Abstract
The disruption of circadian rhythms caused by long-term shift work can cause metabolic diseases such as obesity. Early growth response 3 (EGR3) is a member of early growth response (EGR) family, which is involved in several cellular responses, had been reported as a circadian rhythm gene in suprachiasmatic nucleus. In this research, EGR3 was found to be widely expressed in the different tissue of human and mice, and downregulated in adipose tissue of obese subjects and high-fat diet mice. Moreover, EGR3 was found negatively regulated by cortisol. In addition, EGR3 is a key negative modulator of hADSCs and 3T3-L1 adipogenesis via regulating HDAC6, which is a downstream target gene of EGR3 and a negative regulator of adipogenesis and lipogenesis. These findings may explain how circadian rhythm disorder induced by shift works can cause obesity. Our study revealed a potential therapeutic target to alleviate metabolic disorders in shift workers and may provide better health guidance to shift workers.
Similar content being viewed by others
Introduction
Circadian rhythm can maintain orderly and stable life activities by regulating the glucose and lipid metabolism, hormone secretion and other physiological and metabolic pathways [1,2,3]. Disruption of circadian rhythm such as shift work may increase the risk of obesity, diabetes, metabolic syndrome, cardiovascular disease, and thus affect human health [4, 5].
A stable circadian rhythm is extremely important for human health, but due to the development of society and division of labor, shift work has gradually developed into a common working hour system, which is very common in medical, transportation, manufacturing and service sectors. Disrupting of circadian rhythm due to shift work, complicating metabolic syndrome and tumors have been reported frequently and have attracted extensive attention from researchers [6,7,8,9,10]. Suprachiasmatic nucleus (SCN) acts as a pacemaker for central circadian rhythm regulation, and it is necessary for the circadian rhythm of adrenocorticotropic hormone (ACTH) and glucocorticoid (GC) release [11].
The peripheral circadian rhythm biomarker for adipogenesis was not clearly, variety of circadian rhythm genes, such as Bmal1 and PER3 were reported can directly regulate adipogenesis [12, 13]. Early growth response (EGR) family and regulated by melatonin and are involved in several cellular responses, which are known to be related to circadian rhythm [14,15,16,17]. EGR1 is one of the early downstream nuclear targets for changes in the extracellular stimulation [18, 19], which could be rapidly induced by cytokines, hormones, and stress signals [20]. EGR1 could suppress the transcription of adipose triglyceride lipase and adipogenesis [21, 22]. The expression of uncoupling protein 1 (Ucp1) was restrained in the subcutaneous fat of EGR1 mutant mice, and EGR1 could alter beige adipocyte and promote white adipocyte differentiation in the stem cells of mouse [23]. EGR2 is proved to be an early regulators of the white adipogenic program [24], which can stimulate adipogenesis through C/EBPβ-dependent or -independent mechanisms [25]. EGR3 also has a zinc finger-type transcription factors, however, its behavior in the fat metabolism or adipogenesis is still not clear [26,27,28]. Here we proved the expression of EGR3 is decreased in visceral adipose tissues (VATs) of night shift nurses, and the inhibitor of EGR3 of adipogenesis could be reversed by histone deacetylase 6 (HDAC6).
HDAC6 is one of the subtypes of HDAC family, which is preponderantly localized in the cytoplasm [29, 30]. HDACs has E3 ligase activity and deacetylase activity; beside these functions, HDAC6 has a special C-terminal ubiquitin-binding domain, which can directly interact the proteins should be degraded by proteasome [31]. Nowadays, HDAC6 has been proved to play an important role in the formation of lipid droplet and lipid storage in adipocytes. This indicates that HDAC6 could affect the metabolism of whole body and substantiated the HDAC6 is important regulator of adipose tissue [32]. Mice with specific depletion of hdac6 in adipose showed elevated lipid storage and insulin resistant [33, 34]. EGR3 has been reported to bind to the promoter of HDAC6 to promote its expression [35].
In this research, we attempted to find the relationship between EGR3 abnormalities caused by circadian disorders in night shift nurses and related obesity, and tried to provide a suitable solution to alleviate the metabolic abnormalities caused by circadian disorders.
Results
The body weight gain and metabolic dysfunction in the night shift nurses
To study the relationship between shift work and metabolic parameter, 520 nurse volunteers were recruited. We collected basic information such as frequency of shift work and years of shift work through questionnaires and divided into day shift nurse group and night shift nurse group. The body weight and BMI were significantly increased in night shift nurses compared to day shift nurses; however, average sleep time in a week was found shorter than day shift nurses; the metabolic parameter revealed that cholesterol, 8am ACTH and cortisol levels were increased in night shift nurses, but triglycerides, HDL, FBG, FINS, and 8am melatonin were found statistically indifferent (Table 1). Our results implied that shift work-induced circadian disorder related to metabolic disturbances.
EGR3 was significantly decreased in the VATs of night shift nurses
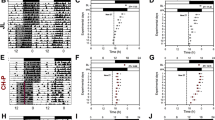
To further investigate the differentially expressed genes (DEGs) in VATs between 6 day shift nurses and 6 night shift nurses, RNA-seq was used. A total 1230 DEGs were identified, of which 339 were upregulated and 891 were downregulated in night shift nurses (Fig. 1A). The heatmap showed the most upregulated 20 genes and the most downregulated 20 genes (Fig. 1B). Through we did not find the change of circadian rhythm pathway in GO analysis, we searched the data base of circadian rhythm genes in public GO (GO:0007623). (http://amigo.geneontology.org/amigo/term/GO:0007623), and intersection with our RNA-seq results, only the expression of EGR3 was found in DEGs of VATs (Fig. 1C). What more, the heatmap showed EGR3 is one of the most downregulated genes (FDR = 9.57E-6) in the results of RNA-seq (Fig. 1B). The pathway analyzed found lipid metabolic and adipogenesis pathways such as lipid synthesis, steroid synthesis, unsaturated acid synthesis and PPAR pathway were enriched, indicating that shift work is a key factor in the development of obesity (Fig. 1D). Next, the expression of EGR3 was further verified that it is downregulated in VATs of 12 night shift nurses compared to 16 day shift nurses (Fig. 1E). Lastly, the STING predicted the EGR3 was closely related to PPARγ, FABP4 and cell death-inducing DFFA-like effector c (CIDEC) (Fig. 1F). We supposed that EGR3 may be the biomarker in the process of obesity caused by rhythm disturbances.
A The volcano plot of RNA-seq. B Heatmap of top 40 representing the DEGs of VATs in day shift nurses and night shift nurses. Red and blue respectively represent upregulation and downregulation expression, n = 6. C Intersection with our RNA-seq with circadian rhythm genes in public GO database (GO007623). D The pathway analysis of RNA-seq. E The PCR results of EGR3 in the VATs of day shift nurses (n = 16) and night shift nurses (n = 12). F The relationship predicted by STING. *p < 0.05 vs day shift nurse group.
The EGR3 was decreased in the SATs and VATs of obese subjects
Although it had been proved that EGR3 as a circadian rhythm gene in central SCN induced by light [36], whether the EGR3 is expressed in adipose tissue is not clear. We found that EGR3 was commonly expressed in the human subcutaneous adipose tissues (SATs), VATs, liver, kidney, muscle small intestine and pancreas (Fig. 2A). Interestingly we found that the protein level of EGR3 expression in the VATs and mRNA expression in SATs and VATs was reduced of obese subjects (Fig. 2B, C). Similarly, we found that EGR3 was highly expressed in mouse brain tissue and commonly expressed in mouse lung, liver, heart, small intestine, kidney, muscle, iWAT, eWAT and brown adipose tissue (BAT) (Fig. 2D). The EGR3 protein expression in iWAT and mRNA expression of iWAT and eWAT was found downregulated in high-fat fed mice (Fig. 2E, F). Our results support that EGR3 may has a physiological role in the function of the peripheral circadian rhythm system and EGR3 may be a potential suppressor of obesity.
The expression of EGR3 mRNA was analyzed in different tissues of human (A) and EGR3 protein was downregulated in VATs of human (B). C The expression of EGR3 mRNA was lower in SATs and VATs of obese people (BMI ≥ 28.0 kg·m2) than that in normal weight subjects (BMI 18.5–23.9 kg m2). D The expression of EGR3 mRNA was analyzed in different tissues of mice. E The expression of EGR3 protein of iWAT and mRNA (F) of iWAT and eWAT was lower in high-fat diet mice. *p < 0.05, **p < 0.01, ***p < 0.001 vs SATs or iWAT.
EGR3 is a key suppressor of adipogenesis in vitro
To explore whether EGR3 can regulate adipogenesis, firstly, the EGR3 mRNA and protein expression were tested during the hADSCs adipogenesis, the results showed that EGR3 was decreased in the early stage of adipogenesis and there was a gradual recovery in middle and later stages of adipogenesis (Fig. 3A, B). Moreover, Oil Red O staining and FFA content in media results indicated that silencing of EGR3 increased hADSCs adipogenesis, however, overexpression of EGR3 decreased hADSCs adipogenesis (Fig. 3C, D). Meanwhile, silencing or overexpression of EGR3 resulted in increasing or decreased markers of adipogenesis, such as, PPARγ, C/EBPα and FABP4, respectively, further demonstrating the adipogenic inhibitory capacity of EGR3 during adipogenesis (Fig. 3D, E). Similar results were verified in 3T3-L1 adipogenesis, where the mRNA and protein expression were only downregulated in early stage of adipogenesis (Fig. 3F, G), and downregulation or upregulation of EGR3 enhanced or inhibited PPARγ, C/EBPα and FABP4 expression and Oil Red O staining in 3T3-L1 adipogenesis (Fig. 3H, I), and elevated FFA or attenuated content, respectively (Fig. 3J).
A PCR and Western blot (B) confirmed that EGR3 was downregulated in the early stage of hADSCs adipogenesis. C Overexpression and silencing of EGR3 affected the expression of related genes for hADSCs adipogenesis on day 6 (PPARγ/CEBPα/FABP4). D Overexpression and silencing of EGR3 affected the early differentiation of hADSCs (Day 6, Oil Red O) and the content of FFA (E). F PCR and Western blot (G) confirmed that EGR3 was downregulated in the early stage of 3T3-L1 adipogenesis. H Overexpression and silencing of EGR3 affected the expression of related genes for 3T3-L1 adipogenesis on day 6 (PPARγ/CEBPα/FABP4). I Overexpression and silencing of EGR3 affected the early adipogenesis of 3T3-L1 (Day 6, Oil Red O) and the content of FFA (J). *p < 0.05 vs LV-shRNA-h/mNC, #p < 0.05 vs LV- h/mNC.
The rhythm of EGR3 is negatively related to cortisol
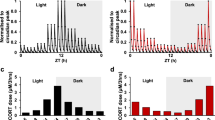
The sleep patterns of day shift nurses and night shift nurses showed obvious difference (Fig. 4A). As a key hormone of the central rhythmic system, cortisol regulates the peripheral rhythmic system, and cortisol secretion is highly rhythmic. Our results revealed that cortisol secretion rhythmicity was not affected in night shift nurses, but cortisol levels were commonly found higher in night shift nurses than day shift nurses at three time points, even cortisol levels still cannot be descended to the lowest levels at 4 pm after night shift nurses sleep for 7 hours at 9 am to 4 pm (Fig. 4B). In addition, we further analyzed the EGR3 mRNA expression in peripheral blood of night shift nurses and day shift nurses. Our results demonstrated that EGR3 expression is downregulated in night shift nurses compared to day shift nurses in three-time points (Fig. 4C).
A Sleep patterns and diagrams of day shift nurses and night shift nurses. B The circadian rhythm of cortisol in the nurses. C The expression of EGR3 in the peripheral blood of day shift nurse and night shift nurse. D Continuous dexamethasone intervention downregulates EGR3 in hADSCs. E Continuous dexamethasone intervention downregulates EGR3 in 3T3-L1. F PCR confirms that dexamethasone continuously downregulated EGR3 expression in continuous group and a 4-h interval (disorder group) simulating a night-shift compare to 12-h interval (interval group) simulating a day-shift in hADSCs and mature adipocytes induced by hADSCs (G). H Western blot confirm that dexamethasone is continuously downregulated EGR3 expression, in a 4-h interval simulating a night-shift in hADSCs and mature adipocytes induced by hADSCs (I). J PCR confirmed that dexamethasone was continuously downregulated EGR3 expression, in a 4-h interval simulating a night-shift in 3T3-L1 and mature adipocytes induced by 3T3-L1 (K). L Western blot confirmed that dexamethasone continuously downregulated EGR3 expression, in a 4-h interval simulating a night-shift in in 3T3-L1 and mature adipocytes induced by 3T3-L1 (M). *p < 0.05.
To further investigate the potential regulation relationship between EGR3 and cortisol in vitro, hADSCs and 3T3-L1 were cultured in 1 μM dexamethasone for 0 h to 72 h, respectively. The EGR3 expression was gradually downregulated during adipogenesis, implying that cortisol negatively regulates EGR3 in vitro (Fig. 4D, E). To explore whether shift work mediated-cortisol dysfunction induced EGR3 down-expression, the hADSCs and 3T3-L1 were culture in 1 μM dexamethasone for 48 h every 12 h interval (interval group) to mimic the normal rhythm, interval dexamethasone cultured for 48 h in every 4 h interval (disorder group) to mimic night shift nurses, and persistent dexamethasone (continuous group) was added to mimic Cushing’s syndrome. Our results showed that the hADSCs treated with 4 h interval dexamethasone could inhibit the mRNA and protein expression of EGR3 and continuous dexamethasone-treated cells more significantly inhibited EGR3 expression than 12 h interval dexamethasone-treated hADSCs and mature adipocytes induced by hADSCs (Fig. 4F–I). Furthermore, in 3T3-L1 and mature adipocytes induced by 3T3-L1, the results also displayed disorder dexamethasone drastically downregulated EGR3 expression compared to interval 12 h dexamethasone intervene (Fig. 4J–M). Our results may explain that adipogenesis and obesity were caused by disorder rhythm secretion of cortisol via negatively regulated EGR3.
EGR3 negatively regulated adipogenesis dependent on HDAC6
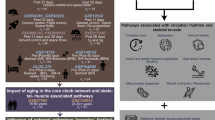
To assess the mechanism of how EGR3 regulates adipogenesis, RNA-seq was used to explore. The DEGs in 3T3-L1 were transfected using EGR3 (FDR = 7.26E-6) overexpression plasmid. There were 324 upregulated and 484 downregulated DEGs in overexpressed EGR3 compared to NC group (Fig. 5A), among them, the heatmap also showed the FABP4 (FDR = 9.73E-14) and PPARγ (FDR = 1.21E-6) were decreased while HDAC6 (FDR = 5.1E-4) was increased in the 3T3-L1, when transfected with LV-EGR3 (Fig. 5B). The pathway analysis showed that DEGs were enriched in fatty acid metabolism and PPAR pathway (Fig. 5C). The western blot results also proved that FABP4, PPARγ and CIDEC were decreased and HDAC6 was increased when EGR3 was overexpressed and increased when EGR3 was silenced (Fig. 6A, B). HDAC6, a critical negative regulatory factor of adipogenesis and lipogenesis, was reported as a target gene of EGR3 through targeting its promoter [32]. In rescue experiments, HDAC6 can partially reverse the adipogenesis and lipogenesis induced by silencing of EGR3 in Oil Red O staining and FFA concentration (Fig. 6C, D). Meanwhile, the expression of FABP4, PPARγ and CIDEC could also be partially reversed in the co-transfected LV-shRNA-mEGR3 and LV-HDAC6 overexpression (Fig. 6E, F). These results revealed the inhibitory effect of EGR3 on adipogenesis and lipogenesis dependent on HDAC6. A proposed biological model showing how the shift work mediated cortisol secretion dysfunction and induced EGR3-HDAC6 downregulation promoting adipogenesis is summarized in Fig. 7.
A Western blot and PCR (B) assays verified the expression of PPARγ, CIDEC, FABP4 and HDAC6 after overexpression or silencing of EGR3 in 3T3-L1. C The rescue experiment using Oil Red O staining assay and FFA content (D) were used to detect the differentiation phenotype after 3T3-L1 was induced for 6 days. E The rescue experiment using western blot and PCR (F) assay verified the changes of lipogenesis and adipogenesis molecules (PPARγ, CIDEC, FABP4) after overexpression of HDAC6 and silencing of EGR3. *p < 0.05.
Discussion
Shift work is a production system that ensures the stable operation of the medical, industrial, transport and many other sectors in modern society. However, the disruption of circadian rhythms caused by shift work should be noticed. To avoid diseases caused by circadian rhythm disorders, the clock gene is receiving more attention from researchers [37, 38]. EGR3, a zinc finger transcription factor, has been proved to be induced by light restricted to the ventral SCN [36, 39]. As the member of EGR family, EGR1 and EGR2 are known to be the critical regulator of peripheral circadian rhythm in human and other animals [40, 41]. Although the rhythm of EGR3 in peripheral tissues had few reports, we speculated that EGR3 might be closely related to peripheral circadian rhythms. In our study, we found EGR3 is universally expressed in most of organisms and for the first time proved that the expression of EGR3 in the VATs is only rhythmically variable gene by transcriptome sequence, indicating that the EGR3 cloud function as peripheral clock gene in peripheral tissues for the universal expression. This implies that the EGR3 could be the critical circadian rhythms regulator for its role in modifying clock gene expression in VATs.
As a critical hormone of neuroendocrine system, cortisol secretion is highly rhythmic, which is highest in the morning and lowest late at night. The circadian rhythm cortisol release was regulated by SCN-hypothalamo-pituitary-adrenal axis (HPA) [42]. Cortisol can promote obesity by influencing leptin sensitivity and insulin resistance, and the abnormally high cortisol in plasma cause abnormal obesity and Cushing’s syndrome [43].
As the human glucocorticoid receptor isoform alpha (GR) contains 2 structural domains of zinc finger, we presumed that the expression of EGR3 could closely related to SCN-ACTH controlled cortisol. Leclerc et al. reported that cortisol inhibits EGR3 in osteoblast [44,45,46]. We primarily explored whether cortisol can affect the expression and rhythms of EGR3 by detecting the plasma in 520 nurses and proved the night shift nurses secreted more cortisol than the day shift nurses, while the expression of EGR3 in the night shift nurses decreased, which help to predicting obesity risk as biomarker. This proved that the cortisol in plasma was negatively related with the expression of EGR3 in peripheral blood.
As some clock genes such as Bmal1 and PER3 can directly regulate lipid adipogenesis [47,48,49,50], there is evidence that the expression of EGR3 is reduced in high-fat fed mice, implying that the EGR3 is negatively related with adipogenesis [51]. EGR3 is the only rhythmically variable gene in VATs between the shift nurses, and so, we conclude that EGR3 may be responsible for causing obesity induced by shift work. To confirm, we transfected EGR3 shRNA or overexpression plasmid to hADSCs and 3T3-L1, and found that EGR3 could negatively regulate adipogenesis, which partially explained cortisol can cause abnormal obesity. Then we further proved hADSCs and 3T3-L1 with 12 hours of alternating dexamethasone treatment expressed most EGR3 as simulating normal rhythm group, while the cells treated with continuous and disorder dexamethasone group presented the least EGR3. These results explain that normal people with rhythm cortisol have low risk of obesity, while continuous and disorder-high cortisol in plasma causes abnormal obesity and decreased EGR3.
EGR3 can negatively regulate adipogenesis through fatty acid metabolism and PPAR pathway, which we confirmed by RNA-Seq, and this function was found cortisol-independent, however, the mechanism of EGR3 regulation of adipogenesis is not clear. To clarify the possibility of the mechanism, bioinformatics tools were used to predict possible pathways and targets for EGR3. Some key transcription factors in adipogenesis and fat metabolism such as PPARγ and FABP4 are closely related to EGR3, and CIDEC, and a recently reported target of HDAC6 was also found to be regulated by EGR3 [32]. As HDAC6 is predominantly localized to the cytoplasm and loss of HDAC6 leads to significant age-dependent ectopic fat accumulation, and there are reports that CIDEC is a specific downstream target of HDAC6 [32], CIDEC contains a PPRE in the region of promoter and can be directly active by PPARγ, which played an important role in the regulation of lipid accumulation in adipocytes [52]. Expression of CIDEC is the most efficacious indicator of strong PPARγ activation [53,54,55]. HDAC6 could abolish the acetylation of CIDEC at K56 through PCAF, which could decrease the stability of CIDEC and elevate lipid droplet fusion and growth [32]. In our study, after overexpression of EGR3 by LV-mEGR3 in 3T3-L1 cells, we found the expression of EGR3 was positively related to HDAC6, and improving the expression of HDAC6 could alleviate the adipogenesis of LV-shRNA-EGR3. These results prove that the adipogenesis inhibitor of EGR3 is positively dependent on HDAC6, and the restorage of HDAC6 could reverse the inhibitory effect of EGR3 on lipogenesis and adipogenesis, which explains the mechanism of EGR3 inhibition of adipogenesis.
In the present study, firstly we identified that EGR3 can function as a cortisol-dependent peripheral clock gene, and we proved that EGR3 can inhibit adipogenesis in vitro, which can explain the continuous high cortisol in plasma causing abnormal obesity. This can also prove that EGR3 can work as an important regulator of rhythm disturbance leading to the development of obesity, which can inhibit adipogenesis through HDAC6. In general, we conclude EGR3 is a peripheral circadian rhythm biomarker for adipogenesis, which can help relieve metabolic disorders caused by shift work, and EGR3 can be a potential target for the abnormal obesity caused by continuous high cortisol in plasma.
Materials and methods
Volunteer recruitment, and collection of data and tissue samples
520 female nurse volunteers (263 day shift nurses and 257 night shift nurses) from the Third Xiangya Hospital of Central South University and The First Hospital of Changsha were recruited. The basic information was obtained by questionnaires; the physical data such as height, body weight was obtained by anthropometric measurement and BMI was calculated according to kilograms divided by the square of height in meters. The VATs in the omentum majus from day shift nurses (n = 16) and night shift nurses (n = 12) were collected; cholesterol, triglyceride and high-density lipoprotein (HDL) were measured by Department of Clinical Laboratory Center, and fasting blood glucose (FBG), fasting insulin (FINS), ACTH, cortisol and melatonin were tested by Department of Endocrine Experiment Center of The Third Xiangya Hospital of Central South University.
In addition, human tissue samples such as abdominal subcutaneous adipose tissues (SATs), VATs, liver, small intestine, kidney, muscle, and pancreas were collected from 4 unrelated patients who suffered severe trauma. Partial SATs and VATs were collected from four normal weight individuals (BMI:18.5–23.9 kg m2) and 4 obese (BMI ≥ 28 kg m2) patients. All human tissue samples were collected from the Department of General Surgery, The Third Xiangya Hospital of Central South University.
All the tissue samples were immediately preserved after surgical collection at −80 °C until analysis. Our study was approved by the Ethics Committee of The Third Xiangya Hospital of Central South University, Changsha, China, (Approval No: 2021-S212), and The First Hospital of Changsha, Changsha, China, (Approval No: 2021-79), and was performed according to the guidelines of Declaration of Helsinki. All participants provided a written informed consent form.
High-fat diet and sample collection in mice
The animal experiments were performed according to the guidelines of the Ethical Committee for Animal Experiments of Central South University. Briefly, C57BL/6 male mice (4 weeks, n = 12) were purchased and fed on a chow diet for suiting the environments. After 2 weeks, 6 mice were fed with high-fat diet (60% fat, 20% carbohydrates, and 20% proteins) (D12492, Research Diets, New Brunswick, NJ), and another 6 mice were continued with chow diet for 12 weeks. The tissue samples including the inguinal white adipose tissue (iWAT), epididymal white adipose tissue (eWAT), liver, pancreas, etc. were collected and preserved at −80 °C to explore the mRNA expression of mEGR3. All mice samples were collected at 8 h of circadian time.
The isolation and differentiation of hADSCs
The isolation and differentiation of human adipose tissue-derived stromal cells (hADSCs) were conducted as described in our previously published study [12]. Briefly, 10 g of fresh abdominal VATs were washed in and cut into 1 mm × 1 mm size, then incubated in collagenase I solution (Gibco, Life Technology, China) for 1.5 h at 37 °C. The treated sample was cultured in DMEM/F12 (Life Technologies, Carlsbad, CA, USA) and supplemented with 10% fetal bovine serum (FBS; Life Technologies). For hADSCs differentiation, confluent hADSCs were cultured in inducing medium (DMEM/F12 supplemented with 1 μM dexamethasone, 10 μM insulin, 0.5 mM isobutylmethylxanthine (IBMX), and 200 μM indomethacin (all provided by Sigma, St. Louis, MO, USA). The medium was replaced every 2 days.
3T3-L1 cell culture and differentiation
3T3-L1 cells were bought from Wellbiology (Changsha, China) and cultured with high glucose DMEM (Life Technologies, Carlsbad, CA, USA) containing 10% FBS and 100 U/ml penicillin and 100 μg/ml streptomycin. When cells reached confluence, the cells were induced by DMEM containing 1 μM dexamethasone, 10 μM insulin and 0.5 mM IBMX. After 48 h, the medium was changed to DMEM containing 10% FBS and 10 μM insulin for every 48 h.
Oil Red O staining and free fatty acid (FFA) measure
The cells were fixed by 4% paraformaldehyde and washed by PBS three times, the Oil Red O (Sigma, St. Louis, MO, USA) was added to the plates and incubated at room temperature for 10 min and washed by water three times. The images were captured by microscope (Leica DM IL LED Fluo, Germany). FFA in an extracellular medium were tested by fluorescent quantification kit (MAK044, Sigma).
Cell transfection
The mouse and human lentiviral EGR3-overexpressing, shRNA, negative control plasmids (LV-shRNA-mEGR3, LV-mEGR3, LV-shRNA-mNC, LV-mNC, LV-shRNA-hEGR3, LV-hEGR3, LV-shRNA-hNC, LV-hNC) and HDAC6-overexpressing plasmid were constructed by Genechem, Shanghai, China. 3T3-L1 or hADSCs were transfected using lentiviral plasmids according to manual when cells were 70%-80% confluent.
RNA-Seq and data analysis
The RNA-Seq analysis was conducted by Tsingke Biotechnology Co. Ltd (Beijing). To extract the RNA of VATs and 3T3-L1, we utilized the TRizol reagent from Life Technologies (Carlsbad, CA, USA). The data were generated on the Illumina Hiseq 2000/2500 platform and subsequently analyzed using an R package [12]. The raw image data files, obtained from high-throughput sequencing, were processed using CASAVA/Basecall_T7_GPU_1.2.0.26_Centos for Base-Calling and converted into raw sequences. To build a reference genome index, we employed Hisat2 (2.2.1) and used it to align paired-end Clean Reads to the reference genome. The differential expression analysis between sample groups was carried out using DEseq2 (1.26.0) package, which allowed us to identify genes that were differentially expressed between the two biological conditions. Additionally, we performed differential gene enrichment analysis using the clusterProfiler (3.14.3) package. To minimize false positives during the detection of differentially expressed genes, we applied the Benjamini & Hochberg method to adjust the P values (FDR). Finally, we set a threshold of |log2 (foldchange)| > 1 and padj < 0.05 for significant differential expression.
Real-time RT-PCR
Real-time RT-PCR was performed as described in our previously published study [56]. The total RNA of cells or tissues were isolated and reversed to cDNA. The primer sequences used for qPCR were shown in Supplementary Table 1.
Western blot assay
Western blotting was performed as described in our previously published study [56]. The total protein of cells was extracted and separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes with a transfer apparatus (Bio-Rad, Hercules, CA, USA). After block, primary antibodies, including EGR3, PPARγ, HDAC6, FABP4, CIDEC, and GAPDH (Proteintech, Wuhan, China) were treated overnight at 4 °C, and second antibody was used for the next ECL (Advansta, Menlo Park, CA, USA) detected.
Statistical analysis
The clinical data of day shift nurses and night shift nurses were summarized and presented as means ± standard deviation (SD) and analyzed using paired Student’s t test. For the data of relative mRNA expression and free fatty acids, Student’s t test were carried out for the two groups, and the multiple comparisons were carried out by analysis of variance and were further carried out for post hoc analysis. Data were statistically analyzed by SPSS 17.0 software. The statistically significant level was defined as p < 0.05.
Data availability
The data are available from the corresponding author on reasonable request.
References
Miyake T, Inoue Y, Shao X, Seta T, Aoki Y, Nguyen Pham KT, et al. Minimal upstream open reading frame of Per2 mediates phase fitness of the circadian clock to day/night physiological body temperature rhythm. Cell Rep. 2023; 112157. https://doi.org/10.1016/j.celrep.2023.112157
Zhong O, Liao B, Wang J, Liu K, Lei X, Hu L. Effects of sleep disorders and circadian rhythm changes on male reproductive health: a systematic review and meta-analysis. Front Physiol. 2022;13:913369. https://doi.org/10.3389/fphys.2022.913369
Liu C, Tang X, Gong Z, Zeng W, Hou Q, Lu R. Circadian rhythm sleep disorders: genetics, mechanisms, and adverse effects on health. Front Genet. 2022;13:875342. https://doi.org/10.3389/fgene.2022.875342
Chen S, Feng M, Zhang S, Dong Z, Wang Y, Zhang W, et al. Angptl8 mediates food-driven resetting of hepatic circadian clock in mice. Nat Commun. 2019;10:3518. https://doi.org/10.1038/s41467-019-11513-1
Miro C, Docimo A, Barrea L, Verde L, Cernea S, Sojat AS, et al. Time” for obesity-related cancer: the role of the circadian rhythm in cancer pathogenesis and treatment. Semin Cancer Biol. 2023;91:99–109. https://doi.org/10.1016/j.semcancer.2023.03.003
Xu J, Ni S, Wang Y, Yan M, Yang X, Ge H, et al. Shift work and nonalcoholic fatty liver disease incidence among Chinese rail workers: a 4-year longitudinal cohort study. Int Arch Occup Environ Health. 2023;96:179–90. https://doi.org/10.1007/s00420-022-01913-0
Wang D, Ruan W, Chen Z, Peng Y, Li W. Shift work and risk of cardiovascular disease morbidity and mortality: a dose-response meta-analysis of cohort studies. Eur J Prev Cardiol. 2018;25:1293–302. https://doi.org/10.1177/2047487318783892
Park JS, Jeong Y, Jung J, Ryu JJ, Lim HK, Jung SK, et al. Shift work sleep disorder is closely associated with an increased risk for periodontal disease. J Clin Periodontol. 2021;48:1066–75. https://doi.org/10.1111/jcpe.13508
Kim K, Lee YJ, Kwon SC, Min YS, Lee HK, Baek G, et al. Correlation between shift work and non-alcoholic fatty liver disease among male workers in the steel manufacturing company of Korea: a cross-sectional study. Ann Occup Environ Med. 2022;34:e33. https://doi.org/10.35371/aoem.2022.34.e33
Cheng M, He H, Wang D, Xu L, Wang B, Ho KM, et al. Shift work and ischaemic heart disease: meta-analysis and dose-response relationship. Occup Med. 2019;69:182–8. https://doi.org/10.1093/occmed/kqz020
Lilley TR, Wotus C, Taylor D, Lee JM, de la Iglesia HO. Circadian regulation of cortisol release in behaviorally split golden hamsters. Endocrinology. 2012;153:732–8. https://doi.org/10.1210/en.2011-1624
Wan X, Zhu L, Zhao L, Peng L, Xiong J, Yang W, et al. hPER3 promotes adipogenesis via hHSP90AA1-mediated inhibition of Notch1 pathway. Cell Death Dis. 2021;12:301. https://doi.org/10.1038/s41419-021-03584-0
Onishi Y, Kawano Y. Rhythmic binding of Topoisomerase I impacts on the transcription of Bmal1 and circadian period. Nucleic Acids Res. 2012;40:9482–92. https://doi.org/10.1093/nar/gks779
Man PS, Evans T, Carter DA. Rhythmic expression of an egr-1 transgene in rats distinguishes two populations of photoreceptor cells in the retinal outer nuclear layer. Mol Vis. 2008;14:1176–86
Abizaid A, Mezei G, Horvath TL. Estradiol enhances light-induced expression of transcription factors in the SCN. Brain Res. 2004;1010:35–44. https://doi.org/10.1016/j.brainres.2004.01.089
Tinarelli F, Garcia-Garcia C, Nicassio F, Tucci V. Parent-of-origin genetic background affects the transcriptional levels of circadian and neuronal plasticity genes following sleep loss. Philos Trans R Soc Lond B Biol Sci. 2014;369:20120471 https://doi.org/10.1098/rstb.2012.0471
He K, Zhang J, Liu J, Cui Y, Liu LG, Ye S, et al. Functional genomics study of protein inhibitor of activated STAT1 in mouse hippocampal neuronal cells revealed by RNA sequencing. Aging. 2021;13:9011–27. https://doi.org/10.18632/aging.202749
Cheong MW, Kuo LH, Cheng YN, Tsai PJ, Ho LC, Tai HC, et al. Loss of Egr-1 sensitizes pancreatic beta-cells to palmitate-induced ER stress and apoptosis. J Mol Med. 2015;93:807–18. https://doi.org/10.1007/s00109-015-1272-4
Kaufmann K, Bach K, Thiel G. The extracellular signal-regulated protein kinases Erk1/Erk2 stimulate expression and biological activity of the transcriptional regulator Egr-1. Biol Chem. 2001;382:1077–81. https://doi.org/10.1515/BC.2001.135
Bongartz H, Seiss EA, Bock J, Schaper F. Glucocorticoids attenuate interleukin-6-induced c-Fos and Egr1 expression and impair neuritogenesis in PC12 cells. J Neurochem. 2021;157:532–49. https://doi.org/10.1111/jnc.15305
Chen W, Jiang Y, Han J, Hu J, He T, Yan T, et al. Atgl deficiency induces podocyte apoptosis and leads to glomerular filtration barrier damage. FEBS J. 2017;284:1070–81. https://doi.org/10.1111/febs.14038
Boyle KB, Hadaschik D, Virtue S, Cawthorn WP, Ridley SH, O’Rahilly S, et al. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. Cell Death Differ. 2009;16:782–9. https://doi.org/10.1038/cdd.2009.11
Bleher M, Meshko B, Cacciapuoti I, Gergondey R, Kovacs Y, Duprez D, et al. Egr1 loss-of-function promotes beige adipocyte differentiation and activation specifically in inguinal subcutaneous white adipose tissue. Sci Rep. 2020;10:15842. https://doi.org/10.1038/s41598-020-72698-w
Jimenez-Preitner M, Berney X, Thorens B. Plac8 is required for white adipocyte differentiation in vitro and cell number control in vivo. PLoS One. 2012;7:e48767 https://doi.org/10.1371/journal.pone.0048767
Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005;1:93–106. https://doi.org/10.1016/j.cmet.2004.12.009
Yamagata K, Kaufmann WE, Lanahan A, Papapavlou M, Barnes CA, Andreasson KI, et al. Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr1/zif268. Learn Mem. 1994;1:140–52.
Ogura Y, Sato S, Kurosaka M, Kotani T, Fujiya H, Funabashi T. Age-related decrease in muscle satellite cells is accompanied with diminished expression of early growth response 3 in mice. Mol Biol Rep. 2020;47:977–86. https://doi.org/10.1007/s11033-019-05189-5
Mookerjee-Basu J, Hooper R, Gross S, Schultz B, Go CK, Samakai E, et al. Suppression of Ca(2+) signals by EGR4 controls Th1 differentiation and anti-cancer immunity in vivo. EMBO Rep. 2020;21:e48904. https://doi.org/10.15252/embr.201948904
Zhou T, Jia D, Han J, Xu C, You X, Ge X. HDAC6 inhibition alleviates acute pulmonary embolism: a possible future therapeutic option. Folia Histochem Cytobiol. 2023. https://doi.org/10.5603/FHC.a2023.0006
Wang L, Sanchez J, Hess D, Matthias P. Immunoprecipitation of HDAC6 and interacting proteins. Methods Mol Biol. 2023;2589:493–508. https://doi.org/10.1007/978-1-0716-2788-4_32
Yan Y, Wang H, Hu M, Jiang L, Wang Y, Liu P, et al. HDAC6 suppresses age-dependent ectopic fat accumulation by maintaining the proteostasis of PLIN2 in drosophila. Dev Cell. 2017;43:99–111.e115. https://doi.org/10.1016/j.devcel.2017.09.001
Qian H, Chen Y, Nian Z, Su L, Yu H, Chen FJ, et al. HDAC6-mediated acetylation of lipid droplet-binding protein CIDEC regulates fat-induced lipid storage. J Clin Investig. 2017;127:1353–69. https://doi.org/10.1172/JCI85963
Lundh M, Petersen PS, Isidor MS, Kazoka-Sorensen DN, Plucinska K, Shamsi F, et al. Afadin is a scaffold protein repressing insulin action via HDAC6 in adipose tissue. EMBO Rep. 2019;20:e48216. https://doi.org/10.15252/embr.201948216
Kumar A, Datta M. H19 inhibition increases HDAC6 and regulates IRS1 levels and insulin signaling in the skeletal muscle during diabetes. Mol Med. 2022;28:81 https://doi.org/10.1186/s10020-022-00507-3
Kwon Y, Kim M, Kim Y, Jeong MS, Jung HS, Jeoung D. EGR3-HDAC6-IL-27 axis mediates allergic inflammation and is necessary for tumorigenic potential of cancer cells enhanced by allergic inflammation-promoted cellular interactions. Front Immunol. 2021;12:680441. https://doi.org/10.3389/fimmu.2021.680441
Morris ME, Viswanathan N, Kuhlman S, Davis FC, Weitz CJ. A screen for genes induced in the suprachiasmatic nucleus by light. Science. 1998;279:1544–7. https://doi.org/10.1126/science.279.5356.1544
Laermans J, Broers C, Beckers K, Vancleef L, Steensels S, Thijs T, et al. Shifting the circadian rhythm of feeding in mice induces gastrointestinal, metabolic and immune alterations which are influenced by ghrelin and the core clock gene Bmal1. PLoS One. 2014;9:e110176 https://doi.org/10.1371/journal.pone.0110176
Hou T, Su W, Guo Z, Gong MC. A novel diabetic mouse model for real-time monitoring of clock gene oscillation and blood pressure circadian rhythm. J Biol Rhythms. 2019;34:51–68. https://doi.org/10.1177/0748730418803719
Porterfield VM, Mintz EM. Temporal patterns of light-induced immediate-early gene expression in the suprachiasmatic nucleus. Neurosci Lett. 2009;463:70–3. https://doi.org/10.1016/j.neulet.2009.07.066
Gast H, Muller A, Lopez M, Meier D, Huber R, Dechent F, et al. CD40 activation induces NREM sleep and modulates genes associated with sleep homeostasis. Brain Behav Immun. 2013;27:133–44. https://doi.org/10.1016/j.bbi.2012.10.004
Torres-Farfan C, Mendez N, Abarzua-Catalan L, Vilches N, Valenzuela GJ, Seron-Ferre M. A circadian clock entrained by melatonin is ticking in the rat fetal adrenal. Endocrinology. 2011;152:1891–900. https://doi.org/10.1210/en.2010-1260
Martocchia A, Curto M, Toussan L, Stefanelli M, Falaschi P. Pharmacological strategies against glucocorticoid-mediated brain damage during chronic disorders. Recent Pat CNS Drug Discov. 2011;6:196–204. https://doi.org/10.2174/157488911796958020
Iwayama H, Hirase S, Nomura Y, Ito T, Morita H, Otake K, et al. Spontaneous adrenocorticotropic hormone (ACTH) normalisation due to tumour regression induced by metyrapone in a patient with ectopic ACTH syndrome: case report and literature review. BMC Endocr Disord. 2018;18:19. https://doi.org/10.1186/s12902-018-0246-2
Leclerc N, Noh T, Cogan J, Samarawickrama DB, Smith E, Frenkel B. Opposing effects of glucocorticoids and Wnt signaling on Krox20 and mineral deposition in osteoblast cultures. J Cell Biochem. 2008;103:1938–51. https://doi.org/10.1002/jcb.21587
Encio IJ, Detera-Wadleigh SD. The genomic structure of the human glucocorticoid receptor. J Biol Chem. 1991;266:7182–8
Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharm Sci. 2013;34:518–30. https://doi.org/10.1016/j.tips.2013.07.003
Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–6. https://doi.org/10.1073/pnas.0502383102
Aggarwal A, Costa MJ, Rivero-Gutierrez B, Ji L, Morgan SL, Feldman BJ. The Circadian clock regulates adipogenesis by a Per3 crosstalk pathway to Klf15. Cell Rep. 2017;21:2367–75. https://doi.org/10.1016/j.celrep.2017.11.004
Sun S, Zhou L, Yu Y, Zhang T, Wang M. Knocking down clock control gene CRY1 decreases adipogenesis via canonical Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2018;506:746–53. https://doi.org/10.1016/j.bbrc.2018.10.134
Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, et al. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26:3453–63. https://doi.org/10.1096/fj.12-205781
Nguyen NM, de Oliveira Andrade F, Jin L, Zhang X, Macon M, Cruz MI, et al. Maternal intake of high n-6 polyunsaturated fatty acid diet during pregnancy causes transgenerational increase in mammary cancer risk in mice. Breast Cancer Res. 2017;19:77. https://doi.org/10.1186/s13058-017-0866-x
Hua L, Wang J, Li M, Sun X, Zhang L, Lei C, et al. An Asp7Gly substitution in PPARG is associated with decreased transcriptional activation activity. PLoS One. 2014;9:e86954 https://doi.org/10.1371/journal.pone.0086954
Kim S, Reed E, Monti S, Schlezinger JJ. A data-driven transcriptional taxonomy of adipogenic chemicals to identify white and brite adipogens. Environ Health Perspect. 2021;129:77006 https://doi.org/10.1289/EHP6886
Shamsi BH, Ma C, Naqvi S, Xiao Y. Effects of pioglitazone mediated activation of PPAR-gamma on CIDEC and obesity-related changes in mice. PLoS One. 2014;9:e106992 https://doi.org/10.1371/journal.pone.0106992
Russell T, Watad A, Bridgewood C, Rowe H, Khan, A, Rao A, et al. IL-17A and TNF modulate normal human spinal entheseal bone and soft tissue mesenchymal stem cell osteogenesis, adipogenesis, and stromal function. Cells. 2021;10. https://doi.org/10.3390/cells10020341
He YF, Hu XM, Khan MA, Yu BY, Sheng YC, Xiao XZ, et al. HSF1 alleviates brain injury by inhibiting NLRP3-induced pyroptosis in a sepsis model. Mediators Inflamm. 2023;2023:2252255. https://doi.org/10.1155/2023/2252255
Acknowledgements
We thank all the sample donors and nurses of the Third Xiangya Hospital of Central South University and The First Hospital of Changsha for their excellent assistance.
Funding
This study was supported in part by grants from the Hunan Province Natural Science Foundation of China (2022JJ70045 and 2022JJ30881).
Author information
Authors and Affiliations
Contributions
All authors were involved in the development of the primary manuscript and have read and approved the final version. Xinxing Wan, Linghao Wang, Keke Zhang, Xiaoying Sun and Xuan Yi performed the experiments, Lin Peng, Zhouqi Wang and Ke Chen analyzed the data and draw the graph. Xinxing Wan, Ke Chen and Md Asaduzzaman Khan. wrote the manuscript. Ke Chen and Lin Peng designed the study, and conceived and supervised the study. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics Statement
Our study was approved by the Ethics Committee of The Third Xiangya Hospital of Central South University, Changsha, China, (Approval No: 2021-S212), and The First Hospital of Changsha, Changsha, China, (Approval No: 2021-79), and was performed according to the guidelines of Declaration of Helsinki. All participants provided written informed consent form.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wan, X., Wang, L., Khan, M.A. et al. Shift work promotes adipogenesis via cortisol-dependent downregulation of EGR3-HDAC6 pathway. Cell Death Discov. 10, 129 (2024). https://doi.org/10.1038/s41420-024-01904-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-024-01904-9