Abstract
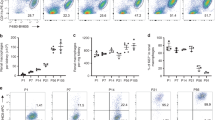
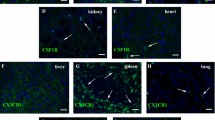
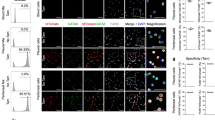
Tissue-resident macrophages are derived from different precursor cells and display different phenotypes. Reconstitution of the tissue-resident macrophages of inflamed or damaged tissues in adults can be achieved by bone marrow-derived monocytes/macrophages. Using lysozyme (Lysm)-GFP-reporter mice, we found that alveolar macrophages (AMs), Kupffer cells, red pulp macrophages (RpMacs), and kidney-resident macrophages were Lysm-GFP−, whereas all monocytes in the fetal liver, adult bone marrow, and blood were Lysm-GFP+. Donor-derived Lysm-GFP+ resident macrophages gradually became Lysm-GFP− in recipients and developed gene expression profiles characteristic of tissue-resident macrophages. Thus, Lysm may be used to distinguish newly formed and long-term surviving tissue-resident macrophages that were derived from bone marrow precursor cells in adult mice under pathological conditions. Furthermore, we found that Irf4 might be essential for resident macrophage differentiation in all tissues, while cytokine and receptor pathways, mTOR signaling pathways, and fatty acid metabolic processes predominantly regulated the differentiation of RpMacs, Kupffer cells, and kidney macrophages, respectively. Deficiencies in ST2, mechanistic target of rapamycin (mTOR) and fatty acid-binding protein 5 (FABP5) differentially impaired the differentiation of tissue-resident macrophages from bone marrow-derived monocytes/macrophages in the lungs, liver, and kidneys. These results indicate that a combination of shared and unique signaling pathways coordinately shape tissue-resident macrophage differentiation in various tissues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
We downloaded the microarray GM-CSF data and M-CSF-induced bone marrow-derived macrophage data from E-MTAB-791 [29]. The raw RNA-seq data generated in our study were deposited in the National Genomics Data Center (NCDC): BioProject PRJCA008365.
Material availability
Methods, including statements of data availability and any further information, are available in the online version of this paper.
References
Bleriot C, Chakarov S, Ginhoux F. Determinants of resident tissue macrophage identity and function. Immunity. 2020;52:957–70.
Knipper JA, Ding X, Eming SA. Diabetes impedes the epigenetic switch of macrophages into repair mode. Immunity. 2019;51:199–201.
Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–49.
Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–51.
Zhao Y, Zou W, Du J, Zhao Y. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J Cell Physiol. 2018;233:6425–39.
Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321.
Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–62.
van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, et al. Yolk Sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–68.
Bonnardel J, T'Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 2019;51:638–54.e9.
Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity. 2020;52:1057–74.e.
Woo YD, Jeong D, Chung DH. Development and functions of alveolar macrophages. Mol Cells. 2021;44:292–300.
Cheah FC, Presicce P, Tan TL, Carey BC, Kallapur SG. Studying the effects of granulocyte-macrophage colony-stimulating factor on fetal lung macrophages during the perinatal period using the mouse model. Front Pediatr. 2021;9:614209.
Lambrecht BN. TGF-beta gives an air of exclusivity to alveolar macrophages. Immunity. 2017;47:807–9.
Yu X, Buttgereit A, Lelios I, Utz SG, Cansever D, Becher B, et al. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity. 2017;47:903–12.e4.
Kucharova K, Stallcup WB. Distinct NG2 proteoglycan-dependent roles of resident microglia and bone marrow-derived macrophages during myelin damage and repair. PLoS ONE. 2017;12:e0187530.
Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–34.e0.
Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, et al. Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol. 2017;57:66–76.
Devisscher L, Scott CL, Lefere S, Raevens S, Bogaerts E, Paridaens A, et al. Non-alcoholic steatohepatitis induces transient changes within the liver macrophage pool. Cell Immunol. 2017;322:74–83.
Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–9.
Lu Y, Basatemur G, Scott IC, Chiarugi D, Clement M, Harrison J, et al. Interleukin-33 signaling controls the development of iron-recycling macrophages. Immunity. 2020;52:782–93.e5.
Sheng J, Ruedl C, Karjalainen K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity. 2015;43:382–93.
Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–28.
Zhu L, Zhao Q, Yang T, Ding W, Zhao Y. Cellular metabolism and macrophage functional polarization. Int Rev Immunol. 2015;34:82–100.
Xu D, Huang SK. IL-33: a key player in the development of iron-recycling red pulp macrophages. Cell Mol Immunol. 2020;17:1218–9.
Umbarawan Y, Enoura A, Ogura H, Sato T, Horikawa M, Ishii T, et al. FABP5 is a sensitive marker for lipid-rich macrophages in the luminal side of atherosclerotic lesions. Int Heart J. 2021;62:666–76.
Moore SM, Holt VV, Malpass LR, Hines IN, Wheeler MD. Fatty acid-binding protein 5 limits the anti-inflammatory response in murine macrophages. Mol Immunol. 2015;67:265–75.
Guo Y, Liu Y, Zhao S, Xu W, Li Y, Zhao P, et al. Oxidative stress-induced FABP5 S-glutathionylation protects against acute lung injury by suppressing inflammation in macrophages. Nat Commun. 2021;12:7094.
Lee MC, Lacey DC, Fleetwood AJ, Achuthan A, Hamilton JA, Cook AD. GM-CSF- and IRF4-dependent signaling can regulate myeloid cell numbers and the macrophage phenotype during inflammation. J Immunol. 2019;202:3033–40.
Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz MG, et al. Defining GM-CSF—and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–65.
Harjes U. Home advantage for tissue-resident macrophages. Nat Rev Cancer. 2021;21:539.
Ma D, Doi Y, Jin S, Li E, Sonobe Y, Takeuchi H, et al. TGF-beta induced by interleukin-34-stimulated microglia regulates microglial proliferation and attenuates oligomeric amyloid beta neurotoxicity. Neurosci Lett. 2012;529:86–91.
Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity. 2019;51:655–70.e8.
Dahik VD, Frisdal E, Le Goff W. Rewiring of lipid metabolism in adipose tissue macrophages in obesity: impact on insulin resistance and type 2 diabetes. Int J Mol Sci. 2020;21:5505.
Zhao Y, Zou W, Du J, Zhao Y. mTOR masters monocyte development in bone marrow by decreasing the inhibition of STAT5 on IRF8. Blood. 2018;131:1587–99.
Tian Q, Zhang Z, Tan L, Yang F, Xu Y, Guo Y, et al. Skin and heart allograft rejection solely by long-lived alloreactive TRM cells in skin of severe combined immunodeficient mice. Sci Adv. 2022;8:eabk0270.
Otogawa K, Kinoshita K, Fujii H, Sakabe M, Shiga R, Nakatani K, et al. Erythrophagocytosis by liver macrophages (Kupffer cells) promotes oxidative stress, inflammation, and fibrosis in a rabbit model of steatohepatitis: implications for the pathogenesis of human nonalcoholic steatohepatitis. Am J Pathol. 2007;170:967–80.
Klei TR, Meinderts SM, van den Berg TK, van Bruggen R. From the cradle to the grave: the role of macrophages in erythropoiesis and erythrophagocytosis. Front Immunol. 2017;8:73.
Shi L, Tian H, Wang P, Li L, Zhang Z, Zhang J, et al. Spaceflight and simulated microgravity suppresses macrophage development via altered RAS/ERK/NFkappaB and metabolic pathways. Cell Mol Immunol. 2021;18:1489–502.
Chu Z, Feng C, Sun C, Xu Y, Zhao Y. Primed macrophages gain long-term specific memory to reject allogeneic tissues in mice. Cell Mol Immunol. 2021;18:1079–81.
Hou Y, Zhu L, Tian H, Sun H, Wang R, Zhang L, et al. IL-23-induced macrophage polarization and its pathological roles in mice with imiquimod-induced psoriasis. Protein Cell. 2018;9:1027–38.
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67.
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5.
Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8.
Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–25.
Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3.
Ferreira MR, Santos GA, Biagi CA, Silva Junior WA, Zambuzzi WF. GSVA score reveals molecular signatures from transcriptomes for biomaterials comparison. J Biomed Mater Res A. 2021;109:1004–14.
Sergushichev AA, Loboda AA, Jha AK, Vincent EE, Driggers EM, Jones RG, et al. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 2016;44:W194–200.
Zhou G, Soufan O, Ewald J, Hancock R, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–41.
Demchak B, Hull T, Reich M, Liefeld T, Smoot M, Ideker T, et al. Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Res. 2014;3:151.
Acknowledgements
We would like to acknowledge Mrs. Qing Meng and Mrs. Xiaoqiu Liu for their expert technical assistance, Mrs. Ling Li for her excellent laboratory management, and Mr. Yiming Jin for his assistance with the animal and cellular experiments. This work was supported by grants from the National Natural Science Foundation for Key Programs (31930041, Y.Z.), National Key Research and Development Program of China (2017YFA0105002, 2017YFA0104402, Y.Z.), and Knowledge Innovation Program of the Chinese Academy of Sciences (XDA16030301, Y.Z.).
Author information
Authors and Affiliations
Contributions
T.L. and Y.Z. designed the methodology and investigation. T.L. and Q.Z. conducted experiments, collected results, and performed analyses. J.Y.Z. performed bioinformatic analysis. T.L., X.R.M., Y.N.X., and Y.Z. performed animal husbandry. T.L., J.Y.Z., and Y.Z. wrote the paper. Y.Z. supervised, directed, performed project administration, and acquired funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lei, T., Zhang, J., Zhang, Q. et al. Defining newly formed and tissue-resident bone marrow-derived macrophages in adult mice based on lysozyme expression. Cell Mol Immunol 19, 1333–1346 (2022). https://doi.org/10.1038/s41423-022-00936-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-022-00936-4
Keywords
This article is cited by
-
Wip1 inhibits neutrophil extracellular traps to promote abscess formation in mice by directly dephosphorylating Coronin-1a
Cellular & Molecular Immunology (2023)