Abstract
Live attenuated vaccines might elicit mucosal and sterilizing immunity against SARS-CoV-2 that the existing mRNA, adenoviral vector and inactivated vaccines fail to induce. Here, we describe a candidate live attenuated vaccine strain of SARS-CoV-2 in which the NSP16 gene, which encodes 2′-O-methyltransferase, is catalytically disrupted by a point mutation. This virus, designated d16, was severely attenuated in hamsters and transgenic mice, causing only asymptomatic and nonpathogenic infection. A single dose of d16 administered intranasally resulted in sterilizing immunity in both the upper and lower respiratory tracts of hamsters, thus preventing viral spread in a contact-based transmission model. It also robustly stimulated humoral and cell-mediated immune responses, thus conferring full protection against lethal challenge with SARS-CoV-2 in a transgenic mouse model. The neutralizing antibodies elicited by d16 effectively cross-reacted with several SARS-CoV-2 variants. Secretory immunoglobulin A was detected in the blood and nasal wash of vaccinated mice. Our work provides proof-of-principle evidence for harnessing NSP16-deficient SARS-CoV-2 for the development of live attenuated vaccines and paves the way for further preclinical studies of d16 as a prototypic vaccine strain, to which new features might be introduced to improve safety, transmissibility, immunogenicity and efficacy.
Similar content being viewed by others
Introduction
The devastating COVID-19 pandemic caused by severe respiratory syndrome coronavirus 2 (SARS-CoV-2) has entered a new stage in which breakthrough infections, particularly those caused by the highly transmissible Delta and Omicron variants, are increasingly prevalent in vaccinees immunized with mRNA, adenoviral vector, protein subunit or inactivated vaccines [1,2,3,4]. In response to the waning of protective immunity in vaccinees over time and the emergence of vaccine-escaped SARS-CoV-2 variants, another booster injection is recommended, and variant-targeted vaccines are being developed [5, 6]. The levels of neutralizing antibodies (NAbs) that target the viral spike (S) protein, particularly the receptor-binding domain (RBD), correlate perfectly with protective immunity against detectable and symptomatic SARS-CoV-2 infection [7, 8]. Although such a correlation has not been observed for immune memory or cytotoxic T lymphocytes, these immune factors might also contribute to the alleviation of severe COVID-19 [9, 10]. Although highly effective in the induction of NAbs, the existing SARS-CoV-2 vaccines fail to elicit the mucosal and sterilizing immunity required for the elimination of breakthrough infections, which often begin in the mucosa of the upper respiratory tract [8]. In this regard, live attenuated vaccines against SARS-CoV-2 will not only provide another powerful tool to combat COVID-19 through the induction of mucosal immunity but might also hold the key to ending the pandemic. This reminds us of the past successes with the eradication of smallpox (variola virus) and near eradication of poliovirus from human populations, as a live attenuated vaccine played a critical role in each success [11]. We have learned new lessons from the COVID-19 pandemic. Emerging evidence suggests that natural infection plus one injection of an mRNA vaccine provides better protection against the Omicron variant than three injections of the mRNA vaccine [12, 13]. On the other hand, the success of the mRNA vaccines might be ascribed, at least in part, to the use of a very high dose [6, 14]. It remains to be seen whether the administration of a high dose of a live attenuated vaccine alone offers sterilizing protection against SARS-CoV-2 and eliminates the possibility of reinfection. Obviously, the chance of establishing sterilizing immunity would be considerably increased when a live attenuated vaccine is used as a booster injection in people who have been naturally infected or immunized with an mRNA vaccine or another type of vaccine. Nevertheless, live attenuated vaccines are known to elicit a much broader range of humoral and cellular immune responses that are cross-reactive and cross-protective [15, 16]. They might contribute substantially to the control of SARS-CoV-2 in the next phase of the pandemic.
Several strategies have been tested in the development of live attenuated vaccines against SARS-CoV [17]. Deletion of E or ORF3a is known to result in attenuation [18, 19]. Two other successful approaches were based on the inactivation of highly conserved nonstructural proteins (NSPs) [20,21,22]. The NSPs form the replication and transcription complex (RTC), which replicates viral genomic RNA and transcribes viral subgenomic RNAs. Two catalytic components of the RTC, NSP14 and NSP16, are critical for the methylation of the 5′ cap of viral mRNAs, a process that allows evasion of immune detection by making the viral RNAs indistinguishable from host mRNA [23]. NSP14 possesses S-adenosylmethionine (SAM)-dependent methyltransferase (MTase) activity, which facilitates the addition of a methyl group to the guanosine at N7 to generate a Cap-0 structure on viral mRNA. In addition, NSP14 is an exoribonuclease that serves as the proofreading enzyme in viral RNA synthesis [24]. The mRNA capping procedure is then completed by NSP16, a SAM-dependent 2′-O-MTase, through further methylation at the ribose 2′-O position of the first nucleotide of the RNA transcript, which produces a Cap-1 structure [25]. Compromising the function of NSPs might have a direct impact on the viral life cycle. Indeed, inactivation of either NSP14 or NSP16 in SARS-CoV has been shown to result in attenuation [20,21,22]. In particular, introducing the D130A mutation in the KDKE motif of SARS-CoV NSP16 gave rise to a severely attenuated but viable mutant virus with augmented type I interferon (IFN) induction but weakened viral replication in macrophages [20, 26]. Introduction of the similar inactivating mutations D330A and Y414A into NSP14 of mouse hepatitis virus also led to attenuation [27]. However, NSP14-deficient mutants of MERS-CoV and SARS-CoV-2 are inviable [28]. A strategy of genome recoding has been used successfully to attenuate SARS-CoV-2 [29,30,31]. Generally consistent with the notion that compromising the expression of NSP16 might attenuate SARS-CoV-2, codon pair deoptimization of the NSP16 gene gave rise to a highly attenuated virus designated sCPD9 [30, 31]. In this study, we generated a recombinant SARS-CoV-2 strain carrying the NSP16 D130A mutation and demonstrated the feasibility of using NSP16 inactivation as a strategy for the development of live attenuated SARS-CoV-2 vaccines.
Results
SARS-CoV-2 d16 is attenuated in vitro
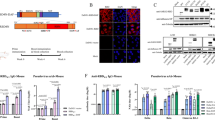
Previous work with mouse hepatitis virus, SARS-CoV and MERS-CoV has shown that catalytic inactivation of NSP16 achieved by disrupting the highly conserved KDKE tetrad gives rise to an attenuated strain sensitive to the activity of IFN-stimulated genes, such as IFIT1 [32]. With this in mind, we sought to construct an NSP16-deficient SARS-CoV-2 strain by starting with the point mutation D130A, which is known to catalytically inactivate the MTase function of NSP16 in other coronaviruses [20, 21]. A point mutation was chosen in view of the risk that deletion of a large fragment might render the SARS-CoV-2 genome less stable. Since NSP14-deficient SARS-CoV-2 is inviable [28], it was of interest to see whether a SARS-CoV-2 mutant virus carrying the D130A mutation in NSP16 could be viable. If it was, we could next ask whether this strain might be attenuated and serve as a candidate for the development of a live attenuated vaccine against SARS-CoV-2. Beginning with the infectious SARS-CoV-2 molecular clone on one bacterial artificial chromosome (BAC) that was made available recently [33,34,35], we generated a recombinant SARS-CoV-2 virus carrying the NSP16 D130A mutation, designated d16 (Fig. 1A, B). A model of the mutant structure generated with the I-TASSER program predicted that the D130A substitution disrupts the interaction between D130 and the methionine moiety of the bound SAM in the methylation reaction center of NSP16 (Fig. 1C). Comparison of the plaque phenotypes of wild-type (WT) and d16 viruses revealed that d16 formed smaller plaques in VeroE6 cells (Fig. 1D). Remarkably, less viral N protein antigen was detected in d16-infected VeroE6 cells by immunofluorescence staining (Fig. 1E). The attenuation of d16 was further demonstrated by its replication kinetics in three different cell lines. Compared to the WT virus, d16 displayed a 1−2 log10 reduction in viral load in VeroE6, A549-ACE2-TMPRSS2 and Calu-3 cells at 48 hpi (Fig. 1F–H). Whereas VeroE6 cells are IFN deficient, A549-ACE2-TMPRSS2 and Calu-3 cells are more physiologically relevant cell lines with an intact IFN response. Importantly, over ten consecutive passages in VeroE6 cells, the d16 virus exhibited genetic stability with no virulence reversion, providing a concrete safety measure (Supplementary Fig. S1).
SARS-CoV-2 NSP16 D130A mutant virus (d16) is attenuated in vitro. A Schematic representation of the SARS-CoV-2 genome and the NSP16 D130A mutant. B Sequencing results for WT SARS-CoV-2 and mutant NSP16 D130A clones. The desired point mutation highlighted by the red box was verified by Sanger sequencing. C Structure of the SAM binding pocket of WT (D130) NSP16 (PDB: 6W4H) compared to that of mutant (A130) NSP16 (I-TASSER prediction). Residues in the catalytic tetrad (K46, D/A130, K170 and E203) are shown in stick representation. SAM, in surface representation, was modeled in the catalytic cleft of mutant NSP16 based on structural alignment to WT NSP16. D Plaque phenotype. VeroE6 cells were infected with the indicated recombinant viruses. After 72 h of incubation at 37 °C, the cells were fixed and stained with 1% crystal violet. Plaque images are shown. Ten plaques were randomly picked, and their sizes were quantified by ImageJ and analyzed by Student’s t-test (***P < 0.001). E Immunofluorescence staining. VeroE6 cells infected with either the WT virus or d16 at an MOI of 5 were fixed at 24 h post-infection, and the SARS-CoV-2 N antigen was detected using rabbit anti-N protein IgG and goat anti-rabbit IgG conjugated with FITC as the primary and secondary antibodies, respectively. The anti-N antisera were raised in-house and reactive to all variants tested. Cellular nuclei were stained with DAPI (blue). Scale bars, 20 μm. F–H VeroE6, A549-ACE2-TMPRSS2 and Calu-3 cells were infected with recombinant WT SARS-CoV-2 (black) or d16 mutant (pink) virus at an MOI of 0.1 at the indicated time points. The cell supernatant was harvested, and SARS-CoV-2 RNA in the RNA-dependent RNA polymerase (RdRP)-coding region was quantitated using RT-qPCR and is plotted as log10 copies per ml. I Transcriptomic analysis of host gene expression in A549-ACE2-TMPRSS2 cells infected with WT SARS-CoV-2 or d16. Bar plots of enriched pathways including GO-CC, GO-MF and GO-BP terms and KEGG pathways between d16 and the WT virus are shown. GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, BP biological process, MF molecular function, CC cellular component. J Heatmap of the enriched differentially expressed genes (false discovery rate <0.05) in the d16-, WT- or mock-infected group in comparison with the other two groups
The IFN antagonism of NSP16 has been described [20, 32]. As a result, disruption of NSP16 might derepress the IFN response, leading to more robust activation of the innate immune response. Thus, the cellular response to d16 infection might have unique features, such as elevated expression of IFNs and IFN-stimulated genes. To verify this and begin to elucidate the landscape of the host transcriptional response to d16 infection, we compared the transcriptomes of human lung A549-ACE2-TMPRSS2 cells infected with d16 or WT viruses. As expected, diminishing SARS-CoV-2 NSP16 MTase activity affected biological processes, such as viral transcription and replication, whereas the innate immune response in d16-infected samples remained robust (Fig. 1I and Supplementary Fig. S2), generally in keeping with the previous finding on the IFN antagonism of NSP16 [20, 32]. KEGG analysis indicated enrichment in several cell cycle-regulating pathways, such as those involved in apoptosis, Hippo signaling and mTOR signaling. In line with this observation, a panel of keratin genes was upregulated in d16 infection compared with either mock or WT virus infection (Fig. 1J and Supplementary Fig. S2). In summary, we generated the replication-competent d16 virus, which is notably less virulent than the WT virus.
Attenuation of SARS-CoV-2 d16 in vivo
To verify the attenuation of d16 in vivo, hamsters were intranasally challenged with 105 plaque-forming units (PFU) of the virus or WT SARS-CoV-2 (Fig. 2A). The hamster model is known to be an excellent model of mild to moderate COVID-19 in humans [36]. Expectedly, hamsters in the WT group developed clinical signs of lethargy, ruffled fur, hunched back posture and rapid breathing starting from 2 days post-infection (dpi), as previously reported [36], whereas such clinical signs were not observed among hamsters subjected to challenge with d16. Approximately 10% loss of body weight was observed in WT hamsters, whereas a steady increase of >10% was recorded in the d16 group over the 14-day post-infection course (Fig. 2B). At 4 dpi, when the viral loads and histopathological changes were expected to be most prominent in this established animal model, we found significantly lower viral loads in the upper (nasal wash, ~1 log10, P < 0.01) and lower (lung, ~1.5 log10, P < 0.05) respiratory tract tissues in the d16 group (Fig. 2C, D). Consistent with this, immunofluorescence staining of lung tissues indicated widespread expression of the viral N protein throughout the alveolar tissues in the WT group, while the expression in the d16 group was restricted to the focal bronchiolar epithelial cells (Fig. 2E). To determine the histopathological condition of the lung tissues, hematoxylin and eosin (H&E) staining was performed. Negligible lung damage was observed in the hamster lungs in the d16 group at 4 dpi compared with those in the WT group (Fig. 2F). Intriguingly, trends toward higher expression of tumor necrosis factor α (TNF-α, P < 0.05), interleukin 4 (IL-4, P < 0.01), forkhead box P3 (FOXP3, P < 0.01), and C-C motif chemokine ligand 17 (CCL17, P < 0.01) were also observed in the d16 group (Fig. 2G). Overall, the d16 virus was attenuated in hamsters in vivo with reduced viral replication and ameliorated tissue damage but enhanced expression of some immunomodulatory cytokines.
SARS-CoV-2 d16 mutant virus is attenuated in vivo. A Viral challenge scheme for the hamster model. Hamsters were intranasally inoculated with 105 PFU (in 50 µl) of recombinant WT or d16 SARS-CoV-2. At 4 dpi, the lungs and nasal wash were harvested for viral yield detection. B Body weight changes of WT or d16 SARS-CoV-2-infected hamsters (n = 6). Viral loads determined assessment of the nasal wash and lung tissues of WT- or d16-challenged hamsters at 4 dpi by RT–qPCR (C) and plaque assay (D) (n = 4/group). The primers have been described elsewhere [67]. E Detection of the SARS-CoV-2 N protein in lung tissues infected with the indicated recombinant virus. The SARS-CoV-2 N protein (green) was labeled with rabbit anti-SARS-CoV-2 N antibodies followed by goat anti-rabbit antibodies conjugated to FITC. Nuclei were counterstained with DAPI (blue). Scale bars, 50 μm. F Effects of the WT virus and d16 on histopathological features of the lungs of hamsters. Representative hamster lung tissue sections harvested at 4 dpi were stained with H&E. Scale bars, 200 μm. G Proinflammatory cytokine and chemokine gene expression in WT virus- or d16-infected hamsters. The relative expression levels of representative chemokines and cytokines in lung tissue homogenates were quantified using RT–qPCR. The results are shown as the mean ± SEM. H Viral challenge scheme for the K18-hACE2 mouse model. Each mouse was intranasally inoculated with 103 PFU (in 20 µl) of recombinant WT virus or d16. At 4 dpi, the viral loads in lung tissues were quantitated using RT–qPCR. Body weight changes (I) and survival rates (J) of WT virus- or d16-infected K18-hACE2 transgenic mice (n = 5/group). K Viral loads in lung tissues at 4 dpi (n = 4/group). L Detection of the SARS-CoV-2 N protein in the lung tissues of WT virus- or d16-infected mice. The SARS-CoV-2 N protein (green) was labeled with rabbit anti-SARS-CoV-2 N antibodies followed by goat anti-rabbit antibodies conjugated to FITC. Nuclei were counterstained with DAPI (blue). Scale bars, 50 μm. M Effects of WT and d16 SARS-CoV-2 on histopathological features of mouse lungs. Lung sections from uninfected mice (mock) were included as a negative control. Representative sections of lung tissues from K18-hACE2 mice harvested at 4 dpi were stained with H&E. Scale bars, 100 μm. N Pathological changes were scored as described in the Materials and Methods. Statistical analyses by Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001)
Likewise, d16 was highly attenuated in transgenic mice in which the expression of human angiotensin-converting enzyme 2 (hACE2) is under the control of the cytokeratin 18 (K18) promoter (Fig. 2H). These K18-hACE2 mice are known to be highly susceptible to SARS-CoV-2 infection, serving as a good model for severe COVID-19 in humans [37, 38]. After the challenge with 103 PFU virus, no detectable body weight loss was observed in d16-infected mice, while a sharp decrease of >10% was recorded in the WT group starting from 4 dpi (Fig. 2I). All mice challenged with the WT virus died on or before 7 days post-challenge (0% survival), whereas all d16-infected mice survived throughout the 14-day course of disease (100% survival, Fig. 2J). At 4 days post-challenge, an average 100-fold decrease in the viral load was detected in the lungs of d16-infected mice compared with those of WT mice (Fig. 2K). Consistent with this finding, the expression of the SARS-CoV-2 N protein was robust in the alveolar and airway epithelial cells of the mouse lungs in the WT group but absent in those in the d16 group (Fig. 2L). Histopathologically, amelioration of the inflammatory effect was observed in d16-infected mice after H&E staining of mouse lung sections (Fig. 2M, N). Our findings from hamsters and K18-hACE2 mice consistently supported the attenuation of d16 in vivo.
d16 as a live attenuated vaccine to prevent SARS-CoV-2 infection and disease in hamsters
To assess the protective effect of d16 as a potential live attenuated vaccine to prevent SARS-CoV-2 infection and pathogenicity, a single-dose vaccination regimen with d16 or WT was performed via intranasal administration, and the animals were monitored for 28 days, followed by a comprehensive evaluation of the vaccine efficacy in preventing infection and disease (Fig. 3A). Equally high levels of antibodies directed against the SARS-CoV-2 RBD were detected in the serum of both WT virus-immunized hamsters and d16-immunized hamsters as early as 7 dpi and maintained for at least 84 days (Fig. 3B and Supplementary Fig. S3). The neutralizing capability of these antibodies at day 28 post-vaccination was investigated using the SARS-CoV-2 surrogate virus neutralization test (sVNT) and the live-virus microneutralization (MN) assay to assess the protein and cellular levels, respectively. Both assays indicated that d16 and the WT virus were able to induce substantial NAb responses (Fig. 3C, D). Interestingly, the results from the MN assay indicated the ability of d16-induced antibodies to neutralize various SARS-CoV-2 variants, including the α, β, θ and κ variants (Supplementary Fig. S4). Generally consistent with a previous report [39], d16-vaccinated hamsters showed higher neutralizing activity against the α variant (Supplementary Fig. S4).
d16 as a live attenuated vaccine to alleviate disease in hamsters infected with SARS-CoV-2. A Vaccination and viral challenge scheme for the hamster model. Hamsters were intranasally inoculated with 105 PFU (in 50 µl) of recombinant WT or d16 SARS-CoV-2 on day 0. Serum samples were collected from each group on days 0, 14 and 28 for ELISA and neutralization tests. On day 29, the immunized hamsters were challenged with 105 PFU (in 50 µl) of clinical isolate HK-13 of SARS-CoV-2. Respiratory tissues (tracheae and lungs) and the nasal wash were collected on day 33, and each respective viral load was detected by a plaque assay. Body weights were recorded daily from day 29 to day 43 after viral challenge. B Antibody responses in hamsters infected with WT SARS-CoV-2 or immunized with d16 SARS-CoV-2 were measured by ELISA to assess RBD-specific antibodies. C The cross-neutralization capability of infected/immunized hamster sera (1:10 dilution) was examined using the SARS-CoV-2 sVNT assay. The dotted line represents the assay cutoff at 30% inhibition. D SARS-CoV-2 neutralization tests to examine the neutralization ability of hamster sera collected from different groups. E Body weight changes of HK-13-infected hamsters (n = 5) vaccinated with d16 viruses or PBS. The WT virus-reinfected group served as a control. F Viral titer determination by a plaque assay for the nasal wash, tracheal tissues and lung tissues of HK-13-challenged hamsters in the WT, d16 or PBS group at 4 dpi (n = 4/group). LOD limit of detection. G Hamster cohousing scheme. Briefly, d16- or sham-vaccinated hamsters (n = 4/group) were infected with SARS-CoV-2 on day 29 post-vaccination and subsequently cohoused with immunologically naïve hamsters for 8 h at 2 dpi, followed by separation and viral load analysis of the recipient hamsters on day 4 post-cohousing. H Viral loads in the nasal wash and lung tissues of recipient hamsters at 4 days post-cohousing. LOD limit of detection. I Histopathological changes in the lungs of SARS-CoV-2-challenged hamsters in the WT, d16 or PBS group at 4 dpi. Representative sections were stained with H&E. Scale bars, 200 μm. J Transcript levels of representative proinflammatory chemokines and cytokines in lung tissue homogenates from the WT, d16 and PBS groups, as measured by RT–qPCR. The results are shown in arbitrary units as the mean ± SEM. Statistical analyses were performed with Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
Hamsters were challenged with a clinical isolate of SARS-CoV-2 on day 29 (105 PFU), which led to ~15% body weight loss followed by a slight rebound at 6 dpi in the PBS group. In stark contrast, the body weights in the WT and d16 groups steadily increased (Fig. 3E). At 4 days after the live-virus challenge, d16 vaccination had decreased the viral titer in lung tissues by >4 log10, and suppression was also observed in the upper respiratory tract (~2 log10 in the nasal wash and trachea) (Fig. 3F). Since the viral loads in the WT and d16 groups were close to the lowest limit of detection of our assay, d16 was considered as efficient as the WT virus in the prevention of SARS-CoV-2 infection (Fig. 3F). Plausibly, the WT and d16 viruses might be equally capable of eliciting sterilizing immunity. To further evaluate the effectiveness of d16 vaccination in blocking viral transmission, immunologically naïve hamsters were cohoused with SARS-CoV-2-infected d16- or sham-vaccinated hamsters for 8 h at 2 dpi and subsequently single housed. Later, viral load analysis was carried out for all recipient hamsters at 4 days post-cohousing (Fig. 3G). Remarkably, live SARS-CoV-2 particles were not detectable in either the nasal wash or the lungs of the hamsters cohoused with the SARS-CoV-2-infected d16-vaccinated hamsters, whereas high titers of infectious virions were detected in those of the hamsters cohoused with the sham-treated hamsters (PBS group, Fig. 3H). In this regard, d16 might have advantages over some existing SARS-CoV-2 vaccines that could be less capable of preventing SARS-CoV-2 transmission than past natural infection.
The results from our pathological examinations were in general agreement with the above findings. The lungs of PBS-treated hamsters showed severe pathological changes, e.g., large areas of consolidation as well as infiltration in the vascular endothelium and peribronchiolar regions. In sharp contrast, lungs from the WT and d16 groups exhibited almost normal morphology and were almost free of infiltration (Fig. 3I). To determine how vaccination with d16 might affect cytokine production, we determined the mRNA expression levels of major proinflammatory cytokines and chemokines, including IL-4, IL-6, IL-10 and CCL17. FOXP3, a key regulator of regulatory T cells [40], was also included in the analysis. Elevated expression of all these transcripts was observed in the WT and d16 groups but not in the PBS group (Fig. 3J), suggestive of heightened cytokine and immune responses consequent to challenge with SARS-CoV-2. Collectively, our results suggest that d16 provides full protection against SARS-CoV-2 challenge by preventing viral replication and the associated lung damage in the hamster model.
d16 elicited robust T-cell immunity in response to SARS-CoV-2 in a K18-hACE2 mouse model
Given the lack of reagents to study T-cell function or antibody responses in the hamster model, we tested the effect of d16 vaccination on T-cell immunity in a K18-hACE2 mouse model and compared this induced immunity with that in PBS-treated mice (Fig. 4A). First, we examined whether d16 vaccination induces a robust T-cell response following SARS-CoV-2 challenge. Two SARS-CoV-2 peptides that were previously shown to contain dominant CD8+ and CD4+ T-cell epitopes, S538–546 and S62–76 [41], were used in the analysis of the T-cell response. Notably, an increased number of SARS-CoV-2-specific S538–546+ CD8+ T cells was detected in the lungs (Fig. 4B). Phenotypic analysis revealed that a significant proportion of short-lived effector cells, a CD8+ T-cell subset responsible for rapid viral clearance, was present in the S538–546+ CD8+ T-cell population in d16-vaccinated mice (Fig. 4C). We further evaluated T-cell functionality by stimulating lung-derived lymphocytes with the S538–546 and S62–76 peptides as well as a peptide pool derived from the SARS-CoV-2 N protein (Fig. 4 and Supplementary Fig. S5). The frequencies of CD8+ and CD4+ T cells expressing functional markers (CD107a and granzyme B) related to cytotoxicity or producing proinflammatory cytokines (IFN-γ, TNF-α and IL-2) were significantly increased in d16-vaccinated mice stimulated with the S538–546 and S62–76 peptides (Fig. 4D, E). A generally consistent but less pronounced trend was seen when mice were stimulated with the peptide pool derived from the SARS-CoV-2 N protein (Supplementary Fig. S5A, B). Similar observations were made for T cells isolated from the spleen of S538–546-stimulated d16-vaccinated mice (Supplementary Fig. S6). Together, these results indicate that d16 elicits robust localized and systemic T-cell immunity against SARS-CoV-2.
T-cell immunity elicited by vaccination with d16 in the K18-hACE2 transgenic mouse model. A Vaccination and viral challenge scheme for d16-vaccinated K18-hACE2 mice assessed with a T-cell assay. K18-hACE2 mice were intranasally vaccinated with 103 PFU (in 20 µl) of d16 or PBS (n = 5 mice/group) and challenged with 104 PFU of clinical isolate HK-13 of SARS-CoV-2 on day 29. Lung-origin T cells were subjected to flow cytometric analysis at 4 days post-infection. B Representative flow tracings and the absolute number of S538–546-specific CD8+ T cells. C Proportions of short-lived effector cells (SLECs, KLRG1+ IL-7R-) among S538–546-specific CD8+ T cells. Lung-origin lymphocytes were stimulated with 1 μg/ml S538–546 (D) or S62–76 (E) peptides for 4 h in the presence of brefeldin A. The percentages of CD107a+ and cytokine-producing CD8+ (D) and CD4+ (E) T cells were also assessed. The results are shown as the mean ± SEM. Statistical analyses were performed with Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001)
Antibody responses elicited by d16 as a candidate live attenuated vaccine
The titers of NAbs directed against the SARS-CoV-2 RBD in the serum of d16-vaccinated mice post-SARS-CoV-2 challenge were higher than the basal levels in naïve mice (Fig. 5A, B). This was generally consistent with the results of the sVNT assay for neutralizing activity (Fig. 5B). Notably, live-virus MN assays indicated prominent neutralizing activity against challenge with the clinical isolate HK-13 of SARS-CoV-2 (Fig. 5C). These results were concordant with a pronounced increase in follicular T helper cells in the spleen (Fig. 5D), which are positive regulators of the germinal center (GC) reaction and fundamental for the generation of high-affinity, long-lasting antibody responses [42]. A higher proportion of GC B cells was also detected in the spleen of d16-vaccinated mice (Fig. 5E). Collectively, the results suggest that d16 induces potent T cell-mediated antibody responses, which are known to confer effective protection against viral infection. Intriguingly, RBD-specific immunoglobulin A (IgA) was detected in the serum and bronchoalveolar lavage fluid (BALF) of d16-vaccinated mice, suggesting stimulation of mucosal immunity (Fig. 5F, G).
Humoral and cell-mediated immune responses in d16-vaccinated mice. A Vaccination and viral challenge scheme for d16-vaccinated or PBS-treated K18-hACE2 mice assessed by antibody analysis. Briefly, K18-hACE2 mice intranasally inoculated with 103 PFU (in 20 µl) of d16 or PBS (n = 5 mice/group) were challenged with 104 PFU of clinical isolate HK-13 of SARS-CoV-2 on day 29. Serum samples were collected from each group on days 21 and 43 for ELISA and neutralization analysis. B Anti-RBD IgG ELISA and sVNT assays for neutralizing activity on day 21 post-vaccination (blue) and 14 days after challenge (green). The dotted line represents the assay cutoff at 30% inhibition for the sVNT assay. Basal levels were obtained for mice before the start of the experiment. C The cross-neutralization ability of d16-immunized mouse serum (1:10 dilution) was examined using a live-virus MN assay for the HK-13 clinical isolate of SARS-CoV-2. D Representative flow charts and percentages of T follicular helper cells (TFH cells, CD4+ PD1+ CXCR5+) in the spleen. E Representative flow charts and percentages of germinal center B cells (GC B cells, B220+ CD38- GL7+ FAS+) in the spleen. Results are shown as the mean ± SEM. Anti-RBD IgA in mouse serum (F) and bronchoalveolar lavage fluid (BALF) (G) collected on day 21 post-vaccination (blue) and 14 days after the challenge (green). Basal levels were obtained for mice before the start of the experiments. Statistical analyses performed with one-way ANOVA with Dunnett’s post-hoc test or Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001)
d16 vaccination confers full protection against lethal challenge with SARS-CoV-2 in K18-hACE2 mice
Finally, we examined the feasibility of applying d16 as a live attenuated vaccine in the K18-hACE2 model (Fig. 6A). A clinical isolate of SARS-CoV-2 (104 PFU) was inoculated intranasally into the mice on day 29 post-vaccination. All mice in the PBS group suffered from rapid weight loss and eventually died over the 14-day course of disease (Fig. 6B, C). In contrast, all mice in the d16-vaccinated group survived with a steady body weight increase. The viral loads in the lungs, the nasal wash and brain tissues were significantly decreased in d16-vaccinated mice at 4 dpi (Fig. 6D, P < 0.05). This was consistent with the SARS-CoV-2 N antigen being barely detectable in the lungs of mice vaccinated with d16 at 4 dpi (Fig. 6E). Concurrently, IL-1β and IFN-γ mRNA expression was significantly elevated (Fig. 6F). In addition, while diffuse alveolar wall thickening, blood vessel congestion, and an absence of alveolar space infiltration were observed in the PBS group, no inflammation was seen in the d16 group (Fig. 6G, H). Overall, our feasibility study performed K18-hACE2 transgenic mice provided strong justifications and paved the way for further assessment of d16 as a candidate live attenuated vaccine against SARS-CoV-2 in nonhuman primates.
d16 confers full protection against SARS-CoV-2 lethal challenge in the K18-hACE2 transgenic mouse model. A Vaccination and viral challenge scheme for K18-hACE2 mice (n ≥ 10). After intranasal vaccination with 103 PFU (in 20 µl) of recombinant d16 SARS-CoV-2 or PBS, each vaccinated mouse was challenged with 104 PFU (in 20 µl) of clinical isolate HK-13 of SARS-CoV-2 on day 29. Body weights were recorded daily for 14 days after the viral challenge. On day 33, the nasal wash, lung tissues and brain tissues were collected for quantification of viral loads using RT–qPCR. Body weight changes (B) and survival rates (C) of SARS-CoV-2-infected mice vaccinated with d16 or PBS (n = 5/group). D Viral loads measured by plaque assays or RT–qPCR for the nasal wash, lung tissues and brain tissues collected from SARS-CoV-2-infected mice vaccinated with d16 or PBS at 4 dpi (n ≥ 5/group). Statistical analyses were performed with Student’s t-test (*P < 0.05; ***P < 0.001). E Detection of the SARS-CoV-2 N protein in the lung tissues of SARS-CoV-2-infected mice vaccinated with d16 or PBS. The SARS-CoV-2 N protein (green) was labeled with rabbit anti-SARS-CoV-2 N antibodies followed by goat anti-rabbit antibodies conjugated to FITC. Nuclei were counterstained with DAPI (blue). Scale bars, 50 μm. F IL-1β and IFN-γ gene expression in SARS-CoV-2-infected mouse lungs. The transcripts of representative chemokines and cytokines in lung tissue homogenates from the indicated groups were quantitated using RT–qPCR. The results are shown as the mean ± SEM. Statistical analyses were performed with Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). G Effect of vaccination on the histopathological changes in the lungs of SARS-CoV-2-infected mice. Representative sections of lung tissue from mice harvested at 4 dpi were stained with H&E. Scale bars, 100 μm. H Pathological changes were scored as described in the Materials and Methods (**P < 0.01)
Although no reversions or other mutations were found in NSP16 of the d16 virus (Supplementary Fig. S1), it will be of great interest to see whether mutations might be present in other areas of the SARS-CoV-2 genome. In this regard, catalytic disruption of NSP16 in SARS-CoV might be compensated by mutations in other regions [21]. With this in mind, we determined the complete sequence of passage-10 d16 (d16-P10) after passaging in VeroE6 cells. Notably, 6 minor nucleotide substitutions were found in d16-P10 (Supplementary Fig. S7A). The plaques produced by d16-P10 were smaller (Supplementary Fig. S7B), but the survival phenotype remained unchanged (Supplementary Fig. S7C) when compared to that of the parental d16 virus (Figs. 1D and 2J). Importantly, the protective efficacies of d16-P10 against SARS-CoV-2 infection and associated lung damage (Supplementary Fig. S7D, E) were similar to those of d16 (Fig. 6D, G). Thus, the nucleotide changes did not seem to influence the attenuation or protective efficacy of d16-P10. In other words, there was no evidence of virulence reversion in d16-P10.
Discussion
Vaccination against SARS-CoV-2 is the most effective measure to control the COVID-19 pandemic. With the development of some degree of immunity against SARS-CoV-2 in increasingly more people in the world achieved through natural infection and/or vaccination, the severity of COVID-19 might decrease to a level at which the associated mortality would be similar to that of influenza or the common cold [43]. However, booster immunization with preferably variant-targeted vaccines is still urgently needed. In addition, vaccines that can activate mucosal and sterilize immunity against SARS-CoV-2 would be another game-changer in the fight against COVID-19 [44]. As the first step toward this goal, we provide proof-of-principle evidence for using NSP16-deficient SARS-CoV-2 as a prototypical candidate strain for live attenuated vaccines that elicit mucosal and sterilizing immunity.
The availability of infectious molecular clones of SARS-CoV-2 has greatly facilitated the rational design and development of live attenuated vaccines [33,34,35]. In addition to the point mutation introduced into d16, several other strategies can be considered for further optimization. First, deletion might be introduced to limit the activation of NSP16 to further reduce the chance for reversion. Whereas a point mutation has the best chance of producing a minimal impact on the structure of the viral genome, deletion should be less likely to be reverted. Indeed, a SARS-CoV-2 virus with deletion of the whole KDKE motif in NSP16 was generated and found to be stable (our unpublished data). Second, other deletions or mutations could be incorporated into d16. For instance, further deletion of ORF3a or E might be considered. This might further prevent reversion and improve safety. Third, a variant-specific mutant might be constructed. For example, an NSP16-deficient Delta or Omicron variant could be created so that the candidate vaccine might be variant targeted in terms of immunogenicity. Since the Delta and Omicron variants are highly transmissible and preferentially replicate to higher titers in the upper respiratory tract [2, 45], it will be of great interest to see whether an NSP16-deficient Delta or Omicron variant might be sufficiently attenuated and nonpathogenic but remain transmissible and replicate well in the nasal mucosa. Such a variant might serve as an ideal candidate vaccine strain with a reasonable viral yield but sufficiently attenuated. In particular, transmission studies will be performed in hamsters with the NSP16-deficient Delta or Omicron variant as described previously [46] to determine whether the vaccine strain might be as highly transmissible as the corresponding parental strain and whether it can be transmitted via aerosols [47]. Fourth, WT NSP16 might be expressed in trans in cultured cells to increase the viral yield of d16. Fifth, a recombination-resistant d16 virus could be made by rewiring the transcriptional regulatory network as described previously [48]. This will render d16 safer. Last but not least, d16 might be tested for use in a booster vaccination context following immunization with an mRNA vaccine. This combination might take advantage of the strengths of both vaccines in the induction of NAbs and mucosal immunity against SARS-CoV-2. In addition, it would also relieve the safety concern for d16. This approach holds the promise of inducing an immune response with a broader spectrum and longer duration. A similar vaccination scheme in which the Salk and Sabin vaccines are sequentially injected was used successfully in the late stage of the eradication of poliovirus [49].
The highly compartmentalized mucosal immune system serves to defend the mucosae, the predominant infection sites of SARS-CoV-2. Mucosa-associated lymphoid tissues, such as the broncho-associated lymphoid tissue within the airway mucosa, function independently of the systemic immune system [50]. Effective mucosal immunity can be induced at the desired sites only when the right vaccination route is used. Particularly, upon nasal vaccination in humans, the strongest secretory IgA (sIgA) response is induced in the upper airway mucosa, saliva and nasal secretions, while the second-best sIgA response takes place in the lungs. Thus, nasal administration might have the best chance of inducing sterilizing immunity against SARS-CoV-2. In our study, the detection of sIgA in the BALF of d16-vaccinated mice (Fig. 5F, G) indicated efficient activation of mucosal immunity against SARS-CoV-2 in the respiratory tract. Moreover, d16 vaccination-induced robust localized T-cell immunity in the lungs (Fig. 4). Plausibly, d16 would be as efficient as the WT virus in the activation of mucosal immunity in the respiratory tract. It has been shown that tissue-resident memory cells but not circulating memory cells provide optimal protection against rechallenge with influenza virus [51]. Therefore, long-term monitoring of systemic and regional immune memory in d16-vaccinated mice is warranted in future studies.
Compared to WT SARS-CoV-2, d16 induced FOXP3 and a few immunomodulatory cytokines more robustly (Fig. 2G). By modulating mucosal immunity and the antigen-specific cytotoxic T-cell response, IL-4 might serve as a vaccine adjuvant [52]. FOXP3 and CCL17 are the defining transcription factor and chemoattractant of regulatory T cells, respectively. These cells play crucial roles in limiting the overt immune response to SARS-CoV-2 [40, 53]. Hence, the induction of these genes by d16 might help to enhance immunogenicity and induce immune responses in a well-controlled manner.
In contrast to that seen before challenge with SARS-CoV-2, more robust neutralizing activity was detected in the live-virus MN assay when d16-vaccinated mice were infected with a clinical isolate of SARS-CoV-2 (Fig. 5C). The increased neutralizing activity might be attributed to antibody affinity maturation when mice were re-exposed to the antigens during SARS-CoV-2 infection [8, 54, 55]. This resulted in B-cell maturation and a substantial increase in the affinity of antibodies for antigens, as seen in prime-boost immunization and natural reinfection [8].
Similar to natural infection, vaccination with d16 apparently conferred sterilizing protection against SARS-CoV-2 infection. No viable virions were detected in the nasal wash of d16-vaccinated hamsters infected with SARS-CoV-2 or hamsters cohoused with them (Fig. 3F, H). No inflammation was seen in the lungs of d16-vaccinated hamsters (Fig. 3I) or K18-hACE2 mice (Fig. 6G) infected with SARS-CoV-2. Breakthrough infections of SARS-CoV-2 following administration of human vaccines often occur in the nasal mucosa [56]. The absence of live virions in the nasal wash is consistent with the sterilizing immunity observed in d16-vaccinated hamsters. Exactly how mucosal immunity contributes to sterilizing protection against SARS-CoV-2 merits further investigation. Nevertheless, our work provides the first evidence for the feasibility of using NSP16-deficient SARS-CoV-2 as a candidate live attenuated vaccine. Further optimization based on this prototypic strain might result in a live attenuated vaccine against SARS-CoV-2 that can ultimately be tested in humans.
Materials and methods
Viruses and cells
The SARS-CoV-2 virus HK-13 isolate (GenBank accession number: MT835140) was isolated from the nasopharyngeal aspirate specimen of a laboratory-confirmed COVID-19 patient in Hong Kong [57]; this isolate was cultured and titrated in VeroE6 cells by plaque assays. All experiments involving live SARS-CoV-2 were performed in a biosafety level 3 facility. A549-ACE2-TMPRSS2 cells were purchased from InvivoGen, and experiments were performed according to the manufacturer’s instructions. VeroE6 cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (P/S). Calu-3 cells were cultured in DMEM-F12 supplemented with 10% FBS and 1% P/S. VeroE6-TMPRSS2 cells were grown in DMEM supplemented with 10% FBS, 1 mg/ml G418 and 1% P/S. A549-ACE2-TMPRSS2 cells were grown in DMEM supplemented with 10% FBS, 1% P/S, 100 μg/ml normocin, 300 μg/ml hygromycin, and 0.5 μg/ml puromycin.
Generation of attenuated SARS-CoV-2 harboring the NSP16 D130A mutation
An infectious molecular clone of SARS-CoV-2 on a BAC, named p-BAC-SARS-CoV-2, was generated and characterized as described previously [33,34,35]. The D130A point mutation was generated in p-BAC-SARS-CoV-2 using λ-Red-mediated homologous recombination as previously described [58]. Briefly, forward and reverse primers containing the D130A nucleotide substitution were synthesized (Integrated DNA Technologies). The primers were annealed and subsequently purified using a QIAquick Gel Extraction Kit (Qiagen). The purified PCR fragment was then electroporated into the BAC clone-incorporated SW105 strain of Escherichia coli. After the initial isolation of galactokinase (galk)-positive clones, successful recombinants were further counterselected using M63 minimal agar with 2-deoxy-D-galactose. The clones were purified and subjected to Sanger sequencing for validation of the D130A mutation in the BAC DNA sequence. The primer sequences are listed in Supplementary Table 1. Validated BAC DNA carrying SARS-CoV-2 d16 with the NSP16 D130A mutation was transfected into VeroE6-TMPRSS2 cells preseeded in a 6-well plate using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. The cells were monitored for cytopathic effects on a daily basis. Virus-containing cells were harvested at 3~4 dpi. SARS2-d16 P0 was further passaged in VeroE6 cells. The titers of the viral stock were determined with a plaque assay using VeroE6 cells.
Plaque assay
Confluent VeroE6 or VeroE6-TMPRSS2 cells seeded in a 12-well plate were incubated with 10-fold serial dilutions of virus for 1 h. After adsorption to VeroE6 cells, the viral supernatant was discarded, followed by rinsing with PBS and overlaying of 1% agarose in DMEM. The cells were incubated for 3 days at 37 °C and then fixed with 10% formaldehyde for 1 day. The overlaid agarose was then removed, and the plaques were stained with 1% crystal violet and counted to measure viral titers.
Growth kinetics
Confluent VeroE6, Calu-3 cells or A549-ACE2-TMPRSS2 cells were infected at a multiplicity of infection (MOI) of 0.1 with the indicated viruses and incubated for 2 days. The virus-containing supernatant was collected at various time points. Viral titers were determined by RT–qPCR as described previously [36, 59].
Transcriptomic analysis
A549-ACE2-TMPRSS2 cells were either mock infected or infected with the WT virus or d16 for 24 h at an MOI of 0.1. Cell lysates were subjected to RNA extraction using an RNeasy Mini Kit. All samples were sequenced on the BGISEQ platform using the constructed library. Paired-end reads were aligned with a reference genome (GRCh38) using HISAT2 and Bowtie2. The levels of gene expression were quantitated as transcripts per kilobase of exon model per million mapped reads (TPM). The limma R package was used to perform differential expression analysis with a cutoff P value of 0.05 [57]. The complex heatmap R package [58] was used to visualize the top 10 differentially expressed genes normalized by z score for the d16 vs. WT + mock, WT vs. d16 + mock, and mock vs. D16 + WT comparisons. Effects on vital biological gene functions were identified by performing enrichment analysis using the clusterProfiler R package [60] with the default parameters of a false discovery rate <0.05 and 1000 permutations. The RNA-seq data were deposited in Mendeley Data under the DOI of 10.17632/d/dnscfxkddd.1.
Infection of Syrian hamsters and K18-hACE2 transgenic mice
All experimental protocols were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) of the University of Hong Kong and were performed according to the standard operating procedures of the biosafety level 3 animal facilities (reference code: CULATR 5754-21) in adherence with the NIH Guide for the Care and Use of Laboratory Animals. Animal infection experiments were performed as previously described with slight modifications [61]. Syrian hamsters (6–8 weeks old) were obtained from the Chinese University of Hong Kong Laboratory Animal Service through the Center for Comparative Medicine Research (CCMR) of the University of Hong Kong. K18-hACE2 transgenic mice were bred, raised in the CCMR and delivered upon reaching 6–8 weeks of age. The animals were raised in a biosafety level 2 facility with ad libitum access to standard pellet feed and water. During vaccination, hamsters were first anesthetized with intraperitoneal ketamine (200 mg per kg) and xylazine (10 mg per kg) administration and then intranasally inoculated with WT or mutant SARS-CoV-2 at 105 PFU. PBS served as a diluent to obtain the desired concentrations from viral stocks. A challenge dose of 105 PFU SARS-CoV-2 (clinical isolate HK-13) was used to challenge vaccinated or sham-vaccinated hamsters [59]. For K18-hACE2 transgenic mice, a dose of 103 PFU recombinant virus (d16) was used for vaccination, whereas a dose of 104 PFU SARS-CoV-2 (clinical isolate HK-13) was inoculated for antiviral evaluation. Body weights were continuously monitored for 14 days after infection or until animal death. For virological and histopathological examinations, animals were sacrificed at 4 dpi, and their organs, tissues, nasal wash and blood were collected for analyses. The two halves of harvested tissues were used for viral titration by RT–qPCR or a plaque assay and for histopathological analysis following fixation in 10% PBS-buffered formaldehyde, as we previously described [59]. For histopathological assessment, a semiquantitative system was employed to evaluate the relative degree of inflammation and tissue damage [62]. Inflammation was scored as follows: 0, no inflammation; 1, perivascular cuff of inflammatory cells; 2, mild inflammation (extending through 25% of the lung); 3, moderate inflammation (25–50% of the lung); and 4, severe inflammation involving over half of the lung. The examination of tissue slides was carried out in a single-blind manner.
Antibody tests
An enzyme-based immunoassay was performed as previously described [59, 63]. Ninety-six-well plates (Nunc, Rochester, NY, USA) were coated with 0.05 μg/well SARS-CoV-2 RBD in 100 μl of 0.05 M NaHCO3 (pH 9.6) overnight at 4 °C. After blocking, 100 μl of heat-inactivated serum samples was serially diluted before being added to the wells and incubated at 37 °C for 1 h. RBD-binding antibodies were detected using a horseradish peroxidase-conjugated goat anti-hamster IgG antibody (A18895 from Invitrogen, Waltham, MA, USA), anti-mouse IgG antibody (Life Technology 626720) or anti-mouse IgA antibody (Sigma–Aldrich SAB5600003), followed by the addition of a diluted 3,3′,5,5′-tetramethylbenzidine (TMB) single solution (Invitrogen) and 1 M HCl. The optical density at 450 nm (OD450) was measured using a microplate reader.
The sVNT was performed using cPass (GenScript) according to the manufacturer’s instructions [64]. Briefly, equal volumes of 1:10 diluted hamster or mouse serum were incubated with a horseradish peroxidase-conjugated RBD for 30 min, followed by an addition to an ACE2-coated ELISA plate for 30 min at 37 °C. Colorimetric signals were developed using the TMB substrate, and the reaction was stopped with 1 M HCl. The OD450 was measured using a microplate reader.
The SARS-CoV-2 MN assay was performed in a certified biosafety level 3 laboratory. Test serum samples were serially diluted, mixed with 50 μL of SARS-CoV-2 at 103 focus forming units/ml in 96-well plates and incubated for 1 h at 37 °C. The virus-serum mixtures were transferred to preseeded 96-well plates (VeroE6 cells, 2 × 104 cells/well) and incubated for 24 h. After the incubation, the culture medium was removed, and the plates were air-dried in a biosafety cabinet (BSC) for 20 min. The cells were then fixed with 10% PBS-buffered formaldehyde for 30 min and air-dried in the BSC again. After permeabilization with 0.2% Triton X-100 in PBS, the cells were incubated with rabbit antiserum raised in-house against the SARS-CoV-2 N protein for 1 h at room temperature, followed by the addition of an Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody (Life Technologies). SARS-CoV-2 foci were quantitated using a Sapphire Biomolecular Imager (Azure Biosystems).
RT–qPCR
RNA extraction, reverse transcription, and qPCR were performed as previously described [65,66,67]. Briefly, total RNA was extracted from hamster or mouse organs (lungs, nasal wash, brain or trachea) with an RNeasy Mini Kit (Qiagen, Germantown, MD, USA) and reverse-transcribed with a Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). Real-time PCR was performed using a CFX96 Touch Deep Well Real-Time PCR Detection System (Bio–Rad, Berkeley, California, USA) according to the manufacturer’s instructions. Relative gene expression was determined either through normalization to the corresponding Ct values for β-actin or glyceraldehyde 3-phosphate dehydrogenase transcripts or by using the 2-ΔΔCt method [68].
Immunostaining and confocal imaging
Immunostaining was performed to visualize the SARS-CoV-2 N protein in hamster lung tissues as we previously described [59]. An in-house-generated rabbit anti-SARS-CoV-2-N protein antibody and a goat anti-rabbit IgG (H+L) antibody conjugated to fluorescein isothiocyanate (FITC; Sigma–Aldrich, AP307F) (Thermo Fisher Scientific, Waltham, MA, USA) were used as the primary and secondary antibodies, respectively. Cell nuclei were stained using the nucleic acid stain 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific). Images were acquired with a Carl Zeiss (Dublin, CA, USA) LSM880 system.
Flow cytometric analysis
Splenocytes were isolated and filtered through a 70-μm cell strainer (BD) and then resuspended in RC-10 medium (RPMI 1640 medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, 50 mM 2-mercaptoethanol, and 10 mM HEPES). Lung tissues were diced and digested in an RPMI solution containing collagenase II (1 mg/ml) (Sigma) and DNase (10 mg/ml) (Roche) for 1 h at 37 °C. Red blood cells were lysed using ACK buffer. Cells were treated with a saturating concentration of the 2.4G2 monoclonal antibody (anti-CD16/32) to block Fc receptors and stained in a PBS solution containing 1% BSA, 2 mM EDTA, and 0.09% NaN3. Gating boundaries were determined with fluorescence minus one controls. The following antibodies against mouse antigens were used: FC block (BioLegend 156604), anti-TCR-β (BV650, BioLegend 109251), anti-CD4 (PerCP-Cy5.5, BioLegend 116012), anti-CD8 (BV421, BioLegend 100753), anti-CD44 (FITC, BioLegend 103005), anti-KLRG1 (APC, BioLegend 138412), anti-IL-7R (BV785, BioLegend 135037), anti-CD62L (PE-Cy7, BioLegend 104418), anti-CD107a (A488, BioLegend 121608), anti-IFN-γ (PE-Cy7, BioLegend 505825), anti-TNF-α (BV785, BioLegend 506341), anti-IL-2 (APC, BioLegend 503810), anti-granzyme B (PE, BioLegend 396406), anti-PD1 (BV785, BioLegend 135225), anti-CXCR5 (biotin, Invitrogen 13-7185-82), anti-streptavidin (AF647, BioLegend 405237), anti-ICOS (A488, BioLegend 313514), anti-B220 (APC/Fire750, BioLegend 103260), anti-GL-7 (Pacific blue, BioLegend 144614), anti-CD38 (PerCP-Cy5.5, BioLegend 102721), anti-CD95 (PE-Cy7, BD Biosciences 557653), anti-CD138 (PE, BioLegend 142504), anti-IgG1 (APC, BioLegend 406610), and anti-IgG2a (FITC, BioLegend 407106). To exclude dead cells from the staining results, a live/dead fixable Aqua stain kit (Molecular Probes, Life Technologies) was used according to the manufacturer’s instructions. Antigen-specific CD8+ T cells were detected using H2-Db S538–546-specific dextramers (Immudex). For intracellular cytokine staining, cells were stimulated in RC-10 medium containing 1 mg/ml S538–546 or S62–76 peptides of 9 or 16 amino acids in length (GL Biochem), a peptide pool derived from the N protein (GenScript) [69], or no peptides as a control. After 1 h, brefeldin A was added, and the cells were incubated for 4 h at 37 °C. A BD Cytofix/Cytoperm™ Plus Fixation/Permeabilization Kit (BD Bioscience) was then used according to the manufacturer’s instructions. All samples were acquired on an LSR Fortessa flow cytometer (BD Biosciences, USA), and the data were analyzed with FlowJo software. Cytokine production was calculated by subtracting the background signal for the no-peptide control.
Protein structural analysis and statistical analysis
The NSP16 amino acid sequence from residue 6718 to residue 7096, with aspartic acid substituted for alanine at 6928 (namely, D130A), was submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/) for structural prediction of the mutant MTase active site. Statistical analysis was carried out using GraphPad Prism software (GraphPad Software). Data are presented as the mean ± S of at least three replicates, unless indicated otherwise. Statistical significance was analyzed by Student’s t-test, one-way analysis of variance with Dunnett’s post-hoc test, or two-way ANOVA with Bonferroni’s post-hoc test. For all tests, asterisks were used to indicate the range of P values: ***P < 0.001; **P < 0.01; *P < 0.05; and ns: not significant.
Data availability
All data are available in the main text or Supplementary materials.
References
Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings – Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2012;70:1059–62. https://doi.org/10.15585/mmwr.mm7031e2
Mishra S, Sören M, Sharma M, Whittaker C, Mellan TA, Wilton T, et al. Changing composition of SARS-CoV-2 lineages and rise of delta variant in England. EClinicalMedicine. 2021;39:101064. https://doi.org/10.1016/j.eclinm.2021.101064
Nanduri S, Pilishvili T, Derado G, Soe MM, Dollard P, Wu H, et al. Effectiveness of Pfizer-BioNTech and Moderna vaccines in preventing SARS-CoV-2 infection among nursing home residents before and during widespread circulation of the SARS-CoV-2 B.1.617.2 (δ) variant – National Healthcare Safety Network, March 1-August 1, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1163–6. https://doi.org/10.15585/mmwr.mm7034e3
Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, et al. Breakthrough infections with SARS-CoV-2 Omicron despite mRNA vaccine booster dose. Lancet. 2022;399:625–6. https://doi.org/10.1016/S0140-6736(22)00090-3
Tré-Hardy M, Cupaiolo R, Wilmet A, Antoine-Moussiaux T, Della Vecchia A, Horeanga A, et al. Six-month interim analysis of ongoing immunogenicity surveillance of the mRNA-1273 vaccine in healthcare workers: a third dose is expected. J Infect. 2021;S0163-4453:00433–3. https://doi.org/10.1016/j.jinf.2021.08.031
Choi A, Koch M, Wu K, Chu L, Ma L, Hill A, et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 2021;27:2025–31. https://doi.org/10.1038/s41591-021-01527-y
Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–11. https://doi.org/10.1038/s41591-021-01377-8
Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–27. https://doi.org/10.1038/s41586-020-2798-3
Zhou R, To KKW, Wong YC, Liu L, Zhou B, Li X, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. https://doi.org/10.1016/j.immuni.2020.07.026
Jarjour NN, Masopust D, Jameson SC. T cell memory: understanding COVID-19. Immunity. 2021;54:14–18. https://doi.org/10.1016/j.immuni.2020.12.009
Minor PD. Live attenuated vaccines: historical successes and current challenges. Virology. 2015;479-480:379–92. https://doi.org/10.1016/j.virol.2015.03.032
Cheng SMS, Mok CKP, Leung YWY, Ng SS, Chan KCK, Ko FW, et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022. https://doi.org/10.1038/s41591-022-01704-7
Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin A, Kawabata H, et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–8. https://doi.org/10.1038/s41586-022-04399-5
Lu L, Mok BW, Chen LL, Chan JM, Tsang OT, Lam BH, et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021:ciab1041. https://doi.org/10.1093/cid/ciab1041
Dangi T, Palacio N, Sanchez S, Park M, Class J, Visvabharathy L, et al. Cross-protective immunity following coronavirus vaccination and coronavirus infection. J Clin Invest. 2021;131:e151969 https://doi.org/10.1172/JCI151969
Gao Y, Cai C, Grifoni A, Müller TR, Niessl J, Olofsson A, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med. 2022. https://doi.org/10.1038/s41591-022-01700-x
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV- a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–36. https://doi.org/10.1038/nrmicro2090
DeDiego ML, Alvarez E, Almazán F, Rejas MT, Lamirande E, Roberts A, et al. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. 2007;81:1701–13. https://doi.org/10.1128/JVI.01467-06
Siu KL, Yuen KS, Castaño-Rodriguez C, Ye ZW, Yeung ML, Fung SY, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–77. https://doi.org/10.1096/fj.201802418R
Menachery VD, Yount BL Jr, Josset L, Gralinski LE, Scobey T, Agnihothram S, et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-O-methyltransferase activity. J Virol. 2014;88:4251–64. https://doi.org/10.1128/JVI.03571-13
Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH III, Leist SR, Yount BL Jr, et al. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J Virol. 2018;92:e00710–18. https://doi.org/10.1128/JVI.00710-18
Graepel KW, Agostini ML, Lu X, Sexton NR, Denison MR. Fitness barriers limit reversion of a proofreading-deficient coronavirus. J Virol. 2019;93:e00711–19. https://doi.org/10.1128/JVI.00711-19
Mandilara G, Koutsi MA, Agelopoulos M, Sourvinos G, Beloukas A, Rampias T. The role of coronavirus RNA-processing enzymes in innate immune evasion. Life (Basel). 2021;11:571 https://doi.org/10.3390/life11060571
Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111:E3900–3909. https://doi.org/10.1073/pnas.1323705111
Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–26. https://doi.org/10.1093/nar/gkw551
Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor MDA5. Nat Immunol. 2011;12:137–43. https://doi.org/10.1038/ni.1979
Zhang Z, Liu Q, Sun Y, Li J, Liu J, Pan R, et al. Live attenuated coronavirus vaccines deficient in N7-methyltransferase activity induce both humoral and cellular immune responses in mice. Emerg Microbes Infect. 2021;10:1626–37. https://doi.org/10.1080/22221751.2021.1964385
Ogando NS, Zevenhoven-Dobbe JC, van der Meer Y, Bredenbeek PJ, Posthuma CC, Snijder EJ. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94:e01246–20. https://doi.org/10.1128/JVI.01246-20
Wang Y, Yang C, Song Y, Coleman JR, Stawowczyk M, Tafrova J, et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy. Proc Natl Acad Sci USA. 2021;118:e2102775118 https://doi.org/10.1073/pnas.2102775118
Trimpert J, Dietert K, Firsching TC, Ebert N, Thi Nhu Thao T, Vladimirova D, et al. Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding. Cell Rep. 2021;36:109493 https://doi.org/10.1016/j.celrep.2021.109493
Trimpert J, Adler JM, Eschke K, Abdelgawad A, Firsching TC, Ebert N, et al. Live attenuated virus vaccine protects against SARS-CoV-2 variants of concern B.1.1.7 (Alpha) and B.1.351 (Beta). Sci Adv. 2021;7:eabk0172 https://doi.org/10.1126/sciadv.abk0172
Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH III, Leist SR, Yount BL Jr, et al. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere. 2017;2:e00346–17. https://doi.org/10.1128/mSphere.00346-17
Almazán F, Dediego ML, Galán C, Escors D, Alvarez E, Ortego J, et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol. 2006;80:10900–6. https://doi.org/10.1128/JVI.00385-06
Ye C, Chiem K, Park JG, Oladunni F, Platt RN II, Anderson T, et al. Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio. 2020;11:e02168–20. https://doi.org/10.1128/mBio.02168-20
Thi Nhu Thao T, Labroussaa F, Ebert N, V’kovski P, Stalder H, Portmann J, et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–5. https://doi.org/10.1038/s41586-020-2294-9
Chan JFW, Zhang AJ, Yuan S, Poon VKM, Chan CCS, Lee ACY, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–46. https://doi.org/10.1093/cid/ciaa325. (2020)
Zheng J, Wong LYR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–7. https://doi.org/10.1038/s41586-020-2943-z
Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21:1327–35. https://doi.org/10.1038/s41590-020-0778-2
Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27:620–1. https://doi.org/10.1038/s41591-021-01270-4
Li Z, Li D, Tsun A, Li B. FOXP3+ regulatory T cells and their functional regulation. Cell Mol Immunol. 2015;12:558–65. https://doi.org/10.1038/cmi.2015.10
Zhuang Z, Lai X, Sun J, Chen Z, Zhang Z, Dai J, et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J Exp Med. 2021;218:e20202187 https://doi.org/10.1084/jem.20202187
Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–42. https://doi.org/10.1016/j.immuni.2014.10.004
Morens DM, Daszak P, Markel H, Taubenberger JK. Pandemic COVID-19 joins history’s pandemic legion. mBio. 2020;11:e00812–20. https://doi.org/10.1128/mBio.00812-20
Lavelle EC, Ward RW. Mucosal vaccines – fortifying the frontiers. Nat Rev Immunol. 2021:1–15. https://doi.org/10.1038/s41577-021-00583-2.
Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022. https://doi.org/10.1038/s41586-022-04474-x
Chan JFW, Yuan S, Zhang AJ, Poon VK, Chan CC, Lee AC, et al. Surgical mask partition reduces the risk of noncontact transmission in a golden syrian hamster model for Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. 2020;71:2139–49. https://doi.org/10.1093/cid/ciaa644
Riediker M, Briceno-Ayala L, Ichihara G, Albani D, Poffet D, Tsai DH, et al. Higher viral load and infectivity increase risk of aerosol transmission for Delta and Omicron variants of SARS-CoV-2. Swiss Med Wkly. 2022;152:w30133 https://doi.org/10.4414/smw.2022.w30133
Graham RL, Deming DJ, Deming ME, Yount BL, Baric RS. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun Biol. 2018;1:179 https://doi.org/10.1038/s42003-018-0175-7
Faden H. Poliovirus vaccination: a trilogy. J Infect Dis. 1993;168:25–28. https://doi.org/10.1093/infdis/168.1.25
Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11:S45–S53. https://doi.org/10.1038/nm1213
Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7:501–10. https://doi.org/10.1038/mi.2013.67
Ranasinghe C, Trivedi S, Wijesundara DK, Jackson RJ. IL-4 and IL-13 receptors: roles in immunity and powerful vaccine adjuvants. Cytokine Growth Factor Rev. 2014;25:437–42. https://doi.org/10.1016/j.cytogfr.2014.07.010
Moriyama S, Adachi Y, Sato T, Tonouchi K, Sun L, Fukushi S, et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity. 2021;54:1841–52. https://doi.org/10.1016/j.immuni.2021.06.015. e4
Muecksch F, Weisblum Y, Barnes CO, Schmidt F, Schaefer-Babajew D, Wang Z, et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853–68. e7 https://doi.org/10.1016/j.immuni.2021.07.008.
Uraki R, Imai M, Ito M, Shime H, Odanaka M, Okuda M, et al. Foxp3+ CD4+ regulatory T cells control dendritic cells in inducing antigen-specific immunity to emerging SARS-CoV-2 antigens. PLoS Pathog. 2021;17:e1010085 https://doi.org/10.1371/journal.ppat.1010085
Kustin T, Harel N, Finkel U, Perchik S, Harari S, Tahor M, et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat Med. 2021;27:1379–84. https://doi.org/10.1038/s41591-021-01413-7
Wong LYR, Ye ZW, Lui PY, Zheng X, Yuan S, Zhu L, et al. Middle East respiratory syndrome coronavirus ORF8b Accessory protein suppresses type I IFN expression by impeding HSP70-dependent activation of IRF3 kinase IKKε. J Immunol. 2020;205:1564–79. https://doi.org/10.4049/jimmunol.1901489
Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29 https://doi.org/10.1186/gb-2014-15-2-r29
Wang P, Lau SY, Deng S, Chen P, Mok BWY, Zhang AJ, et al. Characterization of an attenuated SARS-CoV-2 variant with a deletion at the S1/S2 junction of the spike protein. Nat Commun. 2021;12:2790. https://doi.org/10.1038/s41467-021-23166-0
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. https://doi.org/10.1093/bioinformatics/btw313
Ye ZW, Yuan S, Chan JFW, Zhang AJ, Yu CY, Ong CP, et al. Beneficial effect of combinational methylprednisolone and remdesivir in hamster model of SARS-CoV-2 infection. Emerg Microbes Infect. 2021;10:291–304. https://doi.org/10.1080/22221751.2021.1885998
Yuan S, Wang R, Chan JFW, Zhang AJ, Cheng T, Chik KKH, et al. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat Microbiol. 2020;5:1439–48. https://doi.org/10.1038/s41564-020-00802-x
Zhou D, Chan JFW, Zhou B, Zhou R, Li S, Shan S, et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021;29:551–63. e555 https://doi.org/10.1016/j.chom.2021.02.019.
Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun. 2021;12:972 https://doi.org/10.1038/s41467-021-21240-1
Yuan S, Yin X, Meng X, Chan JFW, Ye ZW, Riva L, et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–23. https://doi.org/10.1038/s41586-021-03431-4
Ye ZW, Yuan S, Poon KM, Wen L, Yang D, Sun Z, et al. Antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin head region of pandemic H1N1 influenza virus play detrimental roles in H1N1-infected mice. Front Immunol. 2017;8:317 https://doi.org/10.3389/fimmu.2017.00317
Chan JFW, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–20. https://doi.org/10.1128/JCM.00310-20
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. https://doi.org/10.1006/meth.2001.1262
Zhou R, To KK, Peng Q, Chan JM, Huang H, Yang D, et al. Vaccine-breakthrough infection by the SARS-CoV-2 Omicron variant elicits broadly cross-reactive immune responses. Clin Transl Med. 2022;12:e720. https://doi.org/10.1002/ctm2.720
Acknowledgements
We thank members of the Jin Laboratory for critically reading the manuscript. This work was supported by the Hong Kong Health and Medical Research Fund grants COVID190121 to JF-WC and COVID190114 to D-YJ and the Hong Kong Research Grants Council grants C7142-20GF and T11-709/21-N to D-YJ.
Author information
Authors and Affiliations
Contributions
Z-WY, SY and D-YJ conceptualized and designed the study. Z-WY, CPO, KT and YF performed most experiments with help from CL, RZ, PL, YC, VG and PW. Z-WY and KT performed the infection study. G-SL performed the T-cell analysis. All authors contributed to the data analysis. Z-WY, SY and D-YJ wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ye, ZW., Ong, C.P., Tang, K. et al. Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell Mol Immunol 19, 588–601 (2022). https://doi.org/10.1038/s41423-022-00855-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-022-00855-4
Keywords
This article is cited by
-
SIgA in various pulmonary diseases
European Journal of Medical Research (2023)
-
Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target
Nature Communications (2023)
-
An interferon-integrated mucosal vaccine provides pan-sarbecovirus protection in small animal models
Nature Communications (2023)
-
Monovalent vaccination with inactivated SARS-CoV-2 BA.5 protects hamsters against Omicron but not non-Omicron variants
npj Vaccines (2023)