDs, a concentrated RBD distribution promoted faster and stronger interaction with the host cell. However, VLPs with even RBD distributions showed much better immune activation effects. DNA origami-based VLPs provide a feasible tool to study viral infection and could facilitate the development of antiviral vaccines. In another interesting case, Angelin et al. combined the bottom-up self-assembly of protein‒DNA structures with the top-down micropatterning of solid surfaces to construct multiscale origami structures as an interface for cells (MOSAIC)161. In the MOSAIC system, the number, stoichiometry, and precise nanoscale orientation could be controlled, thus providing a potential tool to address questions in cell signaling.
a Antigen-DNA hybrid NPs for studying the role of nanoscale antigen organization on B-cell activation. eOG-GT8 antigens are patterned on two DNA origami scaffolds into different spatial arrangements to investigate the impact of the spatial distribution of antigen on B-cell activation. b A laterally mobile DNA origami platform functionalized with antigens for exploring the mechanism of T-cell activation by pMHCs. (Reproduced with permission156, Copyright 2021, Proceedings of the National Academy of Sciences). c DNA origami structure-directed cell‒cell communication. DNA origami structures are used to direct the cell‒cell communications between different cells in defined modes. (Reproduced with permission165, Copyright 2020, American Chemical Society).
In addition to being regulated by ligand binding, cells can communicate with each other by directly connecting to neighboring cells. Cell‒cell communication is a critical process for maintaining biological functions, and constructing artificial cell-cluster models can help us better understand the principles governing multicellular self-organization and communications. Compared to other methods being used to construct artificial cell clusters162,163,164, DNA origami objects provide a convenient approach to organizing cell assemblies with tunable spatial orientation and precisely controlling the cell types. Ge and coworkers designed a DNA origami platform to organize homotypic and heterotypic cells in 3D space165. Multiple DNA origami building blocks were oligomerized to match the size of the cell, and specific ligands were placed at designed sites on the DNA origami platform to bind corresponding cells in the controlled orientation. They investigated different types of cell‒cell communications, including gap junctions, tunneling nanotubes, and immune/tumor cell interactions with this platform (Fig. 5c). Using a similar DNA object, Liu et al. further reconstructed a soma–soma synapse-like junction between cells166. Although the use of DNA origami to direct cell behavior is still in the proof-of-concept stage, features such as the ease of functional modification, nanoscale precision, and hierarchical assembly make DNA origami a promising platform to direct cell behavior. The construction of larger DNA origami platforms with sizes close to those of cells can help us further investigate these processes.
DNA origami-based smart drug delivery systems
An important goal of our exploration of life processes is to understand the causes of disease and develop effective therapies. In past decades, with the development of nanotechnology, various nanocarriers have come into the spotlight and attracted tremendous attention because they can improve the effectiveness and minimize the adverse effects of conventional drugs167,168. The unique features of DNA structures, such as tailored geometries, programmable interactions, and outstanding biocompatibility, make them advantageous nanocarriers for drug delivery. Many well-conducted studies have been performed in this area, especially framework nucleic acid nanostructures. Readers are referred to the following excellent reviews for systematic information169,170,171. In this section, we focus on the recent developments in the design strategies for constructing DNA origami-based smart drug delivery systems.
Targeting and controlled release are two key features for the design of smart drug delivery nanocarriers. The physicochemical properties of nanocarriers have significant effects on their biodistribution and clearance in vivo, including size, hydrophobicity, and charge. A living organism is a highly heterogeneous system, different parts of which have different physical and chemical properties, resulting in different biodistributions of different nanocarriers. For example, compared to small nanocarriers (10–20 nm), which are widespread in various organs, some larger nanocarriers (typical 20 nm < d < 1 μm) tend to be retained in tumor tissues since the pores in tumor vessels are much larger (100 nm to 1 μm) than the tight endothelial junctions of normal vessels (5 to 10 nm size)168,172. In addition, some lipid-soluble drugs require a nanocarrier that provides a hydrophobic environment, while some protein drugs, such as antibodies, require a hydrophilic environment. Therefore, we often customize nanocarriers according to different therapies. Customization is an important feature of DNA origami objects. The typical size of a single DNA origami object is tens to hundreds of nanometers, while we are able to construct tiny ones with shortened scaffolds or larger ones with elongated scaffolds17,18,173. In addition, higher-order DNA origami structures at the micrometer scale could be fabricated, as mentioned above. In 2018, Bastings et al. systematically investigated the uptake of 11 DNA nanostructures of different conformations and sizes in three cell lines174. They found that larger particles with greater compactness were preferentially internalized compared with elongated, high-aspect-ratio particles. In addition, the kinetics of the cellular uptake of nanoparticles is closely related to cell type, and the shape of DNA nanostructures has little effect on it. To further explore the uptake and distribution of DNA origami nanostructures in vivo, Jiang et al. radiolabeled a series of DNA origami nanostructures of different sizes and shapes with Cu-64 for positron emission tomography imaging175. All of them tended to accumulate in the kidneys of healthy mice and mice with acute kidney injury (AKI), while the free M13 scaffold and partially folded DNA objects showed increased liver sequestration. Interestingly, even without drug loading, the rectangular DNA origami object exhibited a therapeutic effect on mice with AKI. In addition, the surface chemistry of nanocarriers may affect their biodistribution. The feasible surface modification of the DNA origami structure allows us to conjugate other materials to change the properties of nanocarriers to meet the requirements of drug delivery. Typically, the electrostatic repulsion between the negatively charged sugar-phosphate backbone and cell membranes prevents the internalization of DNA origami objects. However, after surface modification with cowpea chlorotic mottle virus capsid proteins, the 2D DNA origami plate showed a 13-fold delivery efficiency compared to bare plates176. Ponnuswamy et al. demonstrated that oligolysin-coated DNA origami barrels showed significant resistance to low salt and DNase I and exhibited efficient uptake by endosomal compartments and an increase in pharmacokinetic bioavailability177. In addition, as mentioned above in Section “Regulating lipid membranes”, lipid membranes can be coated onto DNA origami objects as well64. The lipid membrane could improve the internalization of DNA origami nanocarriers and may function as a hydrophobic matrix to load hydrophobic drug molecules. In the future, more studies on these coated DNA origami objects are needed to explore their distribution in organisms and uptake by cells.
Active targeting is another way to construct smart DNA origami drug nanocarriers by adding targeting molecules to the surface. Aptamers are the most commonly used active targeting molecules due to their simple hybridization process, and multiple kinds of aptamers have been used to target DNA origami nanocarriers to tumor cells. Zhao et al. attached MUC1 aptamers to the edges of a 2D DNA nanosheet to selectively deliver cytotoxic protein ribonuclease to execute their cell-killing function178. Similarly, doxorubicin was loaded onto aptamer-modified DNA origami plates for cancer chemotherapy179,180. In addition to tumor-targeted aptamers, other aptamers can be used to achieve drug delivery to different targets. Mela and colleagues designed a perforated DNA origami plate functionalized with aptamers designed to target E. coli and B. subtilis bacterial strains as the vehicle to deliver lysozyme to gram-positive and gram-negative bacteria (Fig. 6a)181. In addition, some small-molecule ligands are also utilized for targeted delivery, such as DUPA (2-[3-(1,3-dicarboxy propyl)-ureido] pentanedioic acid) and folate182,183.
a The E. coli and B. subtilis bacterial strain-targeted aptamer-functionalized DNA origami nanocarriers for antimicrobial delivery. (Reproduced with permission181, Copyright 2020, Wiley-VCH). b A reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo molecules. The pH-responsive latches are DNA triplexes that form at low pH and disassemble at high pH. (Reproduced with permission111, Copyright 2019, American Chemical Society). c A DNA nanorobot for triggered cargo exposure based on AND logic. The sequestered antibody fragments are exposed for cell signaling regulation when both aptamer-based locks are attached to the targeted cell surface markers. (Reproduced with permission189, Copyright 2012, AAAS). d A tubular DNA nanorobot for siRNA/chemo-drug codelivery. TAT peptides are attached to the outer surface of the nanorobot to improve cellular uptake and tumor retention. DNA locks containing disulfide bonds are used to seal the nanotube and release the siRNA cargos upon interacting with glutathione in the cytoplasm. (Reproduced with permission193, Copyright 2021, Wiley-VCH).
Controlled drug release is another way for DNA origami nanocarriers to improve therapeutic effects and reduce toxic side effects. With the development of DNA origami technology, an increasing number of dynamic structures that can transform in response to external signals have been designed, laying the foundation for DNA nanocarriers to achieve controlled release. A typical example is the DNA origami box with a controllable lid constructed by Anderson et al. in 2009109. They constructed the box by folding up six linked DNA origami sheets and functionalized it with a dual lock–key system that could be opened by toehold-mediated strand displacement. Similar designs have been used to control the on/off switching of other DNA containers, such as the DNA nanovault and the DNA nanosphere110,184. Other signals can also be used to control the transformation of DNA origami objects185. Recently, the dual lock-key system of the DNA box was replaced by a protein recognition aptamer for protein sensing186. Using a pH-sensitive DNA triplex structure, Ijäs et al. constructed a pH-responsive nanovault that could be reversibly opened and closed by changing the pH of the surrounding solution (Fig. 6b)111. Light irradiation is another convenient trigger to regulate the conformation of DNA origami carriers, and it can also be used to directly cleave the anchor of the drug molecule to release it from the nanocarrier187,188.
Based on the design strategies above, smart therapeutic DNA origami nanorobots were constructed that can transport molecular payloads to target locations, respond to external simulations, and release the payload in a controlled manner. In 2012, Douglas and colleagues described a logic-gated DNA robot capable of opening and exposing its payloads in response to cell-signaling stimulation189. The large inner chamber of the DNA robot could be used to load various payloads, and the “dual lock-key system” was designed to consist of different logical AND gates that can respond to a wide array of cues (Fig. 6c). Recently, a cleverly designed DNA nanorobot was constructed by Li et al. for cancer therapy in vivo190. They loaded thrombin onto one side of a single-layer DNA origami sheet and rolled it up with nucleolin-binding aptamers. Thus, the thrombin molecules were protected from circulating platelets and plasma fibrinogen. After being intravenously injected into mice with breast tumors, the DNA nanorobots recognized nucleolin on tumor-associated endothelial cells and exposed thrombin to induce intravascular thrombosis, resulting in tumor necrosis and inhibition of tumor growth. In these designs, the aptamer system functioned both as the targeting molecule and as a switch for controlled release. Subsequently, DNA origami carriers were constructed to combine gene therapy and chemotherapy191,192. Wang et al. designed a tubular DNA nanodevice loaded with Dox and small interfering RNAs on its inner wall193. Transactivators of transcription peptides were added to the edge of the DNA nanocarrier to improve cellular uptake and tumor retention. Since the linkers contained disulfide bonds, the nanocarrier would open and release Dox and siRNA in response to glutathione in the tumor (Fig. 6d). Despite showing great potential, there are still many problems plaguing the development of DNA origami-based smart drug delivery systems. For example, the stability of DNA nanostructures in the cellular environment has long been questioned due to native DNase. Recent studies demonstrated that the degradation of DNA origami nanostructures was significantly slower than that of bare double-stranded DNA when exposed to nucleases194 or cell lysate195 and even when injected into mice175. In addition, many groups have developed coating strategies to improve the structural stability of DNA origami196,197,198,199. Much work is still needed to enhance the physiological stability of DNA origami structures, as well as to investigate the metabolic and kinetic processes of DNA origami nanocarriers in vivo.
Summary and prospects
Forty years ago, Professor Seeman proposed the concept of immobile DNA junctions, laying the foundation of DNA nanotechnology. Today, not only can we use the technique of DNA origami to construct various fascinating structures, but we can also use DNA origami objects to fabricate a series of functional nanodevices to undertake complex tasks. In recent years, given their programmability and addressability, many higher-order functional DNA origami structures have demonstrated great promise to manipulate various objects with nanoscale precision and thus reveal the corresponding biological processes at multiple scales. Particular attention is given to utilizing DNA origami scaffolds to study multiple aspects of proteins, including structure, interaction, and function. At larger scales, lipid membranes and cells can also be regulated by higher-order DNA origami structures, providing a powerful platform for probing their biological functions. Despite these success stories, many challenges and opportunities remain. First, it remains challenging to construct micrometer-sized DNA origami structures while retaining full addressability. Such higher-order structures could provide a larger platform to manipulate objects such as organelles and cells while retaining nanoscale precision. Special strategies have been tried to achieve this goal. Tikhomirov et al. employed a fractal assembly strategy to produce a 2D DNA origami array that could be used to render extremely complex images such as the Mona Lisa19. Zhou et al. designed orthogonal and directional bonding to assemble 3D hexagonal prisms into finite-sized hierarchical structures, which maintained the structural integrity and addressability20. However, in these strategies, the multistep assembly process often results in a low final yield and hinders application. Optimized assembly strategies are in great demand to fabricate well-defined higher-order DNA origami structures to bridge the gap between nanoscale and macroscale engineering. Second, recent studies have shown that the DNA origami structure itself affects the state and activity of biomolecules immobilized on the structure. For example, biomolecules display fast folding kinetics with increased mechanical and thermodynamic stabilities in the DNA origami structure due to the confined space200. Negatively charged DNA origami structures can cause the pH environment near their surface to be lower than that of the bulk solution, which affects the state and activity of the protein attached to them122. In addition, some molecules can nonspecifically adhere to the surface of DNA origami structures and alter their surface chemistry. For example, a protein corona was observed to form on DNA nanostructures201. The biomolecules to be studied can be sensitive to these factors; therefore, it is necessary to take these factors into account when applying DNA origami structures to manipulate functional biomolecules. Chemical modification strategies could be helpful to alter the properties of DNA origami structures to enhance their stability and minimize unintended effects, which are of great importance to applications in vivo, although it would be difficult to fully retain the addressability of the surface. Finally, it is important to construct DNA origami nanodevices that can consume certain types of chemical energy (similar to motor proteins using ATP) to do work. Such devices can act like automated nanorobots to perform various tasks in living organisms, extending the application of DNA structures to a previously unimaginable level. Recently, Pumm and colleagues constructed the first nanomotor that can actually perform measurable mechanical work202. The DNA nanomotor can keep rotating in the same direction under an alternating electric field through a mechanism known as a flashing Brownian ratchet and thus store energy by winding up a DNA “spring”. This work is exciting and can shed light on the design of DNA origami-based nanomotors. It is foreseeable that sustained efforts will be dedicated to fabricating DNA origami-based nanodevices to implement sophisticated tasks both in vitro and in vivo and open avenues to explore the fascinating biological world.
References
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Chandrasekaran, A. R. & Zhuo, R. A ‘tile’ tale: Hierarchical self-assembly of DNA lattices. Appl. Mater. Today 2, 7–16 (2016).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Dey, S. et al. DNA origami. Nat. Rev. Dis. Prim. 1, 13 (2021).
Hong, F., Zhang, F., Liu, Y. & Yan, H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017).
Qian, L. et al. Analogic China map constructed by DNA. Chin. Sci. Bull. 51, 2973–2976 (2006).
Andersen, E. S. et al. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano 2, 1213–1218 (2008).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Ke, Y. et al. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 131, 15903–15908 (2009).
Ke, Y., Voigt, N. V., Gothelf, K. V. & Shih, W. M. Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 134, 1770–1774 (2012).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Han, D. et al. DNA gridiron nanostructures based on four-arm junctions. Science 339, 1412–1415 (2013).
Jun, H. et al. Rapid prototyping of arbitrary 2D and 3D wireframe DNA origami. Nucleic Acids Res. 49, 10265–10274 (2021).
Zhang, F. et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 10, 779–784 (2015).
Zhang, H. et al. Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 48, 6405–6407 (2012).
Chen, X. et al. Self-assembly of large DNA origami with custom-designed scaffolds. ACS Appl. Mater. Interfaces 10, 24344–24348 (2018).
Marchi, A. N., Saaem, I., Vogen, B. N., Brown, S. & LaBean, T. H. Toward larger DNA origami. Nano Lett. 14, 5740–5747 (2014).
Tikhomirov, G., Petersen, P. & Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 552, 67–71 (2017).
Zhou, Y., Dong, J., Zhou, C. & Wang, Q. Finite assembly of three-dimensional DNA hierarchical nanoarchitectures through orthogonal and directional bonding. Angew. Chem. Int. Ed. 61, e202116416 (2022).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2018).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Wang, P. et al. Programming self-assembly of DNA origami honeycomb two-dimensional lattices and plasmonic metamaterials. J. Am. Chem. Soc. 138, 7733–7740 (2016).
Ye, J., Helmi, S., Teske, J. & Seidel, R. Fabrication of metal nanostructures with programmable length and patterns using a modular DNA platform. Nano Lett. 19, 2707–2714 (2019).
Tigges, T., Heuser, T., Tiwari, R. & Walther, A. 3D DNA origami cuboids as monodisperse patchy nanoparticles for switchable hierarchical self-assembly. Nano Lett. 16, 7870–7874 (2016).
Wang, Y. et al. DNA origami single crystals with Wulff shapes. Nat. Commun. 12, 3011 (2021).
Lin, Z. et al. Engineering organization of DNA nano-chambers through dimensionally controlled and multi-sequence encoded differentiated bonds. J. Am. Chem. Soc. 142, 17531–17542 (2020).
Yakovchuk, P., Protozanova, E. & Frank-Kamenetskii, M. D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 34, 564–574 (2006).
Woo, S. & Rothemund, P. W. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011).
Wagenbauer, K. F., Sigl, C. & Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science 347, 1446–1452 (2015).
Yao, G. et al. Meta-DNA structures. Nat. Chem. 12, 1067–1075 (2020).
Huang, C.-M., Kucinic, A., Johnson, J. A., Su, H.-J. & Castro, C. E. Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 20, 1264–1271 (2021).
Lan, X. & Wang, Q. DNA-programmed self-assembly of photonic nanoarchitectures. NPG Asia Mater. 6, e97 (2014).
Hannewald, N. et al. DNA origami meets polymers: a powerful tool for the design of defined nanostructures. Angew. Chem. Int. Ed. 60, 6218–6229 (2021).
Kong, G. et al. DNA origami-based protein networks: from basic construction to emerging applications. Chem. Soc. Rev. 50, 1846–1873 (2021).
Stephanopoulos, N. Hybrid nanostructures from the self-assembly of proteins and DNA. Chem. 6, 364–405 (2020).
Zhou, K. et al. Toward precise manipulation of DNA-protein hybrid nanoarchitectures. Small 15, e1804044 (2019).
Duckworth, B. P. et al. A universal method for the preparation of covalent protein–DNA conjugates for use in creating protein nanostructures. Angew. Chem. Int. Ed. 46, 8819–8822 (2007).
Tominaga, J. et al. An enzymatic method for site-specific labeling of recombinant proteins with oligonucleotides. Chem. Commun. 4, 401–403 (2007).
Takahara, M., Hayashi, K., Goto, M. & Kamiya, N. Enzymatic conjugation of multiple proteins on a DNA aptamer in a tail-specific manner. Biotechnol. J. 11, 814–823 (2016).
Meyer, R. & Niemeyer, C. M. Orthogonal protein decoration of DNA nanostructures. Small 7, 3211–3218 (2011).
Nguyen, T. M., Nakata, E., Saimura, M., Dinh, H. & Morii, T. Design of modular protein tags for orthogonal covalent bond formation at specific DNA sequences. J. Am. Chem. Soc. 139, 8487–8496 (2017).
Saccà, B. et al. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. 49, 9378–9383 (2010).
Chen, R. P., Blackstock, D., Sun, Q. & Chen, W. Dynamic protein assembly by programmable DNA strand displacement. Nat. Chem. 10, 474–481 (2018).
Min, D., Arbing, M. A., Jefferson, R. E. & Bowie, J. U. A simple DNA handle attachment method for single molecule mechanical manipulation experiments. Protein Sci. 25, 1535–1544 (2016).
Rosier, B. J. H. M. et al. Incorporation of native antibodies and Fc-fusion proteins on DNA nanostructures via a modular conjugation strategy. Chem. Commun. 53, 7393–7396 (2017).
Marth, G. et al. Precision templated bottom-up multiprotein nanoassembly through defined click chemistry linkage to DNA. ACS Nano 11, 5003–5010 (2017).
Goodman, R. P. et al. A facile method for reversibly linking a recombinant protein to DNA. Chembiochem 10, 1551–1557 (2009).
Shen, W., Zhong, H., Neff, D. & Norton, M. L. NTA directed protein nanopatterning on DNA origami nanoconstructs. J. Am. Chem. Soc. 131, 6660–6661 (2009).
Rosen, C. B. et al. Template-directed covalent conjugation of DNA to native antibodies, transferrin and other metal-binding proteins. Nat. Chem. 6, 804–809 (2014).
Ouyang, X. et al. Docking of antibodies into the cavities of DNA origami structures. Angew. Chem. Int. Ed. 56, 14423–14427 (2017).
Voigt, N. V. et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 5, 200–203 (2010).
Chhabra, R. et al. Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures. J. Am. Chem. Soc. 129, 10304–10305 (2007).
Zhou, K., Ke, Y. & Wang, Q. Selective in situ assembly of viral protein onto DNA origami. J. Am. Chem. Soc. 140, 8074–8077 (2018).
Zhou, K., Zhou, Y., Pan, V., Wang, Q. & Ke, Y. Programming dynamic assembly of viral proteins with DNAorigami. J. Am. Chem. Soc. 142, 5929–5932 (2020).
Praetorius, F. & Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 355, eaam5488 (2017).
Mou, Y., Yu, J. Y., Wannier, T. M., Guo, C. L. & Mayo, S. L. Computational design of co-assembling protein-DNA nanowires. Nature 525, 230–233 (2015).
Buchberger, A., Simmons, C. R., Fahmi, N. E., Freeman, R. & Stephanopoulos, N. Hierarchical assembly of nucleic acid/coiled-coil peptide nanostructures. J. Am. Chem. Soc. 142, 1406–1416 (2020).
Jin, J. et al. Peptide assembly directed and quantified using megadalton DNA nanostructures. ACS Nano 13, 9927–9935 (2019).
Jiang, T. et al. Structurally ordered nanowire formation from co-assembly of DNA origami and collagen-mimetic peptides. J. Am. Chem. Soc. 139, 14025–14028 (2017).
Udomprasert, A. et al. Amyloid fibrils nucleated and organized by DNA origami constructions. Nat. Nanotechnol. 9, 537–541 (2014).
Suzuki, Y., Endo, M. & Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 6, 8052 (2015).
Dong, Y., Yang, Y., Lin, C. & Liu, D. Frame-guided assembly of amphiphiles. Acc. Chem. Res. 55, 1938–1948 (2022).
Engelen, W. & Dietz, H. Advancing biophysics using DNA origami. Annu. Rev. Biophys. 50, 469–492 (2021).
Wang, W. et al. Bioapplications of DNA nanotechnology at the solid–liquid interface. Chem. Soc. Rev. 48, 4892–4920 (2019).
Zhao, Y. & Xu, C. DNA-based plasmonic heterogeneous nanostructures: building, optical responses, and bioapplications. Adv. Mater. 32, 1907880 (2020).
Hu, Y. & Niemeyer, C. M. From DNA nanotechnology to material systems engineering. Adv. Mater. 31, 1806294 (2019).
Wickham, S. F. J. et al. Complex multicomponent patterns rendered on a 3D DNA-barrel pegboard. Nat. Commun. 11, 5768 (2020).
Funke, J. J. & Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 11, 47–52 (2016).
Erben, C. M., Goodman, R. P. & Turberfield, A. J. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. 45, 7414–7417 (2006).
Kahn, J. S., Xiong, Y., Huang, J. & Gang, O. Cascaded enzyme reactions over a three-dimensional, wireframe DNA origami scaffold. JACS Au 2, 357–366 (2022).
Martin, T. G. et al. Design of a molecular support for cryo-EM structure determination. Proc. Natl Acad. Sci. 113, E7456–E7463 (2016).
Fujita, K., Ohmachi, M., Ikezaki, K., Yanagida, T. & Iwaki, M. Direct visualization of human myosin II force generation using DNA origami-based thick filaments. Nat. Commun. 2, 437 (2019).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Zhang, T. et al. 3D DNA origami crystals. Adv. Mater. 30, 1800273 (2018).
Tian, Y. et al. Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels. Nat. Mater. 19, 789–796 (2020).
Nogales, E. The development of cryo-EM into a mainstream structural biology technique. Nat. Methods 13, 24–27 (2016).
Lyumkis, D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 294, 5181–5197 (2019).
Aksel, T., Yu, Z., Cheng, Y. & Douglas, S. M. Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Nat. Biotechnol. 39, 378–386 (2021).
Zhao, Z. et al. DNA-corralled nanodiscs for the structural and functional characterization of membrane proteins and viral entry. J. Am. Chem. Soc. 140, 10639–10643 (2018).
Dong, Y. et al. Folding DNA into a lipid-conjugated nanobarrel for controlled reconstitution of membrane. Proteins Angew. Chem. Int. Ed. 57, 2072–2076 (2018).
Jones, S. & Thornton, J. M. Principles of protein-protein interactions. Proc. Natl Acad. Sci. 93, 13–20 (1996).
Luscombe, N. M., Laskowski, R. A. & Thornton, J. M. Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res 29, 2860–2874 (2001).
Cheng, F. et al. Comprehensive characterization of protein–protein interactions perturbed by disease mutations. Nat. Genet. 53, 342–353 (2021).
Ryan, D. P. & Matthews, J. M. Protein-protein interactions in human disease. Curr. Opin. Struct. Biol. 15, 441–446 (2005).
Garland, W., Benezra, R. & Chaudhary, J. Targeting protein-protein interactions to treat cancer-recent progress and future directions. Annu. Rep. Med. Chem. 48, 227–245 (2013).
Scott, D. E., Bayly, A. R., Abell, C. & Skidmore, J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 (2016).
Berggård, T., Linse, S. & James, P. Methods for the detection and analysis of protein–protein interactions. Proteomics 7, 2833–2842 (2007).
Ke, Y., Meyer, T., Shih, W. M. & Bellot, G. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 7, 10935 (2016).
Shaw, A. et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 14, 184–190 (2019).
Le, J. V. et al. Probing nucleosome stability with a DNA origami nanocaliper. ACS Nano 10, 7073–7084 (2016).
Funke, J. J., Ketterer, P., Lieleg, C., Korber, P. & Dietz, H. Exploring nucleosome unwrapping using DNA origami. Nano Lett. 16, 7891–7898 (2016).
Funke, J. J. et al. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2, e1600974 (2016).
Nickels, P. C. et al. Molecular force spectroscopy with a DNA origami–based nanoscopic force clamp. Science 354, 305–307 (2016).
Kramm, K. et al. DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability. Nat. Commun. 11, 2828 (2020).
Aghebat Rafat, A., Sagredo, S., Thalhammer, M. & Simmel, F. C. Barcoded DNA origami structures for multiplexed optimization and enrichment of DNA-based protein-binding cavities. Nat. Chem. 12, 852–859 (2020).
Dutta, P. K. et al. Programmable multivalent DNA-origami tension probes for reporting cellular traction forces. Nano Lett. 18, 4803–4811 (2018).
Bustamante, C. J., Chemla, Y. R., Liu, S. & Wang, M. D. Optical tweezers in single-molecule biophysics. Nat. Rev. Dis. Prim. 1, 25 (2021).
Pfitzner, E. et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew. Chem. Int. Ed. 52, 7766–7771 (2013).
Kilchherr, F. et al. Single-molecule dissection of stacking forces in DNA. Science 353, aaf5508 (2016).
Gross, S. P. Hither and yon: a review of bi-directional microtubule-based transport. Phys. Biol. 1, R1–R11 (2004).
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Abdellatef, S. A. et al. Oscillatory movement of a dynein-microtubule complex crosslinked with DNA origami. eLife 11, e76357 (2022).
Hariadi, R. F., Sommese, R. F. & Sivaramakrishnan, S. Tuning myosin-driven sorting on cellular actin networks. eLife 4, e05472 (2015).
Hariadi, R. F. et al. Mechanical coordination in motor ensembles revealed using engineered artificial myosin filaments. Nat. Nanotechnol. 10, 696–700 (2015).
Iwaki, M., Wickham, S. F., Ikezaki, K., Yanagida, T. & Shih, W. M. A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads. Nat. Commun. 7, 13715 (2016).
Rai, A. et al. Multimodal regulation of myosin VI ensemble transport by cargo adaptor protein GIPC. J. Biol. Chem. 298, 101688 (2022).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Grossi, G., Dalgaard Ebbesen Jepsen, M., Kjems, J. & Andersen, E. S. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 8, 992 (2017).
Ijäs, H., Hakaste, I., Shen, B., Kostiainen, M. A. & Linko, V. Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 13, 5959–5967 (2019).
Masubuchi, T. et al. Construction of integrated gene logic-chip. Nat. Nanotechnol. 13, 933–940 (2018).
Wilner, O. I. et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 4, 249–254 (2009).
Fu, J., Liu, M., Liu, Y., Woodbury, N. W. & Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 134, 5516–5519 (2012).
Ngo, T. A., Nakata, E., Saimura, M. & Morii, T. Spatially organized enzymes drive cofactor-coupled cascade reactions. J. Am. Chem. Soc. 138, 3012–3021 (2016).
Linko, V., Eerikäinen, M. & Kostiainen, M. A. A modular DNA origami-based enzyme cascade nanoreactor. Chem. Commun. 51, 5351–5354 (2015).
Sun, L. et al. Real-time imaging of single-molecule enzyme cascade using a DNA origami raft. J. Am. Chem. Soc. 139, 17525–17532 (2017).
Chen, Y. et al. A synthetic light-driven substrate channeling system for precise regulation of enzyme cascade activity based on DNA origami. J. Am. Chem. Soc. 140, 8990–8996 (2018).
Fu, J. et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 9, 531–536 (2014).
Ke, G. et al. Directional regulation of enzyme pathways through the control of substrate channeling on a DNA origami scaffold. Angew. Chem. Int. Ed. 55, 7483–7486 (2016).
Yang, Y. R. et al. 2D enzyme cascade network with efficient substrate channeling by swinging arms. Chembiochem 19, 212–216 (2018).
Zhang, Y., Tsitkov, S. & Hess, H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nat. Commun. 7, 13982 (2016).
Zhao, Z. et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 7, 10619 (2016).
Klein, W. P. et al. Enhanced catalysis from multienzyme cascades assembled on a DNA origami triangle. ACS Nano 13, 13677–13689 (2019).
Rosier, B. J. H. M. et al. Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome. Nat. Catal. 3, 295–306 (2020).
Kurokawa, T. et al. DNA origami scaffolds as templates for functional tetrameric Kir3 K+ channels. Angew. Chem. Int. Ed. 57, 2586–2591 (2018).
Kosuri, P., Altheimer, B. D., Dai, M., Yin, P. & Zhuang, X. Rotation tracking of genome-processing enzymes using DNA origami rotors. Nature 572, 136–140 (2019).
Fisher, P. D. E. et al. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano 12, 1508–1518 (2018).
Ketterer, P. et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 9, 902 (2018).
Shen, Q. et al. DNA-origami NanoTrap for Studying the Selective Barriers Formed by Phenylalanine-Glycine-Rich Nucleoporins. J. Am. Chem. Soc. 143, 12294–12303 (2021).
van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Dong, Y. et al. Frame-guided assembly of vesicles with programmed geometry and dimensions. Angew. Chem. Int. Ed. 53, 2607–2610 (2014).
Dong, Y., Yang, Z. & Liu, D. Using small molecules to prepare vesicles with designable shapes and sizes via frame-guided assembly strategy. Small 11, 3768–3771 (2015).
Wang, C. et al. Construction of liposomes mimicking cell membrane structure through frame-guided assembly. Angew. Chem. Int. Ed. 59, 15176–15180 (2020).
Wang, C. et al. pH-responsive Frame-Guided Assembly with hydrophobicity controllable peptide as leading hydrophobic groups. Giant 1, 100006 (2020).
Zhou, C. et al. Precisely controlled 2D free-floating nanosheets of amphiphilic molecules through frame-guided assembly. Adv. Mater. 28, 9819–9823 (2016).
Dong, Y. et al. Cuboid vesicles formed by frame-guided assembly on DNA origami scaffolds. Angew. Chem. Int. Ed. 56, 1586–1589 (2017).
Yang, Y. et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 8, 476–483 (2016).
Zhang, Z., Yang, Y., Pincet, F., Llaguno, M. C. & Lin, C. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 9, 653–659 (2017).
Czogalla, A. et al. Amphipathic DNA origami nanoparticles to scaffold and deform lipid membrane vesicles. Angew. Chem. Int. Ed. 54, 6501–6505 (2015).
Franquelim, H. G., Khmelinskaia, A., Sobczak, J.-P., Dietz, H. & Schwille, P. Membrane sculpting by curved DNA origami scaffolds. Nat. Commun. 9, 811 (2018).
Journot, C. M. A., Ramakrishna, V., Wallace, M. I. & Turberfield, A. J. Modifying membrane morphology and interactions with DNA origami clathrin-mimic networks. ACS Nano 13, 9973–9979 (2019).
Grome, M. W., Zhang, Z., Pincet, F. & Lin, C. Vesicle tubulation with self-assembling DNA nanosprings. Angew. Chem. Int. Ed. 57, 5330–5334 (2018).
Grome, M. W., Zhang, Z. & Lin, C. Stiffness and membrane anchor density modulate DNA-nanospring-induced vesicle tubulation. ACS Appl. Mater. Interfaces 11, 22987–22992 (2019).
Langecker, M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012).
Krishnan, S. et al. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 7, 12787 (2016).
Xing, Y., Dorey, A., Jayasinghe, L. & Howorka, S. Highly shape- and size-tunable membrane nanopores made with DNA. Nat. Nanotechnol. 17, 708–713 (2022).
Diederichs, T. et al. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 10, 5018 (2019).
Fragasso, A. et al. Reconstitution of ultrawide DNA origami pores in liposomes for transmembrane transport of macromolecules. ACS Nano 15, 12768–12779 (2021).
Thomsen, R. P. et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 10, 5655 (2019).
Dey, S. et al. A reversibly gated protein-transporting membrane channel made of DNA. Nat. Commun. 13, 2271 (2022).
Bian, X., Zhang, Z., Xiong, Q., De Camilli, P. & Lin, C. A programmable DNA-origami platform for studying lipid transfer between bilayers. Nat. Chem. Biol. 15, 830–837 (2019).
Xu, W. et al. A programmable DNA origami platform to organize SNAREs for membrane fusion. J. Am. Chem. Soc. 138, 4439–4447 (2016).
Batista, F. D. & Harwood, N. E. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 9, 15–27 (2009).
Veneziano, R. et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 15, 716–723 (2020).
Hellmeier, J. et al. DNA origami demonstrate the unique stimulatory power of single pMHCs as T cell antigens. Proc. Natl Acad. Sci. 118, e2016857118 (2021).
Wang, C.-H., Chen, X.-Q., Su, Y.-Y., Wang, H. & Li, D. Precise regulating T cell activation signaling with spatial controllable positioning of receptors on DNA origami. Chin. J. Anal. Chem. 50, 100091 (2022).
Dong, R. et al. DNA origami patterning of synthetic T cell receptors reveals spatial control of the sensitivity and kinetics of signal activation. Proc. Natl Acad. Sci. 118, e2109057118 (2021).
Kern, N., Dong, R., Douglas, S. M., Vale, R. D. & Morrissey, M. A. Tight nanoscale clustering of Fcγ receptors using DNA origami promotes phagocytosis. eLife 10, e68311 (2021).
Zhang, J. et al. Elucidating the effect of nanoscale receptor-binding domain organization on SARS-CoV-2 infection and immunity activation with DNA origami. J. Am. Chem. Soc. 144, 21295–21303 (2022).
Angelin, A. et al. Multiscale origami structures as interface for cells. Angew. Chem. Int. Ed. 54, 15813–15817 (2015).
Dutta, D., Pulsipher, A., Luo, W. & Yousaf, M. N. Synthetic chemoselective rewiring of cell surfaces: generation of three-dimensional tissue structures. J. Am. Chem. Soc. 133, 8704–8713 (2011).
Guo, F. et al. Controlling cell–cell interactions using surface acoustic waves. Proc. Natl Acad. Sci. 112, 43–48 (2015).
Gartner, Z. J. & Bertozzi, C. R. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. 106, 4606–4610 (2009).
Ge, Z. et al. Programming cell–cell communications with engineered cell origami clusters. J. Am. Chem. Soc. 142, 8800–8808 (2020).
Liu, J. et al. Reconstructing soma–soma synapse-like vesicular exocytosis with DNA origami. ACS Cent. Sci. 7, 1400–1407 (2021).
Patra, J. K. et al. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnology. 16, 71 (2018).
Blanco, E., Shen, H. & Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015).
Jiang, S., Ge, Z., Mou, S., Yan, H. & Fan, C. Designer DNA nanostructures for therapeutics. Chem 7, 1156–1179 (2021).
Li, J., Fan, C., Pei, H., Shi, J. & Huang, Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 25, 4386–4396 (2013).
Hu, Q., Li, H., Wang, L., Gu, H. & Fan, C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 119, 6459–6506 (2019).
Duan, X. & Li, Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 9, 1521–1532 (2013).
Engelhardt, F. A. S. et al. Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano 13, 5015–5027 (2019).
Bastings, M. M. C. et al. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 18, 3557–3564 (2018).
Jiang, D. et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat. Biomed. Eng. 2, 865–877 (2018).
Mikkilä, J. et al. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 14, 2196–2200 (2014).
Ponnuswamy, N. et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017).
Zhao, S. et al. Efficient intracellular delivery of RNase a using DNA origami carriers. ACS Appl. Mater. Interfaces 11, 11112–11118 (2019).
Chaithongyot, S. et al. Selective delivery of doxorubicin using the biomarker-specific, aptamer-functionalized DNA nanosphere. Mater. Lett. 260, 126952 (2020).
Sun, P. et al. Site-specific anchoring aptamer C2NP on DNA origami nanostructures for cancer treatment. RSC Adv. 8, 26300–26308 (2018).
Mela, I. et al. DNA nanostructures for targeted antimicrobial delivery. Angew. Chem. Int. Ed. 59, 12698–12702 (2020).
Ge, Z. et al. DNA origami-enabled engineering of ligand–drug conjugates for targeted drug delivery. Small 16, 1904857 (2020).
Pal, S. & Rakshit, T. Folate-functionalized DNA origami for targeted delivery of doxorubicin to triple-negative breast cancer. Front. Chem. 9, 721105 (2021).
Chaithongyot, S., Chomanee, N., Charngkaew, K., Udomprasert, A. & Kangsamaksin, T. Aptamer-functionalized DNA nanosphere as a stimuli-responsive nanocarrier. Mater. Lett. 214, 72–75 (2018).
Burns, J. R., Lamarre, B., Pyne, A. L. B., Noble, J. E. & Ryadnov, M. G. DNA origami inside-out viruses. ACS Synth. Biol. 7, 767–773 (2018).
Tang, M. S. L. et al. An aptamer-enabled DNA nanobox for protein sensing. Nanomed. J. 14, 1161–1168 (2018).
Kohman, R. E. & Han, X. Light sensitization of DNA nanostructures via incorporation of photo-cleavable spacers. Chem. Commun. 51, 5747–5750 (2015).
Kohman, R. E., Cha, S. S., Man, H.-Y. & Han, X. Light-triggered release of bioactive molecules from DNA nanostructures. Nano Lett. 16, 2781–2785 (2016).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Liu, J. et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 18, 3328–3334 (2018).
Liu, J. et al. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. Int. Ed. 57, 15486–15490 (2018).
Wang, Z. et al. A tubular DNA nanodevice as a siRNA/chemo-drug co-delivery vehicle for combined cancer therapy. Angew. Chem. Int. Ed. 60, 2594–2598 (2021).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Mei, Q. et al. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 11, 1477–1482 (2011).
Perrault, S. D. & Shih, W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–5140 (2014).
Wang, S. T. et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proc. Natl Acad. Sci. 117, 6339–6348 (2020).
Estrich, N. A., Hernandez-Garcia, A., de Vries, R. & LaBean, T. H. Engineered diblock polypeptides improve DNA and gold solubility during molecular assembly. ACS Nano 11, 831–842 (2017).
Anastassacos, F. M., Zhao, Z., Zeng, Y. & Shih, W. M. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 142, 3311–3315 (2020).
Shrestha, P. et al. Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 12, 582–588 (2017).
Smolková, B. et al. Protein corona inhibits endosomal escape of functionalized DNA nanostructures in living cells. ACS Appl. Mater. Interfaces 13, 46375–46390 (2021).
Pumm, A. K. et al. A DNA origami rotary ratchet motor. Nature 607, 492–498 (2022).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21934007), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB36000000), the China Postdoctoral Science Foundation (Grant No. 2022M712325), and the Jiangsu Funding Program for Excellent Postdoctoral Talent.
Author information
Authors and Affiliations
Contributions
Y.Z., J. D., and Q.W. discussed the main topic of the paper; Y.Z. and J.D. drafted the paper; Y.Z. and Q.W. revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, Y., Dong, J. & Wang, Q. Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales. NPG Asia Mater 15, 25 (2023). https://doi.org/10.1038/s41427-023-00470-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41427-023-00470-3
This article is cited by
-
Pros and Cons in Various Immobilization Techniques and Carriers for Enzymes
Applied Biochemistry and Biotechnology (2024)