Abstract
The cell death and survival paradox in various biological processes requires clarification. While spore development causes maternal cell death in Bacillus species, the involvement of other cell death pathways in sporulation remains unknown. Here, we identified a novel ArsR family transcriptional regulator, CdsR, and found that the deletion of its encoding gene cdsR causes cell lysis and inhibits sporulation. To our knowledge, this is the first report of an ArsR family transcriptional regulator governing cell death. We found that CdsR directly repressed lrgAB expression. Furthermore, lrgAB overexpression resulted in cell lysis without sporulation, akin to the cdsR mutant, suggesting that LrgAB, a holin-like protein, induces cell death in Bacillus spp. The lrgAB mutation increases abnormal cell numbers during spore development. In conclusion, we propose that a novel repressor is vital for inhibiting LrgAB-dependent cell lysis.

Similar content being viewed by others
Introduction
Cell death plays a key role in various biological processes. In eukaryotes, cell death encompasses apoptosis, necrosis, oncosis, pyroptosis, and autophagy [1]. These diverse forms of cell death have evolved to eliminate damaged and/or infected cells from affected tissues, enabling the surrounding healthy cells to function more effectively during development [2, 3]. In prokaryotes, cell death is an altruistic process that benefits the entire population [4]. Bacteria undergo altruistic death, conferring advantages on populations, such as vegetative growth [4], sporulation [5], biofilm maintenance [4], and bacteriophage-infection containment [6].
Cell death is reportedly involved in sporulation. Losick et al. proposed a model that elucidates how Bacillus populations delay sporulation through a cell death pathway [7, 8]. This model involves the impact of the killing factor SkfA-H and the spore delay protein SdpABC, which are associated with spore formation in B. subtilis under nutrient-limited conditions, causing the death of some cells within the population [7]. Nutrient limitation induces spore formation in B. subtilis, a process regulated by a gradual increase in the level and activity of the regulatory protein Spo0A [9]. Spo0A is activated by phosphorylation, a process facilitated by a multicomponent phosphoryl group delivery system [10]. This cascade starts with a relay protein, Spo0F, being phosphorylated by histidine protein kinases (KinA, KinB, and KinC) [11, 12]. Spo0F-P then transfers the phosphoryl group to Spo0B, which, in turn, transfers it to Spo0A [10, 13]. The resulting Spo0A~P can activate or repress the transcription of genes involved in sporulation, with its high thresholds activating the expression of spoIIE, spoIIA, and spoIIG [14,15,16,17], and low thresholds activating the expression of skfA-H and sdpABC [18]. Another mechanism that induces the cell death pathway in sporulating cells involves a quality control process that eliminates improperly sporulating cells individually [19]. In B. subtilis, the base layer of the sporulation coat contains a membrane-binding protein, SpoVM, which selectively localizes to the positively curved membrane surrounding the forespore [20]. SpoVM recruits the structural protein SpoIVA (ATPase derived from a family of GTPases), which utilizes the energy from ATP hydrolysis to drive its irreversible polymerization, forming a static platform as a foundation for the assembly of the remaining sporulation coat [19, 21]. CmpA acts as a molecular adaptor that interacts with SpoVM and SpoIVA, crucial proteins that scaffold the coat. This interaction aids in recruiting the protease ClpXP, which specifically degrades SpoIVA. Consequently, the degradation of SpoIVA triggers the lysis of sporulating cells. CmpA plays a role in quality control, overseeing the formation of the spore envelope and eliminates improperly developed spores [22]. This cell death pathway ensures the stable development of sporulating cells within the B. subtilis cell population during the late stages of sporulation. However, the genes related to cell lysis that maintain the fidelity of sporulation remain unknown.
The Cid/Lrg system-mediated bacterial death and lysis occur in the context of developing biofilms in Staphylococcus aureus and represent a well-recognized cell death pathway [23, 24]. The cidABC and lrgAB operons regulate autolysis and extracellular DNA release by controlling peptidoglycan hydrolase activity via holin- and antiholin-like proteins such as CidA and LrgA [24, 25]. However, a recent report concluded that lrgA encodes proteins with bacteriophage holin-like functions, contributing to cell death in S. aureus [26]. Holins are a class of small membrane proteins required for the phage lysis of a host cell, enabling the formation of a membrane hole. Murein hydrolases in the cytoplasm can then escape and degrade peptidoglycans, ultimately reducing the membrane potential and causing cell death [27]. Homologs of lrgAB are widely distributed in bacteria, including both gram-negative and gram-positive bacteria. However, the role of LrgAB in cell lysis, particularly in biological processes like sporulation, still requires further elucidation, besides its established function in S. aureus.
Bacillus thuringiensis is a crucial biocontrol agent widely employed for agricultural pest management, owing to its ability to produce a protein-like crystalline toxin (δ-endotoxin) with specific activity against the gut of certain insect larvae [28]. Interestingly, B. thuringiensis shares a highly similar sporulation program with B. subtilis [29, 30], making it an ideal model for exploring the relationship between cell death and sporulation. In this study, we identified the ArsR family regulatory gene cdsR (HD73_RS03760, cell death and sporulation regulator) involved in cell death in B. thuringiensis subsp. kurstaki HD73 (hereafter referred to as HD73). Deletion of the cdsR gene resulted in a significant autolytic phenotype during liquid cultivation, with CdsR acting as a transcriptional repressor of the lrgAB operon. Overexpressing lrgAB triggered cell lysis and inhibited sporulation in wild-type HD73 cells. Deleting lrgAB resulted in an increased number of abnormal cells. These findings revealed, for the first time to our knowledge, that a novel repressor plays a crucial role in inhibiting cell autolysis mediated by the LrgAB holin-like system.
Results
Mutation of cdsR results in cell lysis and no sporulation
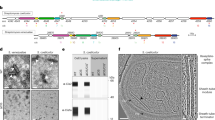
Numerous unidentified regulators are present in B. thuringiensis HD73. To elucidate their functions, we analyzed the expression levels of regulator-encoding genes during the stationary phase using our previous DNA microarray data (Accession No. GSE 48410) [31]. We selected thirty candidate genes with high relative expression levels (Table S1) and their mutants were generated through homologs recombination. Among these mutants, the ΔHD73_RS03760 mutant exhibited autolysis and inability to form endospores (Fig. 1). Therefore, it was designated as cdsR (cell death and sporulation regulator, cdsR).
A Role of CdsR in the growth of Bacillus thuringiensis HD73. Growth curves of HD73 (black circles), ΔcdsR (red squares), and CcdsR (blue triangles) cultured in Schaeffer’s sporulation medium (SSM) at 30 °C with shaking at 220 rpm were plotted by measuring the optical density at 600 nm (OD600) against incubation time. B Autolysis images of HD73, ΔcdsR, and CcdsR in SSM at 21 h. Samples (5 mL) were collected from a 500 mL triangular bottle and photographed in clear glass test tubes. C The autolytic rate, expressed as the percentage increase in the OD580, was determined for hourly samples up to T6. Mean values (SD, n = 3) are represented by symbols: HD73 (black circles), ΔcdsR (red squares), and CcdsR (blue triangles). D Imaging of cell death in B. thuringiensis cell populations using SYTOXTM Green at T3 and T17 for HD73 and ΔcdsR, with a scale bar of 5 μm. E Statistical analysis of cell death rate in B. thuringiensis cells, calculated by comparing the number of dead cells to the total cell count in photographs. The cell death rate is expressed as the ratio of the number of cells exhibiting green fluorescence to the number of total cells, multiplied by 100. Values display the means of at least three independent replicates. The cell death rates of ΔcdsR and CcdsR were compared with HD73 and analyzed using a T-test (*P < 0.05; **P < 0.01). F Polar septum formation during spore initiation was observed using laser confocal microscopy. Strains grown to T2 and T3 represent HD73, ΔcdsR, and CcdsR, with red lines indicating membranes stained with FM4-64. Transmission electron microscopic (TEM) observation was conducted to determine the cell shape and sporulation structure of HD73, ΔcdsR, and CcdsR strains at T24. G Construction of a transcriptional reporter by fusing the spo0A gene promoter with lacZ. The β-galactosidase activities of three clones were determined at specified time points following cell growth in SSM at 30 °C. Each value represents the mean and standard error of at least three independent replicates. H Fusion of the spoIIE gene promoter with lacZ to construct a transcriptional reporter.
To determine the effect of cdsR mutation on cell growth, we plotted a growth curve, which showed a significant reduction in the cell density of ΔcdsR from 14 to 24 h compared to that of the wild-type HD73 strain (Fig. 1A). However, the cell density was restored in the genetically complementary strain CcdsR (Fig. 1A). In a test tube, the OD600 values of HD73, ΔcdsR, and CcdsR were 3.166, 1.226, and 2.699 at 21 h, respectively (Fig. 1B). This suggests that cdsR deletion causes autolysis during the late stationary phase. To determine the autolytic rate of the ΔcdsR and HD73, we collected cells at T6, induced autolysis with Triton X-100, and measured the OD580 value every hour. The results showed that the autolytic rate of ΔcdsR mutant was significantly higher than that of HD73 and CcdsR from A1 to A8 (Fig. 1C). These results strongly indicate that CdsR acts as a repressor of cell lysis in HD73 cells.
To determine the effect of cdsR on cell death, cells were stained with a sensitive nucleic acid dye, SYTOXTM Green, to visualize dead cells using laser confocal microscopy. The cell death rate was calculated as the ratio of the number of dead cells (stained with SYTOXTM Green) to the total number of cells. Fewer cells with green fluorescence were observed in HD73 or ΔcdsR at T3, but they became more apparent at T17 (Fig. 1D). At T17, 47.85(±6.45)% of ΔcdsR cells were dead, whereas only 18.22(±5.00)% and 19.09(±5.53)% were dead in HD73 and CcdsR, respectively (Fig. 1E). This indicated that the deletion of the cdsR gene increases cell death.
It is noteworthy that the ΔcdsR strain did not produce spores and parasporal crystals, in contrast to the wild-type HD73 and CcdsR (Fig. 1Fg, Fi). To elucidate the reason for the absence of spores in ΔcdsR, the formation of a polar septum during sporulation was observed by laser confocal microscopy. The cell membranes of HD73, ΔcdsR, and CcdsR cells were stained with FM4-64, which labels the plasma membranes of living cells. At T2 in SSM, vegetative cells were observed in HD73 (Fig. 1Fa), ΔcdsR (Fig. 1Fb), and CcdsR (Fig. 1Fc). At T3 in SSM, the polar septum was observed in the HD73 (Fig. 1Fd) and CcdsR (Fig. 1Ff), while ΔcdsR (Fig. 1Fe) showed no polar septum formation at all. Furthermore, forespore engulfment was visualized through staining with FM4-64 and MitoTracker Green FM (MTG). HD73 and CcdsR cells completed the engulfment process by T7 (Fig. S1b and S1h). In contrast, ΔcdsR cells still lacked a polar septum at T7 (Fig. S1e). This suggests that the effect of cdsR deletion on sporulation occurs before polar septum formation.
Spo0A is the master regulator of sporulation initiation in Bacillus [9, 32]. To determine whether cdsR affects the transcription of Spo0A, the transcriptional activity of the spo0A promoter was analyzed in HD73 and ΔcdsR strains. The β-galactosidase assay showed no significant difference in the transcriptional activity of spo0A between HD73 and ΔcdsR (Fig. 1G). The activity of Spo0A is determined by its phosphorylation level, and the phosphorylated Spo0A protein can activate the transcription of spoIIE gene in B. subtilis [15] and B. thuringiensis [29, 33]. Therefore, the promoter activity of spoIIE can serve as an indicator of the activity of Spo0A [32]. The β-galactosidase assay showed high transcriptional activity of spoIIE in HD73 from T2 to T6, whereas no transcriptional activity of spoIIE was detected in ΔcdsR (Fig. 1H). This suggests that CdsR modulates sporulation by affecting Spo0A activity.
CdsR is negatively autoregulated
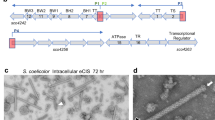
CdsR consists of 95 amino acids and belongs to the ArsR family of transcriptional regulators, which include a single helix-turn-helix (HTH) DNA-binding domain ranging from 15 to 90 amino acid residues. The genes upstream and downstream of cdsR include HD73_RS03755 (istA), HD73_RS03765 (sspH), and HD73_RS03770 (dtpA), which encode the IS21-like element IS232 family transposase, acid-soluble spore protein H, and peptide MFS transporter, respectively (Fig. 2A). To determine the transcriptional levels of the cdsR gene, the cdsR promoter (PcdsR) was fused with the lacZ reporter gene, and the activity of PcdsR in HD73 and ΔcdsR were measured. The β-galactosidase assay showed that in HD73, the activity of the PcdsR remained low from T0 to T2, increased significantly from T2 to T5, and remained high from T5 to T10 (Fig. 2B). However, in ΔcdsR, the transcriptional activity of PcdsR was significantly higher than in HD73 from T4 to T10 (Fig. 2B). To confirm whether the CdsR protein could bind to its own promoter, it was expressed and purified in E. coli (Fig. S2), and an electrophoretic mobility shift assay (EMSA) was conducted. Various concentrations of the CdsR protein were used to bind to 0.29 nM of the PcdsR-labeled probe. At low concentrations (1.28 μM and 2.13 μM), the CdsR protein was unable to effectively bind to the PcdsR-labeled probe. However, at a higher concentration of 3.84 μM, CdsR was able to bind and completely shift 0.29 nM of PcdsR-labeled probe. Notably, a 200-fold excess of unlabeled probe competed with the labeled probe, confirming specific binding (Fig. 2C). These results collectively indicate that CdsR undergoes negative autoregulation.
A Genetic loci of the cdsR gene in HD73. In the genome region of the cdsR gene (encoding cell death and survival repressor, indicated by the red arrow), the upstream and downstream genes are HD73_RS03755 (encoding IS21-like element IS232 family transposase) and HD73_RS03765 (encoding acid-soluble spore protein H), and HD73_RS03770 (encoding peptide MFS transporter), respectively. The black arrow represents the cdsR promoter region. B Transcription analysis of the cdsR promoter in HD73 and ΔcdsR. The β-galactosidase activities of three clones were determined at specified time points after cultivating B. thuringiensis cells in SSM at 30 °C. Each value represents the mean and standard error of at least three independent replicates. C Detection of the interaction between the cdsR promoter and increasing concentrations of the CdsR protein through electrophoretic mobility shift assay (EMSA). The lanes contained 0, 1.28, 2.13, 2.99, and 3.84 μM of the CdsR protein. The final lane contained 200-fold unlabeled probes to compete with the combined FAM-labeled probes.
CdsR directly regulates lrgAB and cwlD expression
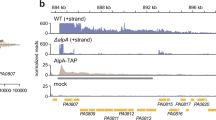
Previous findings showed that the cdsR gene was transcribed at a low level (Fig. 2B) and had an impact on the onset of the stationary phase (Fig. 1H). To identify potential autolysis and sporulation-related target genes of CdsR, the transcriptomes of HD73 and ΔcdsR at T0 were compared. Differentially expressed genes were screened based on a fold-change>2 and a false discovery rate cutoff of P < 0.05, using RNA sequencing (RNA-Seq) data. The RNA-Seq data were deposited in the NCBI Gene Expression Omnibus database (accession no. GSE216307). The analysis revealed 728 genes with differential expression in ΔcdsR relative to the HD73. Among these genes, 514 were upregulated, and 214 were downregulated. To investigate the functional roles of these differentially expressed genes, they were classified into groups related to amino acid metabolism, signaling, cellular processing, and virulence, according to the KEGG function (Table S2). While CdsR was shown to modulate sporulation primarily by affecting Spo0A activity (Fig. 1G, H), the RNA-Seq data showed that the transcription of genes involved in Spo0A activity (such as kinA-E, spo0F, spo0B, phr-rap, and spo0E, etc.) exhibited no significant change between HD73 and ΔcdsR (Table S3). Notably, RNA-Seq analysis identified only four upregulated genes associated with autolysis, including HD73_RS29150 (lrgA, encoding a holin-like protein), HD73_RS29145 (lrgB, encoding an antiholin-like protein; lrgA and lrgB compose the lrgAB operon), HD73_RS20125 (encoding N-acetylmuramoyl-L-alanine amidase, named cwlD), and HD73_RS04330 (encoding N-acetylmuramoyl-L-alanine amidase, named cwlE). The RNA-Seq data revealed that the FPKM values of lrgA and lrgB were higher than those of the cwlD and cwlE genes in ΔcdsR at T0 (Fig. 3A).
A Analysis of the ΔcdsR transcriptome at T0. The differentially expressed genes associated with cell autolysis (lrgA, lrgB, cwlD, and cwlE) were analyzed according to fragments per kilobase of transcript per million mapped reads (FPKM). B Transcriptional analysis of the promoter regions of autolysis-related genes (lrgAB and cwlD) fused to lacZ in HD73 and ΔcdsR, respectively. Black circles indicate the HD(PlrgAB) strain, red squares indicate the ΔcdsR(PlrgAB) strain, blue triangles indicate the HD(PcwlD) strain, and yellow inverted triangles indicate the ΔcdsR(PcwlD) strain. C Examination of the interaction between the CdsR and PlrgAB promoter regions via electrophoretic mobility shift assay (EMSA), with increasing quantities of purified CdsR protein incubated with the PlrgAB promoter regions. The last lane was employed for the 200-fold unlabeled probe. D Increasing amounts of purified CdsR protein incubated with the PcwlD promoter regions. The last lane was used for the 200-fold unlabeled probe.
To detect the transcription dynamics of these genes, the lrgAB (PlrgAB), cwlD (PcwlD), and cwlE promoters (PcwlE) were fused to lacZ reporter gene. Subsequently, we investigated the expression of these three genes/operons in both HD73 and ΔcdsR through β-galactosidase assays. The result showed that the activities of the PlrgAB and PcwlD were significantly higher in ΔcdsR than in HD73. Notably, the transcriptional activities of PlrgAB and PcwlD were similar in HD73 cells (Fig. 3B). However, PcwlE activity was extremely low in both HD73 and ΔcdsR mutant (Fig. S3). These results clearly indicate that CdsR represses the transcription of lrgAB and cwlD.
To determine whether CdsR binds to the promoter regions of lrgAB, cwlD, and cwlE, we conducted an EMSA. Various concentrations of the CdsR protein were used to bind to 0.12 nM of the PlrgAB-labeled probe. Notably, low concentrations of the CdsR protein (3.84 and 7.68 μM) were able to effectively bind to the PlrgAB-labeled probe. The PlrgAB-labeled probe at a concentration of 0.12 nM was completely shifted with 15.36 μM of CdsR. However, a 200-fold excess of unlabeled probe successfully competed with the labeled probe, indicating specific binding (Fig. 3C). In another EMSA, 1.75 μM of CdsR was used to bind to 0.39 nM PcwlD-labeled probe. Remarkably, low concentrations of CdsR protein (0.89 and 1.34 μM) were able to effectively bind to the PcwlD-labeled probe. The PcwlD-labeled probe at a concentration of 0.39 nM was completely shifted with 1.75 μM of CdsR. A 200-fold excess of unlabeled probe successfully competed with the labeled probe, confirming specific binding (Fig. 3D). However, when 0.3 nM of the PcwlE-labeled probe was exposed to CdsR concentration ranging from 1.49 μM to 5.95 μM, no binding occurred, indicating that CdsR cannot interact with the cwlE promoter region (Fig. S3). These results strongly support the direct repression of lrgAB and cwlD by CdsR.
Overexpression of lrgAB results in cell lysis without sporulation
Holins act in conjunction with endolysins to cause cell lysis [34, 35]. Although lrgAB was initially annotated as an antiholin protein, a recent study confirmed that LrgAB can indeed act as a holin, inducing cell lysis in S. aureus [26]. The heightened transcriptional activity of lrgAB and cwlD in ΔcdsR in comparison to HD73 suggests that LrgAB, acting as a holin, and CwlD, serving as an endolysin, may collaborate to induce cell lysis. However, it is imperative to ascertain the impact of the overexpression of the lrgAB operon, lrgA, or lrgB on cell lysis.
OlrgAB, OlrgA, and OlrgB strains were generated by employing the pHT304 vector to overexpress the lrgAB operon, lrgA, and lrgB as described in “Materials and methods” section. An RT-qPCR analysis revealed that the transcriptional level of lrgA increased by approximately 100- and 30-fold in OlrgA and OlrgAB strains, respectively, compared to HD73. Similarly, lrgB exhibited increased transcription in both OlrgB and OlrgAB strains compared to HD73 cells (Fig. 4A), unequivocally confirming the successful overexpression of lrgA and lrgB in the corresponding strains.
A Analysis of lrgA expression in HD73, OlrgAB, and OlrgA strains and lrgB expression in HD73, OlrgAB, and OlrgB strains by RT-qPCR at T5. Experiments were conducted in triplicate. Error bars represent standard deviations (****P < 0.0001). B Role of LrgAB in the growth of B. thuringiensis. Growth curves were plotted for HD73 (black circles), OlrgAB (red squares), OlrgA (blue triangles), and OlrgB (yellow diamond) strains grown in SSM at 30 °C with shaking at 220 rpm. C Autolytic rates of HD73, OlrgAB, OlrgA, and OlrgB in Tris-HCl and Triton X-100 mixed buffer, expressed as a percentage increase in OD580. D Cell death imaging in B. thuringiensis cell populations using SYTOXTM Green at T3 and T17 for HD73 and OlrgAB. Bar, 10 μm. E Statistical analysis of the cell death rates of the HD73 and OlrgAB strains, obtained by counting the number of dead and live cells from microscopic images. The cell death rate of OlrgAB was compared to that of HD73, and the data were analyzed using a T-test (**P < 0.01). F Observation of polar septum formation during spore initiation by laser confocal microscopy. The strains grown in SSM at T2 and T3 correspond to HD73 and OlrgAB, respectively.
The growth curve showed a significant reduction in cell density for OlrgAB from 14 to 24 h compared to the HD73, OlrgA, and OlrgB strains (Fig. 4B). Similarly, the autolytic rate of OlrgAB was significantly higher than that of HD73, OlrgA, and OlrgB from A1 to A8 (Fig. 4C), indicating an increased autolytic activity in the OlrgAB strain. Taken together, the simultaneous expression of lrgA and lrgB led to cellular autolysis, a phenomenon that did not occur when lrgA and lrgB were overexpressed alone. These findings suggest that LrgAB is a critical complex responsible for cell lysis in HD73. The proportion of dead cells was determined using SYTOXTM Green staining, with fewer cells exhibiting green fluorescence in HD73 cells compared to OlrgAB cells at both T3 and T17 (Fig. 4D). A quantitative analysis revealed that the proportion of dead cells was 16.93(±4.63)% in HD73 and 50.63(±9.31)% in OlrgAB at T17 (Fig. 4E). Collectively, these results indicate that the overexpression of lrgAB induces cell lysis.
When HD73 and OlrgAB cells cultured in SSM were stained with FM4-64, polar septa were observed in HD73 cells (Figs. 4Fa and 4Fb), whereas OlrgAB cells exhibited no polar septa at T3 (Fig. 4Fd). In additon, HD73 cells completed the process of engulfment at T7, whereas OlrgAB cells showed no polar septum formation (Fig. S1k). In summary, OlrgAB and ΔcdsR exhibit similar phenotypes, characterized by cell lysis and the absence of sporulation (Figs. 1 and 4). This indicates that CdsR plays a critical role in cell lysis and sporulation by regulating lrgAB.
Deletion of lrgAB leads to a greater number of abnormal cells
We demonstrated that LrgAB induces cell lysis without sporulation, and this regulation is mediated by CdsR. This highlights the essential role of the cell death pathway regulated by LrgAB in the sporulation process and emphasizes its tight regulation. Further investigation into the effects of disrupting this pathway on sporulation would be highly intriguing. Optical microscopy revealed the presence of two distinct cell types: sporulating cells and abnormal cells lacking spore formation ability, displaying a deformed cell shape in both HD73 and the ΔlrgAB mutant (Fig. 5A). A quantitative analysis revealed that the proportions of abnormal cells in the total cell population at T12, T17, and T22 were 7.13(± 1.72)%, 0.61(± 0.28)%, and 1.38(± 0.3)%, respectively, in HD73, and 12.87(± 0.75)%, 34.35(± 7.43)%, and 37.94(± 7.86)%, respectively, in the ΔlrgAB mutant (Fig. 5B). The ΔlrgAB mutant exhibited a significant accumulation of abnormal cells compared to HD73. These findings suggest that lrgAB deletion leads to a greater number of abnormal cells.
A Morphological modification of ΔlrgAB by optical microscopy. Bar, 10 μm. Red arrows indicate spore cells, and yellow arrows indicate abnormal cells. B Statistical analysis of the abnormal cell rates of HD73 and ΔlrgAB, obtained by counting the number of sporulating and abnormal cells from microscopic images. The abnormal cell rate of ΔlrgAB was compared to that of HD73, and the data were analyzed using the T-test (***P < 0.001. ****P < 0.0001). C Laser confocal microscopy was performed to determine the expression of lrgAB in ΔlrgAB. PlrgAB promoter-directed gfp gene expression was examined in ΔlrgAB and HD73. Bar, 20 μm. D The GFP-expressing cell rate of the ΔlrgAB was compared to that of HD73. The percentage of GFP-expressing cells in HD73 and ΔlrgAB was statistically analyzed by counting the number of total cells and GFP-expressing cells through micrographs. E The sporulating cell rate of the ΔlrgAB was compared with that of HD73. The percentage of sporulating cells in ΔlrgAB and HD73 was statistically analyzed by counting the number of total cells and sporulating cells through micrographs. Data were obtained from three random microscopy images and analyzed by T-test (**P < 0.01. ***P < 0.001. ****P < 0.0001).
To clarify lrgAB expression in abnormal cells, a fusion expression vector harboring the PlrgAB promoter and the gfp gene was introduced into HD73 and ΔlrgAB mutant cells, respectively. Laser confocal microscopy revealed that the number of cells exhibiting green fluorescence at T12, T17, and T22 remained extremely low and did not accumulate in HD73 cells over time. In contrast, while very few cells exhibited green fluorescent at T12, a higher number of non-sporulating cells expressing green fluorescent protein appeared in the ΔlrgAB mutant at T17 and T22 (Fig. 5C). Quantification of cells showed that in HD73, GFP-expressing cells accounted for 3.50(± 0.31)%, 4.84(± 1.07)%, and 0.44(± 0.22)% of the total cells at T12, T17, and T22, respectively. However, in the ΔlrgAB mutant, GFP-expressing cells represented 6.92(± 0.71)%, 32.27(± 1.55)%, and 35.24(± 1.84)% of the total cells at T12, T17, and T22, respectively (Fig. 5D). This suggests that LrgAB is activated in abnormal cells.
To determine the role of the LrgAB system in sporulation, the number of total cells and sporulating cells were quantified. The result of the plate count showed that the total number of HD73 (1.358 × 107 CFU/mL) and ΔlrgAB (1.352 × 107 CFU/mL) cells were similar (Fig. S4). However, the result of quantification through microscopy images showed that the sporulating cell rate of HD73 (95.23(± 1.03)%, 94.92(± 0.62)%, and 97.26(± 1.17)%) was higher than that of ΔlrgAB (90.42(± 0.64)%, 62.29(± 6.12)%, and 63.78(± 2.14)%) at T12, T17, and T22, respectively (Fig. 5E). This suggests that the LrgAB system plays a crucial role in the sporulating cell population. In summary, deletion of lrgAB leads to an accumulation of abnormal cells and the PlrgAB promoter is more active in abnormal cells.
Discussion
In this study, we identified a novel transcriptional regulator, CdsR, which inhibits cell lysis and promotes sporulation in B. thuringiensis (Fig. 1). We analyzed CdsR homologs with >30% similarity and >60% coverage. We examined 75 sequences from annotated and publicly available bacterial genomes that represent species diversity in terms of evolutionary distance using the Basic Local Alignment Search Tool BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Subsequently, the sequences corresponding to the best hits were subjected to multiple sequence alignment using ClustalW (https://www.genome.jp/tools-bin/clustalw). Thereafter, we constructed phylogenetic trees based on the alignments and found that CdsR homologs are present in a variety of bacterial species. Further analysis revealed that CdsR homologs were primarily distributed in the B. cereus group, Clostridium spp., Paenibacillus spp., and Selenomonas spp. (Fig. S5). CdsR homologs with 100% similarity were predominantly found in the B. cereus group, which included B. thuringiensis known for its insecticidal properties, B. cereus associated with emetic and/or diarrheal foodborne illnesses as food poisoning agents, and B. anthracis responsible for anthrax [36, 37]. This suggests that the mechanisms underlying CdsR-dependent inhibition of cell lysis and promotion of sporulation might be similar within the B. cereus group. CdsR belongs to the ArsR family of regulators, which are known to be involved in various functions such as metal ion homeostasis [38], biofilm formation [39], and virulence [40]. Generally, the N-terminus of ArsR family regulators contains a winged helix-turn-helix DNA-binding domain that represses transcription by binding to downstream target promoters [41]. In this study, we found that a novel ArsR family regulator plays an important role in cell death and sporulation.
Homologs of lrgAB are found across a wide range of bacteria, including gram-negative bacteria (Fusobacterium spp., Bradyrhizobium spp., Brucella spp., Rhodobacter spp., Burkholderia spp., Chromobacterium spp., Desulfuromonas spp., Escherichia spp., Salmonella spp., and Yersinia spp.), gram-positive bacteria (Kineococcus spp., Streptomyces spp., Deinococcus spp., Bacillus spp., Exiguobacterium spp., Staphylococcus spp., Clostridium spp., Enterococcus faecalis, Pediococcus spp., Lactococcus spp., and Streptococcus spp.), and archaea (Methanosarcina spp. and Pyrococcus spp.) [24]. These LrgAB homologs were initially annotated as antiholin-like proteins until the discovery of LrgA in S. aureus, which was identified as a bacteriophage holin-like protein that plays a role in cell death [26]. In Bacillus spp., LrgAB proteins have various functions across different strains. For instance, the lrgA homolog, ysbA, is not involved in cell lysis but is essential for pyruvate transport or utilization in B. subtilis [42]. In B. cereus, LrgAB is involved in extracellular DNA release and biofilm formation [43]. Mutations in lrgAB affect stationary-phase survival and sporulation efficiency in B. anthracis [5]. A comparison of the amino acid sequences of available LrgA homologs revealed that LrgA (HD73_RS29150) of B. thuringiensis HD73 shares 62%, 99%, and 100% similarity with LrgA (YsbA, BSU28910) of B. subtilis 168, LrgA (BC5439) of B. cereus ATCC 14579, and LrgA (BA5690) of B. anthracis Ames, respectively. In our previous study, we found that deletion of lrgAB did not significantly affect sporulation efficiency by the method of heating cells for elimination of non-sporulating cells [44]. In this study, we directly counted the number of sporulating cells in the microscopy images. We found that the total number of cells in HD73 and ΔlrgAB mutant was comparable (Fig. S4), but the rate of sporulating cell in HD73 was significantly higher than that in ΔlrgAB mutant (Fig. 5E). The difference may be due to the heating method in which some non-sporulating cell also showed thermal resistance. The aforementioned studies on Bacillus species have predominantly focused on the effects of deleting lrgAB. However, in this study, we found that in B. thuringiensis, the overexpression of LrgAB plays a role in both cell death and sporulation, functioning as a holin-like protein (Fig. 4). Notably, only the OlrgAB strain showed cell lysis, whereas the OlrgA, OlrgB and HD73 strains showed no cell lysis (Fig. 4). A previous study by Bayles and Endres suggested that LrgA alone can oligomerize and form pores within the cytoplasmic membrane, leading to cell lysis. However, our findings indicate that these functions must be performed in the context of LrgB [26]. Therefore, we speculate that lrgAB overexpression disrupts the cell membrane structure, and that lrgA and lrgB expressed alone are incapable of performing such biological functions in B. thuringiensis. To our knowledge, this is the first report on LrgAB acting as a holin-like protein that induces cell death in Bacillus spp.
Within the B. cereus group, there are three additional homologs of lrgA, namely cidA, clhA1, and clhA2 [5, 45]. While disrupting of B. anthracis clhAB2 had a minor impact on murein hydrolase activity, it was found to be crucial for sporulation [5]. However, the molecular mechanisms underlying the roles of cidAB and clhAB1 (not transcription) in B. anthracis cell death remain unclear [5]. The amino acid sequence similarity of homologs CidA (HD73_RS19705), ClhA1 (HD73_RS20090), and ClhA2 (HD73_RS27290) comparisons with LrgA (HD73_RS29150) were 24% for CidA, 29% for ClhA1, and 26% for ClhA2 in B. thuringiensis, respectively. Among the four lrgA homologs in the HD73 strain, only lrgA exhibited upregulated expression with a 2.36-fold in ΔcdsR, and other three genes had no change (Table S5). This suggests that CdsR is involved in cell lysis and sporulation via regulation of the lrgAB operon. CdsR inhibits lrgAB expression via the cell death pathway. Although activation of lrgAB by LytSR and CidR positive regulation of lrgAB expression in B. anthracis [5] and S. aureus [24] has been well studied, very few repression regulators have been reported. lrgAB is negatively regulated by CcpA in B. subtilis and Streptococcus mutans during the exponential phase [46]. We report, for the first time, that CdsR represses LrgAB during the stationary phase.
Cell death can help eliminate unwanted or damaged cells and maintain the health of a population [47,48,49]. For example, cell death as a critical player in establishing a properly formed central nervous system by removing genetically compromised cells. The AIM2 inflammasome monitors DNA damage accumulation in neurons through its regulation of Gasdermin-D-mediated cell death, contributing to the elimination of genetically compromised central nervous system cells [47]. Physiological apoptosis is mediated by VAB-1/EphR receptor signaling, which promotes apoptosis in the Caenorhabditis elegans germline. In C. elegans, as germ cells progress through meiosis, about half of them undergo physiological apoptosis, in which their cytosolic components are redistributed to ‘nurse’ the remaining oocytes [48]. Furthermore, cell death is activated in response to DNA damage in a subset of Pseudomonas aeruginosa cells [49]. DNA damage results in the derepression of alpA expression, leading to the cleavage of genes, such as alpBCDE, which are activated for expression [50]. Thus, some lysed P. aeruginosa cells provide nutrients or liberate toxins or other bacterial factors that promote colonization of the lung by the remaining cells in the population [49]. In this study, the absence of lrgAB resulted in the inability to remove abnormal cells, which gradually accumulated in the B. thuringiensis population (Fig. 5). This is most likely a new example of a cell death pathway increases the benefits of a cell population.
In previous studies, a series of quality control systems have been identified to maintain the fidelity of sporulation program [19, 22, 51]. Sda, a suppressor of DnaA, and SirA, a sporulation inhibitor of replication A, control cells to enter sporulation in response to DNA replication defects. DisA is a DNA integrity scanning protein that functions as part of a DNA damage checkpoint immediately following the initiation of sporulation [21]. CmpA persists and acts as an adaptor of ClpXP to mediate degradation of SpoIVA in a σK-dependent manner, and SpoVID monitors the polymerization state of the coat basement layer via an extension to a functional intracellular LysM domain [52], which eradicate sporulating cells that inaccurately assemble the envelope through cell lysis. However, the checkpoint-mediated cell death pathway in B. subtilis remains unclear. In this study, we found that LrgAB acts as a holin-like protein and that LrgAB-dependent cell death pathways maybe increase the benefit of sporulating cells (Fig. 5). It is one possibility that this cell death pathway followed the checkpoint system result in cell lysis. Therefore, the relationship between quality control systems and LrgAB-dependent cell death requires further investigation.
In summary, we propose a model for the LrgAB-dependent cell death pathway, which is repressed by the novel regulator, CdsR (Fig. 6). CdsR is negatively autoregulated, functioning as a repressor that significantly inhibits the expression of the lrgAB operon. The lrgAB operon is repressed by CdsR in sporulating cells, allowing spore development. However, the CdsR repression of lrgAB is released by unknown intracellular signals to activate the expression of LrgAB, which acts as a holin, leading to cell lysis in abnormal cells (Fig. 6). In conclusion, we propose that the CdsR-LrgAB cell death system may be beneficial for the sporulation of Bacillus species. This study provides new insights into the relationship between cell survival and death.
Left: The HD73 cell population comprises two types of cells: sporulating cells (fully developed) and abnormal cells (abnormally developed). Right (top): CdsR serves as an autoregulator that negatively regulates lrgAB expression in sporulating cells. Right (bottom): In abnormal cells, CdsR relieves its inhibition of lrgAB through an unknown signal. LrgAB acts as a holin that induces cell lysis. Therefore, the CdsR-LrgAB cell death system is beneficial for the sporulation.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Bacillus thuringiensis HD73 and its derivatives were cultured under 30 °C in Luria-Bertani (LB) medium containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl or on solid LB medium with 1.5% agar. The growth of bacterial cells was monitored using Schaeffer’s sporulation medium [53] (SSM; 0.8% nutrient broth, 0.012% MgSO4, 0.1% KCl, 0.5 mM NaOH, 1 mM Ca(NO3)2, 0.01 μM MnCl2, and 1 μM FeSO4). The Escherichia coli strains TG1 and ET were, respectively, used for molecular cloning and generation of non-methylated plasmid DNA for subsequent transformation into B. thuringiensis strains. Both strains were cultured at 37 °C in LB medium, supplemented with 5 μg/mL of erythromycin and 50 μg/mL of kanamycin for B. thuringiensis and with 100 μg/mL of ampicillin for E. coli, when necessary. The bacterial strains and plasmids used in this study are listed in Table 1.
DNA manipulation and transformation
Plasmid DNA was extracted from E. coli cells using the Plasmid Miniprep Kit (Axygen, Beijing, China). Restriction enzymes and T4 DNA ligase (Takara Biotechnology Corporation, Dalian, China) were used according to the manufacturer’s instructions. PCR was performed using either the high-fidelity PrimeStar HS DNA polymerase (Takara Biotechnology Corporation, Beijing, China) or Taq DNA polymerase (BioMed, Beijing, China). DNA fragments were purified from 1% agarose gels using the AxyPrep DNA Gel Extraction Kit (Axygen). Standard transformation procedures for E. coli [54] and B. thuringiensis [55] have been previously described. The primers used in this study are listed in Table 2.
Construction of the cdsR deletion mutant
The primers used for gene deletion were designed based on the HD73 genome sequence. A 1032-bp fragment preceding the start codon of cdsR (cdsR fragment A) was amplified by PCR, utilizing HD73 genomic DNA as the template and cdsR-AF and cdsR-AR as the forward and reverse primers, respectively. Furthermore, the primers cdsR-BF and cdsR-BR were used to amplify a 1015-bp fragment downstream of the 96th codon of cdsR (cdsR fragment B), whereas the primers cdsR-KmF and cdsR-KmR were utilized to amplify a 1473-bp kanamycin resistance gene cassette (kana) derived from pMAD-ΔlrgAB [44] plasmid. The cdsR A, kana, and cdsR B fragments were ligated through overlapping PCR using the primers cdsR-AF and cdsR-BR. The 3568-bp overlapping fragment was inserted between the BamHI and EcoRI restriction sites of the temperature-sensitive and erythromycin-resistant plasmid pMAD [56], resulting in the generation of the plasmid pMAD-ΔcdsR, which was then electroporated into HD73 using the Gene Pulser II apparatus (Bio-Rad, Hercules, CA, USA). Subsequently, transformants were selected for erythromycin resistance at 30 °C. The positive transformants were transferred to 37 °C and screened for the colonies lacking erythromycin resistance but resistant to kanamycin to obtain the mutant ΔcdsR, which was characterized by PCR.
Construction of multiple strains
The cdsR gene fragment and cdsR promoters were amplified with the CcdsR-F/CcdsR-R primers using HD73 genomic DNA as a template. The resulting 969-bp PCR product was inserted into plasmid pHT304 [57] using EcoRI and BamHI to obtain pHT304-PcdsR-cdsR. This recombinant construct was replicated in E. coli and subsequently introduced into the ΔcdsR mutant, resulting in a genetically complementary strain CcdsR.
Two fragments were amplified using HD73 genomic DNA as a template: one was a 1676-bp fragment containing the lrgAB operon and its promoter region, amplified with the ClrgAB-F/ClrgAB-R primers, and the other was a 948-bp fragment containing the lrgA gene and its promoter region, amplified with the ClrgA-F/ClrgA-R primers. The lrgAB operon’s promoter region (513-bp) was amplified with the ClrgAB-F/CPlrgAB-lrgB-R primers and the lrgB gene fragment (731-bp) was amplified with the CPlrgAB-lrgB-F/ClrgAB-R primers, using HD73 genomic DNA as a template. The above two fragments were combined to create a 1244-bp fragment through overlapping PCR using the ClrgAB-F/ClrgAB-R primers. The fragments of sizes 1676, 948, and 1244 bp were individually ligated into plasmid pHT304, using the EcoRI and BamHI sites, to obtain the recombinant plasmids pHT304-PlrgAB-lrgAB, pHT304-PlrgAB-lrgA, and pHT304-PlrgAB-lrgB, respectively. These recombinant plasmids were then introduced into the HD73 strain to obtain the OlrgAB, OlrgA, and OlrgB strains for overexpression, respectively.
The strains in which the promoter of the lrgAB operon directs gfp gene expression were constructed as follows: HD73 genomic DNA served as a template to amplify the lrgAB promoter fragment (PlrgAB, 528-bp) with the PlrgAB-gfp-F/PlrgAB-gfp-R primers. The gfp gene (735-bp) was amplified using the plasmid p35′gfp [58] as a template with the lrgAB-gfp-F/lrgAB-gfp-R primers. The PlrgAB and gfp fragments were used as templates to jointly amplify the PlrgAB-gfp gene fusion fragment (1226-bp) using the PlrgAB-gfp-F/lrgAB-gfp-R primers. The resulting PCR product was inserted into the plasmid pHT304 through the EcoRI and PstI sites, leading to the generation of the plasmid pHT304-PlrgAB-gfp, which, in turn, was introduced into both the HD73 and ΔlrgAB mutant strains [44].
Autolysis observation and autolytic activity
To determine the bacterial culture density, a 5 mL sample was cultured in 50 mL SSM medium for 21 h and then transferred to a glass test tube. The strains HD73, ΔcdsR, and CcdsR were measured by the OD600 value, using three independent biological replicates.
The 5 mL samples collected from the 50 mL SSM at T6 (T0 indicates the end of the exponential growth phase, and Tn indicates the number of hours before (-) or after T0) were centrifuged at 12,000 × g for 10 min at 4 °C. The resulting pellets were washed thrice with 50 mL of sterile water and resuspended in 30 mL of 0.05 M Tris-HCl (pH 7.2) containing 0.05% (vol/vol) Triton X-100 (Sigma Chemical Company, St. Louis, MO, USA) [59]. Subsequently, the bacteria cells were incubated in a shaking incubator at 220 rpm and 30 °C. The OD580 values of the samples were measured every 1 h from A1 to A8 (with A0 denoting the initial value of cells at T6 and An indicating the number of hours after time A0).
Microscope observation
Bacillus thuringiensis cells were cultured in SSM, and samples were collected at specified time points for analysis. Transmission electron microscopy (TEM) was used to observe the bacterial cell and spore morphology [60]. Optical microscopy was performed to characterize cell types using an Olympus BX63 microscope. Confocal microscopy (Carl Zeiss 880) was performed to examine polar septum formation, spore engulfment processes, and cell viability. Various fluorescent dyes, including FM4-64 (100 μM), MitoTracker Green (MTG) (100 nM), and SYTOXTM Green (5 nM), were used for these observations.
Determination of cell number
Samples of B. thuringiensis cells, including HD73 and ΔlrgAB mutant cultured for T24, were collected for dilution and plating. The total number of cells was determined by counting the total number of colony-forming units (CFUs) on LB plates.
The number of total cells and dead cells in the microscopic images were counted. Optical or laser confocal microscopy images were used to quantify at least 500 cells of each strain type. The cell death rate was defined as the ratio of the number of dead cells (stained with SYTOXTM Green) to the total number of cells. The cell death rate was determined from at least three independent cultures. Moreover, the abnormal and GFP-expressing cell rates were statistically calculated in a similar manner.
Transcriptome sequencing and analysis
The HD73 and ΔcdsR mutant strains were cultured in SSM at 30 °C and harvested at T0. Total RNA was extracted using the RNAprep Pure Bacteria Kit (Aidlab, Beijing, China). RNA sequencing (RNA-Seq) was carried out by SinoBiocore (Beijing, China). Ribosomal RNA was removed using the Ribo Zero™ rRNA Removal Kit (Epicentre, Madison, WI, USA), and rRNA-free residue was removed by ethanol precipitation. Subsequently, the ribosome-depleted mRNA was fragmented and reverse transcribed. The first-strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase, and the second-strand cDNA was synthesized using DNA Polymerase I and RNase H. For the dNTPs in the reaction buffer, the dTTPs were replaced with dUTPs. After adenylation of 3′ ends of DNA fragments, the NEBNext Adaptors (New England BioLabs, Ipswich, MA, USA) with a hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments of preferentially 250–300 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA, USA). Subsequently, 3 µL of the USER Enzyme (New England BioLabs) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. The PCR was performed using the Phusion High-Fidelity DNA Polymerase, universal PCR primers, and index (X) primers. The resulting PCR products were purified using the AMPure XP system, and the quality of the libraries was assessed using an Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). The clustering of index-coded samples was carried out on a cBot Cluster Generation System (Illumina, USA). Library preparations were sequenced on an Illumina HiSeq X Ten platform with a 150-bp paired-end module. The sequence reads were mapped to the HD73 reference genome (NC_020238.1).
The relative transcript abundance was calculated as fragments per kilobase of exon sequence per million mapped sequence reads (FPKM). Genes were considered differentially expressed only if they met the criteria of having an adjusted P-value ≤ 0.05 and absolute value of log2 fold-change ≥1. The differentially expressed genes were subjected to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using an online gene function analysis tool. All RNA-Seq data were uploaded to the Gene Expression Omnibus database of the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov) (accession no. GSE216307).
β-galactosidase activity assays
The promoter sequence of cdsR (614-bp upstream of the cdsR translational start codon) was amplified from HD73 genomic DNA using the PcdsR-F and PcdsR-R primers. The resulting PcdsR fragments were then digested with PstI and BamHI and ligated into the linearized pHT304-18Z plasmid, which harbors a promoter-less lacZ gene [61], to obtain the recombinant plasmid pHT304PcdsR. Subsequently, the recombinant plasmid was introduced into both HD73 and ΔcdsR strains, leading to the generation of the strains HD(PcdsR) and ΔcdsR(PcdsR), respectively, which were validated by erythromycin resistance and PCR identification. To assess the transcription of spo0A, spoIIE, lrgAB, cwlD, and cwlE in HD73 and ΔcdsR, a similar methodology was employed to construct the desired strains. The recombinant plasmids pHT304Pspo0A [32], pHT304PspoIIE [44], and pHT304PlrgAB [44] were introduced into both HD73 and ΔcdsR strains. The PcwlD promoter (592-bp) and PcwlE promoter (418-bp) were amplified from HD73 genomic DNA using the primers PcwlD-F/PcwlD-R and PcwlE-F/PcwlE-R, respectively. Following amplification, these promoter fragments were digested with PstI and BamHI and subsequently ligated into the linearized pHT304-18Z plasmid to obtain recombinant plasmids pHT304PcwlD and pHT304PcwlE, which were then introduced into HD73 and ΔcdsR, respectively. The β-galactosidase activities were measured as previously described [62] and expressed as Miller units. Samples were collected at 1-h intervals from T−2 to T6. For each sample, 2 mL of culture was centrifuged (12,000 × g, 1 min), and the resulting pellets were stored at −20 °C until testing. The reported values represent the mean and standard error of at least three independent assays.
Expression and purification of CdsR
The expression plasmid pETcdsR, containing the cdsR gene from HD73, was constructed by amplifying with the primers pETcdsR-F and pETcdsR-R, followed by cloning into BamHI/SalI-digested pET21b. The resulting pETcdsR plasmid was transferred into E. coli BL21 (DE3), and the transformants were grown to the exponential phase in LB medium supplemented with ampicillin at 37 °C. Subsequently, the expression of CdsR-His protein was induced by adding 1 mM isopropyl-β-D-thiogalactopyranoside (final concentration) to the culture for 12 h at 18 °C. The resulting CdsR-His protein was purified as previously described [63].
Electrophoresis mobility shift assays
The DNA fragments were PCR-amplified from HD73 genomic DNA using specific primers labeled with a 5′-end fluorescein amidite (FAM) modification and confirmed by DNA sequencing. Electrophoretic mobility shift assays (EMSA) [31] were performed as previously described to analyze the binding between the purified CdsR protein and PlrgAB, PcdsR, and PcwlD DNA fragments.
Real-time quantitative PCR assay
Cells expressing OlrgAB, OlrgA, and OlrgB were harvested to analyze the expression levels of lrgA and lrgB at T6. Total RNA was extracted from the collected cells using the RNAprep Pure Kit (Aidlab, Beijing, China). Subsequently, cDNA was synthesized by reverse transcription using the HiScript II Q RT SuperMix (Vazyme, Nanjing, China). The cDNA concentration was diluted to 200 ng/μL. The transcriptional levels of the target genes were determined by real-time quantitative PCR (RT-qPCR) using the ChamQ Universal SYBR qPCR master mix (Vazyme). The 16S rRNA gene was used as an internal control. The primers used for RT-qPCR analysis are listed in Table 2.
Bioinformatics analysis
The CdsR homologous protein sequences obtained from NCBI were used for the phylogenetic tree analysis. Multiple sequence alignments were performed and visualized using the software programs ClustalW and GeneDoc 3.2, respectively, and a phylogenetic tree was constructed using the neighbor-joining method in the MEGA 7.0 software. Furthermore, the homologous protein sequences of LrgAB were retrieved from the NCBI to analyze interspecies distributions and perform similarity comparisons.
Statistical analysis
Data are means ± sem. Significant differences were determined with a Student’s T-test with *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001 (n ≥ 3). Statistical analysis was performed using Excel and GraphPad Prism 9 software. A P-value of less than 0.05 was considered significant for all tests.
Data availability
We have uploaded the DNA microarray and RNA-Seq data, which will be available in the NCBI Gene Expression Omnibus database with the accession no. GSE48410 and GSE216307. All data generated or analyzed during this study are included in this published article.
References
Qiu S, Liu J, Xing F. ‘Hints’ in the killer protein gasdermin D: unveiling the secrets of gasdermins driving cell death. Cell Death Differ. 2017;24:588–96.
Ndhlovu A, Durand PM, Ramsey G. Programmed cell death as a black queen in microbial communities. Mol Ecol. 2021;30:1110–9.
Morris JJ. Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 2015;31:475–82.
Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–14.
Chandramohan L, Ahn JS, Weaver KE, Bayles KW. An overlap between the control of programmed cell death in Bacillus anthracis and sporulation. J Bacteriol. 2009;191:4103–10.
Durand PM, Sym S, Michod RE. Programmed cell death and complexity in microbial systems. Curr Biol. 2016;26:R587–R593.
González-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–3.
Engelberg-Kulka H, Hazan R. Microbiology. Cannibals defy starvation and avoid sporulation. Science. 2003;301:467–8.
Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–44.
Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–52.
LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–3.
Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–42.
Phillips ZE, Strauch MA. Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci. 2002;59:392–402.
Wu JJ, Howard MG, Piggot PJ. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989;171:692–8.
York K, Kenney TJ, Satola S, Moran CP Jr, Poth H, Youngman P. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992;174:2648–58.
Baldus JM, Green BD, Youngman P, Moran CP Jr. Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol. 1994;176:296–306.
Molle V, Fujita M, Jensen ST, Eichenberger P, González-Pastor JE, Liu JS, et al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–701.
Fujita M, González-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–68.
Decker AR, Ramamurthi KS. Cell death pathway that monitors spore morphogenesis. Trends Microbiol. 2017;25:637–47.
Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–7.
Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, Ramamurthi KS. A versatile nano display platform from bacterial spore coat proteins. Nat Commun. 2015;6:6777.
Tan IS, Weiss CA, Popham DL, Ramamurthi KS. A quality-control mechanism removes unfit cells from a population of sporulating bacteria. Dev Cell. 2015;34:682–93.
Allocati N, Masulli M, Di Ilio C, De Laurenzi V. Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis. 2015;6:e1609.
Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–6.
Sadykov MR, Bayles KW. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol. 2012;15:211–5.
Endres JL, Chaudhari SS, Zhang X, Prahlad J, Wang SQ, Foley LA, et al. The Staphylococcus aureus CidA and LrgA proteins are functional holins involved in the transport of by-products of carbohydrate metabolism. mBio. 2022;13:e0282721.
Laabei M, Duggan S. CidA and LrgA: a “hole” lot more than programmed cell death. mBio. 2022;13:e0076122.
Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806.
Cardoso P, Fazion F, Perchat S, Buisson C, Vilas-Bôas G, Lereclus D. Rap-Phr systems from plasmids pAW63 and pHT8-1 act together to regulate sporulation in the Bacillus thuringiensis serovar kurstaki HD73 strain. Appl Environ Microb. 2020;86:e01238–20.
Cardoso PF, Perchat S, Vilas-Boas LA, Lereclus D, Vilas-Bôas GT. Diversity of the Rap-Phr quorum-sensing systems in the Bacillus cereus group. Curr Genet. 2019;65:1367–81.
Peng Q, Wang G, Liu G, Zhang J, Song F. Identification of metabolism pathways directly regulated by sigma(54) factor in Bacillus thuringiensis. Front Microbiol. 2015;6:407.
Hou S, Zhang R, Lereclus D, Peng Q, Zhang J, Slamti L, et al. The transcription factor CpcR determines cell fate by modulating the initiation of sporulation in Bacillus thuringiensis. Appl Environ Microbiol. 2022;88:e0237421.
Perchat S, Talagas A, Poncet S, Lazar N, Li de la Sierra-Gallay I, Gohar M, et al. How quorum sensing connects sporulation to necrotrophism in Bacillus thuringiensis. PLoS Pathog. 2016;12:e1005779.
Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205.
Saier MH Jr, Reddy BL. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol. 2015;197:7–17.
Carroll LM, Cheng RA, Wiedmann M, Kovac J. Keeping up with the Bacillus cereus group: taxonomy through the genomics era and beyond. Crit Rev Food Sci. 2022;62:7677–702.
Ehling-Schulz M, Lereclus D, Koehler TM. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr. 2019;7:0032–2018.
Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27:668–80.
Guimarães BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, et al. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286:26148–57.
Shi C, Wang S, Han J, Xi L, Li M, Li Z, Zhang H. Functional insights into Brucella transcriptional regulator ArsR. Microbiol Pathogenesis. 2022;168:105557.
Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol Rev. 2003;27:131–43.
Van den Esker MH, Kovács ÁT, Kuipers OP. YsbA and LytST are essential for pyruvate utilization in Bacillus subtilis. Environ Microbiol. 2017;19:83–94.
Zhang J, Meng L, Zhang Y, Sang L, Liu Q, Zhao L, et al. GapB is involved in biofilm formation dependent on LrgAB but not the SinI/R system in Bacillus cereus 0-9. Front Microbiol. 2020;11:591926.
Peng Q, Wu J, Chen X, Qiu L, Zhang J, Tian H, et al. Disruption of two-component system LytSR affects forespore engulfment in Bacillus thuringiensis. Front Cell Infect Microbiol. 2017;7:468.
Huillet E, Bridoux L, Wanapaisan P, Rejasse A, Peng Q, Panbangred W, et al. The CodY-dependent clhAB2 operon is involved in cell shape, chaining and autolysis in Bacillus cereus ATCC 14579. PLoS ONE. 2017;12:e0184975.
Van den Esker MH, Kovács ÁT, Kuipers OP. From cell death to metabolism: Holin-antiholin homologues with new functions. mBio. 2017;8:e01963–17.
Lammert CR, Frost EL, Bellinger CE, Bolte AC, McKee CA, Hurt ME, et al. AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature. 2020;580:647–52.
Li X, Johnson RW, Park D, Chin-Sang I, Chamberlin HM. Somatic gonad sheath cells and Eph receptor signaling promote germ-cell death in C. elegans. Cell Death Differ. 2012;19:1080–9.
McFarland KA, Dolben EL, LeRoux M, Kambara TK, Ramsey KM, Kirkpatrick RL, et al. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc. Natl Acad. Sci. USA. 2015;112:8433–8.
Peña JM, Prezioso SM, McFarland KA, Kambara TK, Ramsey KM, Deighan P, et al. Control of a programmed cell death pathway in Pseudomonas aeruginosa by an antiterminator. Nat Commun. 2021;12:1702.
Meneghini MD. Spore no more: quality control during bacterial development. Dev Cell. 2015;34:611–2.
Delerue T, Anantharaman V, Gilmore MC, Popham DL, Cava F, Aravind L, et al. Bacterial developmental checkpoint that directly monitors cell surface morphogenesis. Dev. Cell. 2022;57:344–360.e6.
Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–11.
Sambrook BJ, Russell, DW. Molecular cloning. A laboratory manual. 3rd edn. Cold Spring Harbor Laboratory Press; 2015.
Lereclus D, Arantes O, Chaufaux J, Lecadet M. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;51:211–7.
Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–91.
Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–9.
Deng C, Slamti L, Raymond B, Liu G, Lemy C, Gominet M, et al. Division of labour and terminal differentiation in a novel Bacillus thuringiensis strain. ISME J. 2015;9:286–96.
Mani N, Tobin P, Jayaswal RK. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J Bacteriol. 1993;175:1493–9.
Zhang X, Gao T, Peng Q, Song L, Zhang J, Chai Y, et al. A strong promoter of a non-cry gene directs expression of the cry1Ac gene in Bacillus thuringiensis. Appl Microbiol Biotechnol. 2018;102:3687–99.
Agaisse H, Lereclus D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol. 1994;13:97–107.
Yang H, Wang P, Peng Q, Rong R, Liu C, Lereclus D, et al. Weak transcription of the cry1Ac gene in nonsporulating Bacillus thuringiensis cells. Appl Environ Microbiol. 2012;78:6466–74.
Peng Q, Yang M, Wang W, Han L, Wang G, Wang P, et al. Activation of gab cluster transcription in Bacillus thuringiensis by γ-aminobutyric acid or succinic semialdehyde is mediated by the Sigma 54-dependent transcriptional activator GabR. BMC Microbiol. 2014;14:306.
Munson RS Jr, Sasaki K. Protein D, a putative immunoglobulin D-binding protein produced by Haemophilus influenzae, is glycerophosphodiester phosphodiesterase. J Bacteriol. 1993;175:4569–71.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (Grant No. 2022YFE0116500) and National Natural Science Foundation of China (General Program, Grant Nos. 32372623 and 32072499).
Author information
Authors and Affiliations
Contributions
Fuping Song, Xin Zhang, and Qi Peng designed the experiments. Xin Zhang, Hengjie Wang, Yuhan Chen, and Tinglu Yan performed the experiments. Xin Zhang, Qi Peng, and Fuping Song analyzed the results. Xin Zhang and Qi Peng drafted the manuscript. Fuping Song revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Chen, Y., Yan, T. et al. Cell death dependent on holins LrgAB repressed by a novel ArsR family regulator CdsR. Cell Death Discov. 10, 173 (2024). https://doi.org/10.1038/s41420-024-01942-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-024-01942-3