Abstract
The pandemic of COVID-19 caused by SARS-CoV-2 continues to spread around the world. Mutant strains of SARS-CoV-2 are constantly emerging. At present, Omicron variants have become mainstream. In this work, we carried out a systematic and comprehensive analysis of the reported spike protein antibodies, counting the epitopes and genotypes of these antibodies. We further comprehensively analyzed the impact of Omicron mutations on antibody epitopes and classified these antibodies according to their binding patterns. We found that the epitopes of the H-RBD class antibodies were significantly less affected by Omicron mutations than other classes. Binding and virus neutralization experiments showed that such antibodies could effectively inhibit the immune escape of Omicron. Cryo-EM results showed that this class of antibodies utilized a conserved mechanism to neutralize SARS-CoV-2. Our results greatly help us deeply understand the impact of Omicron mutations. Meanwhile, it also provides guidance and insights for developing Omicron antibodies and vaccines.
Similar content being viewed by others
Introduction
The pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has lasted for three years1,2. The spike (S) protein on the surface of the virus particle is the key protein for the virus to invade cells3,4. The S protein is a trimer containing multiple domains, of which the domain that directly binds to the receptor angiotensin-converting enzyme 2 (ACE2) is called the receptor-binding domain (RBD)3,4,5. The RNA genome of SARS-CoV-2 is prone to mutate in the replication process, resulting in the constant emergence of mutant strains6. So far, several mutant strains have been identified as mutants worthy of attention by the World Health Organization, including Omicron and the previous Alpha7, Beta8, Gamma9, and Delta10 mutants. Among these mutant strains, Omicron contains the largest number of mutations and has stronger transmissibility than other mutant strains11,12. Omicron strains include several subtypes, such as BA.1–BA.513,14. Mutations in the S protein confer stronger ACE2 affinity and immune escape ability15,16,17,18,19. Among them, 30–36 mutations are located in the S protein, including 15–17 on the RBD. Some of these mutations can enhance the binding of the virus and the receptor, resulting in stronger viral infectivity19,20. Some other mutations can change the immunogenicity of the virus and give the virus the ability to escape11,21. This makes the Omicron strain, especially BA.5, quickly replace the original prevalent strain and cause rapid and widespread transmission in the population14.
The neutralizing antibody is an important protective barrier against viral infection22,23. Antibodies against SARS-CoV-2 can be divided into RBD antibodies, N-terminal domain (NTD) antibodies and other antibodies according to their action sites24,25,26. These antibodies can also be identified as ordinary antibodies or nanobodies according to their types25,26. The complex structure of many antibodies with the viral S protein or RBD domain has been resolved5,27,28,29. The S proteins in these complexes are diverse, including wild-type (WT) and various mutant proteins. A comprehensive and systematic analysis of the epitopes and modes of action of these antibodies can help us deeply understand the working mechanism of antibodies.
In order to study the immune escape of Omicron in more detail, we comprehensively and systematically studied the interaction between the antibodies reported in PDB and current Omicron strains. Our results showed that Omicron mutations affected the epitopes of most of the existing antibodies in Protein Data Bank (PDB). Based on the binding mode of antibodies, we classified these antibodies and found that the epitopes of the H-RBD class antibodies were significantly less affected by Omicron mutations than other classes. Binding experiments and neutralization experiments showed that such antibodies could effectively inhibit the immune escape of Omicron. In addition, antibodies developed for Omicron BA.1 strain can effectively inhibit the other Omicron subtypes. Our work provides important insights into developing antibodies and a new generation of vaccines.
Results
Analysis of antibodies
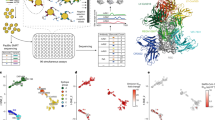
We chose the antibodies of which complex structures with the S protein of SARS-CoV-2 have been resolved (Table 1). We found 518 complex structures of the antibody of SARS-CoV-2 with the S protein from the PDB database. Most of these complexes contain only one antibody (430, accounting for 83.01%), and the rest contain multiple antibodies as a cocktail combination. There are 82 complexes containing two antibodies (accounting for 15.83%), 5 complexes containing three antibodies (accounting for 0.97%), and 1 complex containing four antibodies (accounting for 0.19%) (Fig. 1a). In order to analyze the interaction between the antibody and the S protein in detail, we extracted subcomplexes from these complex structures. Each subcomplex contains an ordinary antibody or nanobody and its binding domain in the S protein. A complex structure may contain multiple subcomplexes. A total of 613 subcomplex structures were obtained. Among them, 514 subcomplexes bind to the S protein of WT SARS-CoV-2, accounting for 83.85% of the total, followed by Beta, Omicron, Delta, Kappa, Alpha, and Gamma, with the number of antibodies being 44, 33, 8, 6, 3, 3, and 2, respectively (Fig. 1b). There is still a huge demand for developing antibodies against mutant strains such as Omicron.
a Distribution of the antibody number in the available structures. The antibody and corresponding binding domain are extracted as a subcomplex. Most structures contain only one antibody. b Strain distribution of the S protein in subcomplexes. Most of the S proteins are WT. c Statistics of ordinary antibodies and nanobodies and their corresponding targeting domains in the S protein. d Cluster distribution of the 10 classes of ordinary antibodies targeting RBD, named Ab-A-RBD to Ab-J-RBD, where Ab represents antibody and can be omitted as A-RBD to J-RBD without causing confusion, and structural scheme of the structural classes of some representative RBD antibodies. e is the same as d for 4 classes of NTD antibodies (named Ab-A-NTD to Ab-D-NTD, where Ab represents antibody and can be omitted as A-NTD to D-NTD without causing confusion). f is the same as d for the 5 classes of RBD nanobodies. For the t-SNE analysis of NTD antibodies, all of subcomplexes were used for analysis. For RBD antibodies, only the representative subcomplexes were used. g Epitope distributions of 10 classes of ordinary antibodies targeting RBD. The color depth represents the frequency of the residues as epitopes. The PDB ID for RBD of the S protein is 7QUS.
Considering that some antibodies contained in the subcomplex structures are duplicated, after removing these duplicated antibodies, we found a total of 293 ordinary antibodies and 60 nanobodies. Among them, there are 233 ordinary antibodies or nanobodies, each corresponding to only one subcomplex. Antibodies with multiple subcomplexes indicate that they have attracted more research interest than other antibodies. Two antibodies, CR3022 and EY6A, have the largest number of subcomplexes at present (13 subcomplexes) (Supplementary Fig. S1a). Among them, 5 single chain variable fragments (scFv) are included in the statistics when counting the number of epitope residues mutated in Omicron (NERMO) but not included in the genotypes. Among 293 ordinary antibodies, 256 bind the RBD domain, 29 bind the NTD domain, and 8 bind the other regions of the S protein. While 60 kinds of nanobody bind to RBD, there is a lack of nanobodies that bind to NTD (Fig. 1c).
We analyzed the genotype of the antibodies. From the perspective of genotype, the heavy chains of antibodies are mainly encoded by IGHV3, accounting for about half of the number of heavy chains. The numbers of antibodies coded by IGHV1 and IGHV4 are also relatively large, accounting for 29.97% and 11.50%, respectively. The light chains of antibodies are mainly encoded by IGKV1, IGKV3, IGLV2, and IGLV1, accounting for more than 75% (Supplementary Fig. S1b). Due to the diversity of species sources of nanobodies, we did not conduct genotype analysis and statistics of nanobodies.
We classified antibodies according to their spatial positions binding to RBD or NTD by comparing the structures of the subcomplexes (Fig. 1d–g; Supplementary Fig. S1c–e). For antibody with multiple structures, we selected the subcomplex deposited earliest in the PDB database as the representative for analysis. The ordinary antibodies binding to RBD can be divided into 10 categories (Fig. 1d), named Ab-A-RBD to Ab-J-RBD, where Ab represents antibody and can be omitted as A-RBD to J-RBD without causing confusion. These antibodies covered almost all of the RBD surface, where B-RBD accounts for 25.39%, A-RBD accounts for 16.02%, and C-RBD class accounts for 14.84%, the three types of RBD antibodies with the largest proportion. We showed the epitopes of different antibodies structurally (Fig. 1g). The epitope patterns of A-RBD, B-RBD, and G-RBD are similar, while the epitope patterns of C-RBD, E-RBD, and I-RBD are relatively concentrated and distributed on the other side. The epitopes of these two patterns are mainly located in the receptor-binding motif (RBM) region, covering a residues range of RBD from S438 to G502. Group A-RBD, B-RBD, and G-RBD antibodies share an epitope range from T470 to P490 with groups C-RBD, E-RBD, and I-RBD. In addition to these shared epitopes, the antibody groups of A-RBD, B-RBD, and G-RBD occupy epitope regions from R495 to W505 and from L455 to N414, whereas groups of C-RBD, E-RBD, and I-RBD bind to the region from S443 to N450 (Fig. 1d). All of these six classes of antibodies can interact with the antiparallel beta-sheet on RBD. The epitopes of the remaining four classes of antibodies are far away from the antiparallel beta-sheet, where D-RBD and H-RBD bind to the loop region of R454–L492, F-RBD binds to the loop region of N437–W449, and J-RBD binds to the loop region of W495–P507. The structural comparison found that all 10 classes of RBD antibodies can bind to the up conformation of RBD. A-RBD, B-RBD, G-RBD, H-RBD, and J-RBD may cause steric hindrance to other protomers of S protein or NTD domain when RBD is in the down conformation (Fig. 1d; Supplementary Fig. S1c).
Ordinary antibodies binding NTD can be divided into four categories, named Ab-A-NTD to Ab-D-NTD, where Ab represents antibody and can be omitted as A-NTD to D-NTD without causing confusion. Because the number of NTD antibodies is small, all subcomplexes were used for analysis rather than using the representative subcomplex only (Fig. 1e). Based on the structure information, there are five loops in NTD, designated N1 (residues 14–26), N2 (residues 67–79), N3 (residues 141–156), N4 (residues 177–186), and N5 (residues 246–260). These antibodies bind to different regions of NTD (Fig. 1e; Supplementary Fig. S2a), and the largest number of classes, A-NTD, account for 41.38% of the total NTD antibodies. B-NTD class is the only class of antibodies binding the interface without an N-glycosylation site. Its epitope is close to the N2 region, accounting for 27.59%. C-NTD account for 24.14% of the total NTD antibodies. A-NTD and C-NTD classes bind to the N3 region of NTD, and D-NTD class mainly binds to the N4 loop (Fig. 1e; Supplementary Fig. S1d).
The nanobodies against RBD can be divided into five categories (Fig. 1f), named Nano-A-RBD to Nano-E-RBD, where nano represents nanobody. Nano-A-RBD, Nano-C-RBD, and Nano-E-RBD classes are all bound to the RBM region, accounting for 61.67% (Supplementary Figs. S1e, S2b). For nanobodies, the RBM region is a hot area for antibody research and development; and the binding of these three kinds of nanobodies is not affected by RBD conformation. The other two types of Nano-B-RBD and Nano-D-RBD are bound in the core region of RBD, and when RBD is deeply closed, it will collide with the RBD domain of the adjacent protomer in space.
We further analyzed the genotypes corresponding to the heavy and the light chains of the antibodies of various structural types. Some genotypes can encode multiple structural types of heavy and light chains, such as IGHV3-30, which encodes 10 structural types of antibody heavy chains, and IGLV2-14, which encodes 9 structural types of antibody light chains, both of which cover RBD and NTD antibodies. While some genotypes encode only one structural type, for example, IGHV2-70 encodes only the heavy chain of J-RBD, and IGKV3D20 encodes only the light chain of A-RBD (Supplementary Fig. S3a, b). From the perspective of structural types, some structural types have obvious genotype preferences, such as the heavy chain of B-RBD antibodies is mainly encoded by IGHV3-53 and IGHV3-66, and the light chain is mainly encoded by IGKV1-9, IGKV3-20, and IGKV1-33. Some classes have genotype preference only for heavy chains, such as the heavy chain of A-NTD is mainly encoded by IGHV1-24, and the light chain has no obvious genotype preference. Some heavy chain and light chain classes lack genotype preference, such as E-RBD, J-RBD, and D-NTD, etc. From the perspective of genotype, some genotypes preferentially encode some structural types, such as IGHV3-53, which is mainly involved in the heavy chain coding of B-RBD and C-RBD antibodies, while IGKV3-20 is mainly involved in the light chain coding of B-RBD, A-RBD, and I-RBD (Supplementary Fig. S3c).
Effects of Omicron mutations
We analyzed the average number of epitope residues mutated in Omicron (ANERMO), including BA.1, BA.2, BA.3, BA.4, BA.5, and BA.2.12.1 subtypes, of all 353 ordinary antibodies or nanobodies. We counted the ANERMO of these antibodies. There are 44 antibodies with the ANERMO < 0.5, accounting for 12.46% of the total. The ANERMO is large than 2 for most antibodies (229, accounting for 64.87%), which indicates that most antibodies cannot inhibit immune evasion of Omicron (Supplementary Fig. S4a). We counted and analyzed the strain type of the S protein in the 613 subcomplexes and found that antibodies developed against Omicron are least affected by Omicron mutations (the ANERMO is about 0.43), and epitopes of antibodies developed against Kappa are less affected by Omicron mutations than the other strains. However, the antibodies developed against Alpha, WT, Gamma and Beta strains have a relatively higher ANERMO (Fig. 2a).
a Epitope mutations in Omicron of antibodies targeting different strains. The ANERMO of the antibodies targeting each strain are listed at the top of the columns. b The ANERMO from different RBD antibody structural classes. c The ANERMO from different NTD antibody structural classes. d The ANERMO from different RBD nanoantibody structural classes. e The ANERMO from different heavy chain genotypes. f Statistics of the potential broad-spectrum antibodies. Red points stand for ANERMO; blue squares stand for average number of epitope residues mutated in WT, Alpha, Lambda, Beta, Delta, Kappa, Gamma, and Omicron strains.
The extent affected by Omicron mutations vary for antibodies of different structural classes. The ANERMO of ordinary antibodies targeting RBD is 3.47, but it varies for different structural classes. The ANERMO of the H-RBD class is 0, significantly less than other types of antibodies (Fig. 2b; Supplementary Fig. S4b–d). The ANERMO of B-RBD antibodies reaches 5.24 (Fig. 2b).
NTD antibodies are less affected than RBD antibodies in general (Fig. 2b, c; Supplementary Fig. S4b). The ANERMO of all NTD antibodies is 0.78. The D-NTD antibodies have no mutated epitopes in Omicron strains, which bind the N4 domain and may be a good target for developing the Omicron neutralizing antibodies (Fig. 2c; Supplementary Fig. S4b, e). It is worth noting that the B-NTD class contains antibodies that bind to two infection-enhancing sites30. These antibodies can help the RBD domain maintain an open conformation, which facilitates the receptor binding of SARS-CoV-2. The RBD nanobodies are generally more affected by Omicron mutations than ordinary antibodies (Fig. 2d), and the ANERMO of the RBD nanobodies is 3.94 (Supplementary Fig. S4b–f).
From the perspective of antibody genotype, the heavy chains encoded by IGHV3-13, IGHV3-9, and IGHV1-24 are least affected by Omicron mutations with the ANERMO < 1 (Fig. 2e). The heavy chains encoded by IGHV2-70 are greatly affected by Omicron mutations. The light chains are generally less affected by Omicron mutations than the heavy chains, while the light chains are less involved in binding to the S protein. In term of the light chain genotypes, the antibodies encoded by IGLV8-61, IGKV2-24, and IGLV3-1 are least affected by Omicron mutations, while the epitopes of the antibodies encoded by IGKV1-9, IGLV5-37, and IGKV9-49 are mutated more in Omicron strains (Supplementary Fig. S3d), which may not be able to effectively inhibit Omicron immune evasion.
Evaluation of neutralization ability of potential broad-spectrum antibodies
We selected antibodies with few ANERMO (ANERMO ≤ 0.5) as potential broad-spectrum antibodies for further research. These potential broad-spectrum antibodies with few epitope residue mutations are likely to be able to maintain their binding ability and neutralizing activity against Omicron. A total of 43 potential broad-spectrum ordinary antibodies or nanobodies were selected (38 ordinary antibodies and 5 nanobodies) (Fig. 2f). There were 23 kinds of antibodies whose epitope residues were not affected by Omicron mutations (ANERMO = 0), accounting for 6.5% of the total number of antibodies. Of the 43 potential broad-spectrum antibodies, 9 antibodies target the NTD, 28 antibodies are bound to the RBD, and the remaining 6 antibodies are bound to the S2 region. The total number of antibodies bound to the S2 region enrolled in this work is 7, which indicates that the antibodies developed against the S2 region are more likely to be used as broad-spectrum antibody drugs to avoid the immune escape problem caused by Omicron mutations.
The proportion of NTD antibodies among potential broad-spectrum antibodies (20.93%) is higher than that of NTD antibodies among all antibodies (8.22%) with the ANERMO of 0.22. We also selected antibodies with more affected epitope residues (ANERMO > 0.5), including 4-18 and CR3022.
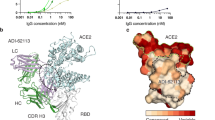
We synthesized the gene of some representative potential broad-spectrum antibodies from different groups and some antibodies with high ANERMO as control and expressed these antibodies. We first determined the binding ability of these antibodies to the S proteins of WT, Delta strain and Omicron subtypes. Among the 38 expressed antibodies, there are 12 antibodies whose EC50 values of binding to the S protein of WT, Delta strain or Omicron strain remain within the same range. Twelve antibodies whose binding ability to one or two Omicron subtypes is at least one order of magnitude lower than that of WT, and 13 antibodies whose binding ability to three to five Omicron subtypes is at least one order of magnitude lower than that of WT. The binding ability of antibody 2G12 to Delta strain or Omicron subtypes is stronger than that of WT strain (Fig. 3a). In E-RBD, H-RBD, I-RBD, and S2 groups, there are half antibodies whose binding ability to S protein is not affected by Delta or Omicron mutations (Fig. 3a). Antibodies with less ANERMO and good broad-spectrum binding ability are mainly from the E-RBD, F-RBD, H-RBD and S2 groups (Fig. 3b).
a Heatmap showing the binding EC50 against S fragments of SARS-CoV-2 variants and neutralization IC50 of pseudotyped SARS-CoV-2 variants. b The average binding EC50 against the WT, Delta, and the five indicated Omicron variants, and the average number of epitope residues mutated in Omicron (ANERMO) of different classes of antibodies. c The average neutralization IC50 against the WT, Delta, and the five indicated Omicron variants, and the ANERMO of different classes of antibodies. d The Pearson correlation coefficients analysis between the average binding EC50 and the average neutralization IC50 values of the expressed antibodies. e The average binding EC50 fold change between the five indicated Omicron variants and WT, and the ANERMO of different classes of antibodies. f The average neutralization IC50 fold change between the five indicated Omicron variants and WT, and the ANERMO of different classes of antibodies. g The Pearson correlation coefficients analysis between the average binding EC50 fold change and the ANERMO of the expressed antibodies. Samples were tested in triplicate. Data are shown as mean ± SD. h The neutralization activities of antibodies from H-RBD class against pseudotyped XBB and XBB.1.5 variants.
We measured the neutralization activity of these antibodies against WT, Delta, and Omicron pseudoviruses. The neutralization IC50 values against WT, Delta and Omicron subtypes are <100 µg/mL for 17 antibodies (Fig. 3a), and the antibodies with high average neutralization activity and with less ANERMO are mainly from the E-RBD, F-RBD, H-RBD classes (Fig. 3c). There is a statistical correlation between the average IC50 of neutralizing activity of antibodies against Omicron subtypes and the average EC50 of these antibodies binding Omicron (r = 0.5592, P = 0.0003) (Fig. 3d), indicating that antibodies with a good binding ability to mutant strains often have good neutralizing activity against mutant strains.
We calculated the average fold change between the EC50 bound to Omicron and that to the WT (average EC50 fold change) and the average fold change between the IC50 of the antibodies to Omicron and that to WT (average IC50 fold change) for the 38 expressed antibodies. The results show that the E-RBD, H-RBD, and S2 groups have a low ANERMO (< 0.5), low average EC50 fold change, and low average IC50 fold change, indicating that the antibodies in these three groups are mainly good broad-spectrum antibodies against a variety of Omicron strains (Fig. 3e, f). We calculated the relationship between the average EC50 fold change and the ANERMO for the antibodies with ANERMO < 3 and found a significant correlation between them (r = 0.5605, P = 0.0003) (Fig. 3g). Impressively, three (COVOX-45, ION-300, and WRAIR-2057) of four antibodies of H-RBD class tested in this work have NERMO of 0 against XBB and other variants and retain good neutralization capacity against pseudotyped XBB and XBB.1.5 variants (Fig. 3h). The NERMO against XBB of the antibody N-612-056 is 2, which has a relative lower neutralization capacity against pseudotyped XBB and XBB.1.5 variants. Interestingly, the ANERMO of S2X259 is high (ANERMO = 3.67); however, it can maintain good binding and neutralizing activity against Omicron, which shows that our calculation method can effectively predict broad-spectrum antibodies with relatively low ANERMO (< 3.0).
Structural analysis using cryo-EM
To further investigate the mechanism of these antibodies, we selected several antibodies with conserved epitopes, including A-RBD class antibody 10-28, E-RBD class antibodies XGv282 and XGv289, H-RBD class antibodies COVOX-45, ION-300, S2H97, WRAIR-2057 and N-612-056, C-NTD class antibody S2L20, and S2 antibody CV3-25, and tried to solve the cryo-EM structures of these antibodies in complex with the S protein of Omicron BA.5. We have successfully solved the cryo-EM complexes of antibodies XGv282, XGv289, and S2L20 with the S protein in high-resolution (Fig. 4a; Supplementary Figs. S5, S6 and Table S1).
a The domain-colored cryo-EM structures of Omicron BA.5 S protein in complex with XGv282, XGv289 and S2L20 are shown in two perpendicular views. The domains (RBD or NTD) bound with antibodies are colored cyan, with the other part of the S protein shown in gray. The heavy and light chains are colored green and palegreen for XGv282, marine and blue for XGv289, and orange and yellow for S2L20, respectively. b The binding interface between Omicron BA.5 RBD and XGv282. Extensive hydrophobic and hydrophilic interactions on the interface are shown in three gray boxes. The red-framed right panel is the binding interface between Omicron BA.1 RBD and XGv282 (PDB ID: 7WLC). Black dashed lines indicate polar interactions. c, d Structural comparison of the Omicron BA.5 RBD–XGv282 complex and the Omicron BA.1 RBD–XGv282 complex (PDB ID: 7WLC). The RBD is superimposed, with the circled regions shown in detail in d. The heavy and light chains of XGv282 are colored green and pale green, respectively. RBD is colored cyan and gray for Omicron BA.5 and BA.1, respectively. e is the same as (b), but for XGv289. The red-framed right panel is the binding interface between Omicron BA.1 RBD and XGv289 (PDB ID: 7WE9). f Structural comparison of the Omicron BA.5 RBD–XGv289 complex and the Omicron BA.1 RBD–XGv289 complex (PDB ID: 7WE9). The RBD is superimposed. The boxed region is shown in inset in details. g The RBD/COVOX–45 complex (PDB ID: 7BEL) is superimposed on the RBD of the trimeric S protein (PDB ID: 7DWZ). COVOX-45 clashes with trimeric S protein in both the “up” and “down” conformation of RBD.
E-RBD class antibodies XGv282 and XGv289
Both XGv282 and XGv289 have a strong neutralizing ability against each Omicron variant, the WT and Delta variants (Fig. 3a). The neutralization ability of XGv282 to BA.2 and BA.4/BA.5 is more potent than that of BA.1 and BA.3. And the neutralization ability of XGv289 to BA.1 is stronger than that to other strains. The XGv282 antibody binds to the BA.1 S protein through hydrogen bonds, cationic π bonds, and numerous hydrophobic interactions31 (Fig. 4b). Compared with the XGv282/BA.1 S protein complex, the XGv282 antibody bound to the BA.5 S protein is rotated by 4.5° (Fig. 4c), which increases the interaction interface area between the antibody and the S protein (the interface area of XGv282 with BA.5 or BA.1 S protein is 750 Å2 or 550 Å2, respectively) (Supplementary Fig. S7a). The L452R mutation exists in the BA.5 S protein, considered to evade HLA-A24-restricted cellular immunity32, which disrupts the hydrophobic interaction between XGv282 and the BA.5 S protein (Supplementary Fig. S7b). In addition, compared with the previous structure (XGv282/BA.1 S protein), residue F490 is further away from R74, and the distance between the two residues has reached 4 Å, affecting the binding ability between XGv282 and S protein (Fig. 4d). The critical mutation S446G of the BA.5 S protein (Ser in BA.1 and BA.3) results in a conformational change in the loop region, reshaping the interaction network with XGv282 and inducing the formation of six hydrogen bonds (R346–S31, K444–S101, G446–R50, Y449–R50, N450–S31, and R498–N97) and a cation–π interaction (Y449–K57) (Fig. 4b). Moreover, K440 forms a salt bridge with D104 on HCDR3. The rotation of the F106 side chain extends the hydrophobic interaction interface, all of which further stabilize the interaction between XGv282 and the BA.5 S protein (Fig. 4b).
The epitopes of XGv289 on the BA.5 S protein are mainly located in two loop regions (N439–G447 and F497–Q506) of high mutation frequency (Supplementary Fig. S7c). Similar to XGv282 in complex with the BA.5 S protein, the loop containing S446G and L452R mutations exhibits slight conformational changes, resulting in a rotation of XGv289 by 9.1° (Fig. 4f) and increasing the interface area from 550 Å2 for XGv289/BA.1 to 650 Å2 for XGv289/BA.5 mainly through hydrogen bonds (R498–A57, K440–N32, Q506–D94, T500–S58 and T500–Y59) and hydrophobic interactions (Fig. 4e). The binding experiment also shows that the affinity of XGv289 to the BA.5 S protein is higher than that to the BA.1 S protein.
H-RBD antibody utilizes a conserved mechanism to neutralize SARS-CoV-2
H-RBD is a class of antibodies that binds at the site of the RBD flank, with a high affinity for S proteins of the SARS-CoV-2 variants, such as WT, Delta, and Omicron variants (Fig. 3a). In our present work, six antibodies are classified into the H-RBD class, including FD20, COVOX-45, ION-300, S2H97, WRAIR-2057, and N-612-056. Such antibody binding sites are hidden away from the RBM, but FD20 can still inhibit the interaction of the RBD with the receptor ACE233. In addition, the binding sites of FD20, COVOX-45, ION-300, WRAIR-2057, and N-612-056 are highly conserved among the mainstream strains that have appeared so far.
To further characterize the commonalities of these antibodies, we investigated five H-RBD antibodies using cryo-EM. We show that these antibodies could induce the dissociation of the trimeric S protein after incubation with the S protein (Supplementary Fig. S8), which is consistent with the previous report about the antibodies FD2033 or N-612-05634. In addition, the neutralization ability of N-612-056 is related to incubation time, which may be due to the poor accessibility of its targeted epitopes and its dependence on the conformational change of the S protein34. We also make a superposition between the H-RBD antibody/RBDs and the antibody/S protein trimer with different conformational structures from the PDB database (PDB IDs: 7DWZ and 7BEL, respectively), indicating the epitopes of H-RBD antibodies are hidden in the trimeric S protein, regardless of whether the RBD domain of the S protein is up or down. When the RBD is in the “down” state, the binding site of H-RBD antibodies is close to the NTD of the adjacent monomer, causing a significant steric hindrance between the antibodies (Fig. 4g). Even when the RBD is in the “up” state, the clash between the antibodies and the NTD, or other parts of the S protein, can still be observed. Thus, we suggest that the antibodies of the H-RBD class are more likely to inhibit virus infection by destabilizing the S protein and inducing S protein dissociation.
C-NTD class antibody S2L20
S2L20, a member of the C-NTD class antibodies, has a strong binding affinity to the S protein and broad neutralization efficiency against the WT, Delta and Omicron variants (Fig. 3a). In this work, we solved the cryo-EM structure of the S2L20/BA.5 S protein complex at 3.1 Å (Supplementary Fig. S9a). Each subunit binds to one S2L20 antibody with a highly conserved epitope near the glycan flank at position N234 of NTD, close to the RBD side of the same monomer; and the interface is stabilized by numerous polar interactions (Supplementary Fig. S9b), which are consistent with the previously reported structure of the S2L20/S protein of Delta variant35,36,37.
Discussion
Extensive efforts have been made to classify antibodies targeting the S protein using various methods. The classification of ordinary antibodies or nanobodies described previously was based on the mutant profile of epitopes38 or whether they can block the binding with the ACE2 and recognize different states (up or down or both) of the RBD39,40 or biochemistry evidence of pairwise competition between antibodies41,42 or binding to different epitope bins43,44,45. The accumulation of 3D structures of the S protein in complex with antibody/nanobody allowed us to characterize a panel of over 300 antibodies/nanobodies based on a new, systematic, and unbiased method that classifies antibodies or nanobodies utilizing the structures. There are several advantages of this method. Firstly, this method is explicit and accurate. Secondly, it requires no library screening for the mutant profile or competitive binding assay. Finally, the antibody distribution relative to the S protein is tightly linked to the epitope bins, so the classification of antibodies based on structure information is also coupled with different types of epitope bins. This method allows for a more detailed classification of antibodies or nanobodies, which provides new insights into the classification of antibodies.
In this work, we obtained several antibodies with relatively conserved epitopes, mainly from E-RBD, H-RBD, and C-NTD classes. Among them, the E-RBD antibodies XGv289 and XGv282 have the most potent ability to neutralize the current mainstream Omicron BA.5 strain, as these two antibodies were initially developed for Omicron BA.1 strain. In addition, mutations of S446G and L452R in BA.5 remodeled the interactions between XGv282 and RBD, increasing the interaction surface and affinity. The H-RBD antibodies have a high affinity for the S protein of various strains, including WT, Delta and Omicron variants, which targets highly conserved epitopes. Previous studies have revealed the epitopes of H-RBD at a cryptic site in the RBD. The binding of the H-RBD class antibodies will induce the trimeric S protein to dissociate due to the steric hindrance. This may explain why there are no complex structures of this class of antibodies with trimeric S protein reported.
We note some potential broad-spectrum antibodies with fewer ANERMO, such as 4-8, 5-24, and A19-46.1. However, when considering more strains, the number of epitope residue mutations of these antibodies is relatively large. Among them, 4-8 and 5-24 have a large number of epitope residue mutations in the C.37 strain, while A19-46.1 has epitope mutations in multiple non-Omicron strains.
With the continuous emergence of new mutant strains, such as the XBB, XBB.1.5, BQ.1.1, BU.1, BR.2, CA.2, and CJ.1, the problem of immune escape has become more severe over time. We focused on 20 antibodies (S309, S3H3, C1717, 10-40, 35B5, XGv347, C1520, XGv282, XGv289, S2K146, LY-CoV1404, BD-515, BD55-5840, Omi-18, Omi-3, P2G3, SP1-77, XGv051, XGv264, and ZCB11). The first 12 antibodies are included in Table 1. The PDB structures of the last 8 antibodies are released after the beginning of this work, whose neutralizing activity targeting Omicron BQ and XBB mutants have been tested46. Two of these antibodies (S3H3, XGv289) have low ANERMO (< 0.5, making up 10%). The antibody S3H3 has an ANERMO of 0, showing neutralizing activity against BQ.1 and XBB variants46. The other antibodies of higher ANERMO (> 0.5, making up 90%) lost greatly or completely their neutralizing activity against BQ.1 and XBB variants46. We also observed that the NERMO of H-RBD class against XBB was 0 except for the antibody N-612-056, suggesting that the H-RBD antibodies could neutralize XBB. Our investigation of the neutralizing activity of H-RBD antibodies against XBB and XBB.1.5 pseudoviruses verified our prediction (Fig. 3h).
Some broad neutralizing antibodies have been released or published after the beginning of this work. Among them, SA55 belongs to the J-RBD group, and SA58 belongs to the F-RBD group. S2X25947 and SA5548 maintain good binding and neutralizing activity against Omicron strains. Thus, mutation of epitope residues may increase or decrease affinity. This work does not predict the specific effects of epitope mutations but only analyzes and counts the number of epitope mutations. Our method is useful for preliminary screening and prediction. Among the antibodies expressed and verified in this work, the antibody with a number of epitope mutations < 3 has a significant correlation between its activity change fold and the number of epitope mutations. To predict the effect of antibody epitope mutations and screen more accurately for broad-spectrum antibodies, changes of parameters such as surface binding Gibbs free energy can be calculated based on analyzing the number of epitope mutations. In the follow-up work, a reliable method for predicting the effect of mutations on antibody activity can be realized so that the broad-spectrum activity of antibodies against virus strains harboring a high number of mutations can be predicted more accurately.
Materials and methods
Statistics and analysis of antibodies
Structures of SARS-CoV-2 S protein in complex with antibody or nanobody were obtained from the PDB (https://www.rcsb.org/). We manually labeled chains corresponding to the S protein and the associated heavy chain and light chain of antibody or nanobody in these structures. Subcomplexes were extracted from structures as separated PDB files utilizing UCSF Chimera.
Subsequently, the name and amino acid sequence of heavy chain and light chain of antibody or nanobody were obtained from EMBL-EBI (https://www.ebi.ac.uk/pdbe/api/pdb/entry/status/). Genotype analysis was performed using NCBI IgBlast (v1.17.1) with the IMGT reference (https://www.ncbi.nlm.nih.gov/igblast/) with default parameters except that the number of germline gene was set to 1. The strain type of the S protein of each structure was determined using sequence alignment.
The epitopes were defined as residues that contact antibody within a distance of 4 Å in the spike protein. Subsequently, domains of S protein recognized by the antibody or nanobody were figured out using epitope residues. By comparing the mutant profiles of Omicron strains (BA.1, BA.2, BA.2.12.1, BA.3, BA.4/5) with the epitopes recognized by the antibody, the number of epitope residues mutated in Omicron was computed. Noted that one antibody may form complexes with multiple strains of S protein, resulting in multiple versions of epitopes under which condition the mean value was used.
The same classification method is used for antibodies binding to different domains of S protein. In order to clearly describe this process, we take RBD_antibody subcomplex as an example. Firstly, all structures were aligned to a template structure to ensure that all structures are of the same initial position. The rotation angle between two aligned structures is measured in UCSF Chimera using the command “measure rotation” after superimposing the antibody chain of these two structures using the command “matchmaker”. After that, an NxN matrix was generated for all of structures, whose values represent the rotation angle between the different structures. A KxN feature matrix (N is the number of structures, K is the number of groups) was generated using unsupervised k-means clustering. Lastly, 2D t-SNE embeddings were performed with the Python3 TSNE function for visualization. 2D t-SNE plots were generated using the Pyhton3 matplotlib package and cluster heatmaps were generated using the Python3 seaborn package.
Generation and purification of monoclonal antibodies
For ordinary antibodies, the sequences of their variable region were codon-optimized and synthesized in Azenta and then inserted into pcDNA3.4 vector plasmid containing human IgG1 heavy chain constant region sequence or light chain constant region sequence (κ or λ chain). To express these antibodies, 15 μg heavy chain plasmid and 15 μg light chain plasmid of the same antibody were cotransfected into 30 mL Expi293F cells using ExpiFectamine™ 293 Transfection Kit (Thermo Fisher) according to the manufacturer’s instructions. The Expi293F cells were then cultured for 120 h in a shaker under 37 °C and 5% CO2. The expressed products were centrifuged at 4000× g at 4 °C for 20 min, and the supernatants were filtered through a 0.22 μm filter. The filtered supernatant was then subjected to ProteinG column (Cytiva), equilibrated with phosphate-buffered saline (PBS), pH 7.4. After washing with 5 column-volume (CV) PBS, the antibody was eluted with 5 CV 0.1 M Glycine, pH 2.7 and immediately neutralized to pH 7.4 using 1 M Tris-HCl, pH 9.0. The eluted antibodies were then exchanged into PBS and concentrated using a 30 kDa ultrafiltration centrifugal tube. The concentration of purified antibodies was measured using Nanodrop Plus (Thermo Fisher). The antibodies were then aliquoted and stored at –80 °C until use.
Enzyme-linked immunosorbent assay (ELISA)
The binding activities of purified antibodies to the S protein of SARS-CoV-2 variants were tested using ELISA. Recombinant proteins of the extracellular domain of the S protein (S-ECD) of SARS-CoV-2 WT (Sino Biological, 40589-V08H4), B.1.617.2 (Sino Biological, 40589-V08B16), BA.1 (Acro Biosystems, SPN-C52Hz), BA.2 (Acro Biosystems, SPN-C5223), BA.2.12.1 (Acro Biosystems, SPN-C522d), BA.3 (Acro Biosystems, SPN-C5225), BA.4 (Acro Biosystems, SPN-C5229) were coated onto 96-well plates at the concentration of 2 μg/mL overnight at 4 °C. The plates were washed three times with PBS plus 0.2% Tween (PBST), followed by incubation at 37 °C for 1 h with PBST containing 2% bovine serum albumin (BSA). Following washing with PBST, antibodies four-fold serial-diluted in PBST containing 0.2% BSA starting at 1 μg/mL were added to the wells in triplicates and allowed to incubate at 37 °C for 1 h. After washing with PBST, horseradish peroxidase (HRP)-conjugated anti-human IgG antibody (Abcam) diluted in PBST containing 0.2% BSA at the dilution of 1:10,000 was added to each well and incubated at 37 °C for 1 h. After washing with PBST, TMB substrate solution (Solarbio) was added to the plates and incubated for 6 min at room temperature. ELISA stop solution (Solarbio) was added to stop the reaction. The absorbent at 450 nm was measured using a microplate reader (TECAN) with the absorbent at 630 nm as a reference. The data were processed using GraphPad Prism v8.3, and the EC50 values were calculated using a four-parameter nonlinear regression model.
Neutralizing assay with pseudotyped SARS-CoV-2 variants
The pseudotyped SARS-CoV-2 WT, B.1.617.2, BA.1, and BA.2 variants were packaged as previously described. Briefly, HIV backbone plasmid pNL4-3.Luc.R-E- was cotransfected into HEK293T cells with pCAGGS vector carrying full-length S protein gene sequence belonging to SARS-CoV-2 WT (QHD43416.1), B.1.617.2 (EPI_ISL_2029113), BA.1 (EPI_ISL_6640917) or BA.2 (YP_009724390.1) at the different ratios, respectively. Supernatants containing pseudotyped virus were harvested at 48 h, 60 h, and 72 h post-transfection, filtered through a 0.45-μm filter, aliquoted and stored at −80 °C. The pseudotyped viruses of SARS-CoV-2 BA.2.12.1 (DD1777), BA.3 (DD1774), BA.4 (DD1776), XBB (DD1796), and XBB.1.5 (DD1797) variants were purchased from Vazyme.
To determine the neutralizing efficacy of antibodies, 50 μL monoclonal antibody diluted in DEME medium containing 10% fetal bovine serum (FBS) (starting concentration at 100 μg/mL) was incubated with 50 μL pseudotyped virus in 96-well cell culture plates at 37 °C for 1 h. ACE2-293T cells at the concentration of 2.5 × 105 cells/mL in 100 μL DMEM medium supplemented with 10% FBS were then added into each well. The plates were incubated at 37 °C and 5% CO2 condition for 48 h. After incubation, 100 μL cell culture medium was discarded, and 100 μL Brite-Lite Luciferase reagent (Vazyme) was added to each well and incubated for 2 min to avoid from light. Each well was then mixed 10 times by pipetting, and the 150 μL mixture was transferred to a white plate to measure the luciferase activity using a microplate reader (TECAN Spark). The inhibition percent was determined by comparing the relative luminescence units to the cell control (cells without pseudotyped virus or antibody) and virus control (cells with the pseudotyped virus but without antibody). The data were processed using GraphPad Prism v8.3, and the IC50 values were calculated using a three-parameter nonlinear regression model.
Protein expression and purification
The S-ECD (1–1208 aa) of Omicron BA.5 was cloned into the pCAG vector (Invitrogen) with substitution of six prolines at residues 817, 892, 899, 942, 986, and 98749, a “GSAS” substitution at residues 682 to 685 and a C-terminal T4 fibritin trimerization motif followed by one Flag tag. The recombinant S-ECD protein was overexpressed using HEK 293F mammalian cells (Invitrogen) at 37 °C under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm). The plasmid was transiently transfected into the cells when the cell density reached 2.0 × 106 cells/mL. To transfect one liter of cell culture, about 1.5 mg of the plasmid was premixed with 3 mg of polyethylenimines (PEIs) (Polysciences) in 50 mL of fresh medium for 15 min before adding to cell culture. Cells were removed by centrifugation at 4000× g for 15 min after 60 h of transfection. The secreted S-ECD protein was purified using anti-FLAG M2 affinity resin (Sigma Aldrich). After loading two times, the anti-FLAG M2 resin was washed with the wash buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl. The protein was eluted with the wash buffer plus 0.2 mg/mL Flag peptide. The eluent was then concentrated and subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in a buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl. The peak fractions were collected and concentrated to incubate with antibody. The purified S-ECD was mixed with the antibody at a molar ratio of about 1:5 for 1 h. Then the mixture was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl. The peak fractions were collected for cryo-EM analysis.
Cryo-EM sample preparation
The peak fractions of the complex were concentrated to about 3.5 mg/mL and applied to the grids. Aliquots (3.5 μL) of the protein complex were placed on glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3). The grids were blotted for 3.5 s and flash-frozen in liquid ethane cooled by liquid nitrogen with Vitrobot (Mark IV, Thermo Scientific). The prepared grids were transferred to a Titan Krios operating at 300 kV equipped with a Gatan K3 detector and GIF Quantum energy filter. Movie stacks were automatically collected using EPU software (Thermo Fisher Scientific), with a slit width of 20 eV on the energy filter and a defocus range from –1.2 µm to –2.2 µm in super-resolution mode at a nominal magnification of 81,000×. Each stack was exposed for 2.56 s with an exposure time of 0.08 s per frame, resulting in a total of 32 frames per stack. The total dose rate was ~50 e-/Å2 for each stack.
Data processing
The movie stacks were motion corrected with MotionCor250 and binned twofold, resulting in a pixel size of 1.077 Å/pixel. Meanwhile, dose weighting was performed51. The defocus values were estimated with Gctf52. Particles of Omicron BA.5 S in complex with XGv289 were automatically picked using Relion 3.0.653,54,55,56 from manually selected micrographs. After 2D classification with Relion, good particles were selected and subject to 2D classification and multiple cycles of heterogeneous refinement without symmetry using cryoSPARC57. The good particles were selected and subjected to non-uniform refinement, local CTF refinement and local refinement with C1 symmetry, resulting in the 3D reconstruction for the whole structures, which was further subject to local refinement with an adapted mask on the interface between RBD of Omicron BA.5 S and XGv289 to improve the map quality on RBD-XGv289 subcomplex.
For Omicron BA.5 S (spike protein) in complex with XGv282 and S2L20, the CTF of stacks were estimated by patch CTF estimation (multi), and particles were automatically picked by template picker and extracted with cryoSPARC. The subsequent processing methods are consistent with the above description of Omicron BA.5 S in complex with XGv289.
The resolution was estimated with the gold-standard Fourier shell correlation 0.143 criterion58 with high-resolution noise substitution59. Refer to Supplementary Figs. S5, S6 and Table S1 for details of data collection and processing.
Model building and structure refinement
For the model building of Omicron BA.5 S in complex with XGv282, XGv289, and S2L20, the atomic model of the S-ECD (PDB ID: 7DWZ) were used as templates, which were molecular dynamics flexible fitted60 into the whole cryo-EM map and manually adjusted with Coot61 to obtain the atomic model of Omicron BA.5 S protein. The reported models of XGv282 (PDB ID: 7WLC), XGv289 (PDB ID: 7WE9), and S2L20 (PDB ID: 7SO9) were manually refined based on the focused-refined cryo-EM map. Each residue was manually checked with the chemical properties taken into consideration during model building. Several segments, whose corresponding densities were invisible, were not modeled. Structural refinement was performed in Phenix62 with secondary structure and geometry restraints to prevent overfitting. To monitor the potential overfitting, the model was refined against one of the two independent half maps from the gold-standard 3D refinement approach. Then, the refined model was tested against the other map. Statistics associated with data collection, 3D reconstruction and model building were summarized in Supplementary Table S1.
Data availability
Atomic coordinates and cryo-EM density maps of the S protein of Omicron BA.5 SARS-CoV-2 in complex with antibodies (PDB ID: 8GTO, 8GTP, and 8DTQ; EMDB ID: EMD-34259, EMD-34260, EMD-34261, EMD-34262, EMD-34263 and EMD-34264) have been deposited to the Protein Data Bank (http://www.rcsb.org) and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/), respectively. Materials and data will be shared upon request.
References
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Gorbalenya, A. E. et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020).
Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020).
Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020).
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H. & Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021).
Leung, K., Shum, M. H., Leung, G. M., Lam, T. T. & Wu, J. T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eur. Surveill. 26, 2002106 (2021).
Makoni, M. South Africa responds to new SARS-CoV-2 variant. Lancet 397, 267 (2021).
Faria, N. R. et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372, 815–821 (2021).
Singh, J., Rahman, S. A., Ehtesham, N. Z., Hira, S. & Hasnain, S. E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 27, 1131–1133 (2021).
Hoffmann, M. et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185, 447–456.e11 (2022).
Flemming, A. Omicron, the great escape artist. Nat. Rev. Immunol. 22, 75 (2022).
Huang, M. et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity 55, 1501–1514.e3 (2022).
Malato, J. et al. Risk of BA.5 infection among persons exposed to previous SARS-CoV-2 variants. N. Engl. J. Med. 387, 953–954 (2022).
Hong, Q. et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 604, 546–552 (2022).
Mannar, D. et al. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 375, 760–764 (2022).
Cui, Z. et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185, 860–871.e13 (2022).
Liu, L. et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022).
Harvey, W. T. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021).
Ou, J. et al. V367F mutation in SARS-CoV-2 spike RBD emerging during the early transmission phase enhances viral infectivity through increased human ACE2 receptor binding affinity. J. Virol. 95, e0061721 (2021).
Arora, P. et al. The spike protein of SARS-CoV-2 variant A.30 is heavily mutated and evades vaccine-induced antibodies with high efficiency. Cell. Mol. Immunol. 18, 2673–2675 (2021).
Ledford, H. Antibody therapies could be a bridge to a coronavirus vaccine - but will the world benefit? Nature 584, 333–334 (2020).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Hwang, Y. C. et al. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 29, 1 (2022).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Tang, Q., Owens, R. J. & Naismith, J. H. Structural biology of nanobodies against the spike protein of SARS-CoV-2. Viruses 13, 2214 (2021).
Chi, X. et al. Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination. Signal Transduct. Target. Ther. 7, 139 (2022).
Wang, Y. et al. Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses. Cell Discov. 8, 36 (2022).
Chi, X. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020).
Liu, Y. et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell 184, 3452–3466 (2021).
Wang, K. et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 603, 919–925 (2022).
Deng, X. et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 184, 3426–3437 (2021).
Li, T. et al. Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody. EMBO Mol. Med. 13, e14544 (2021).
Tanaka, S. et al. Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display. Cell Rep. 38, 110348 (2022).
McCallum, M. et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021).
McCallum, M. et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021).
McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375, 864–868 (2022).
Cao, Y. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Mittal, A., Khattri, A. & Verma, V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 18, e1010260 (2022).
Brouwer, P. J. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020).
Dejnirattisai, W. et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183–2200 (2021).
Rogers, T. F. et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020).
Sun, D. et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 12, 1–14 (2021).
Yuan, M. et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 373, 818–823 (2021).
Wang, Q. et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8 (2022).
Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021).
Cao, Y. et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602 (2022).
Hsieh, C. L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife 4, e06980 (2015).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
We thank the cryo-EM facility and the High-Performance Computing Center of Westlake University for providing technical support. This research was supported by grants from the National Natural Science Foundation of China (32022037), Hangzhou agricultural and social development scientific research project (202204B14), the Young Elite Scientists Sponsorship Program by CAST, the Leading Innovative and Entrepreneur Team Introduction Program of Hangzhou.
Author information
Authors and Affiliations
Contributions
Q.Z., W.C., and C.Y. conceived and supervised the project. Xiangyang C., L.X., G.Z., Y.Z., J.H., L.W., Z.L., H.S., and P.H. did the experiments. Ximin C., B.H., and Z.C. analyzed the PDB structures. Xiangyang C., L.X., G.Z., Ximin C., B.H., Y.Z., and Z.C. performed the visualization. All authors contributed to the data analysis. Xiangyang C., L.X., G.Z., Ximin C., B.H., Y.Z., Z.C., and Q.Z. wrote the draft of the manuscript. Q.Z., W.C., and C.Y. reviewed and edited the manuscript. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chi, X., Xia, L., Zhang, G. et al. Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants. Cell Discov 9, 37 (2023). https://doi.org/10.1038/s41421-023-00535-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-023-00535-1