Abstract
NSUN2 is a nuclear RNA methyltransferase which catalyzes 5-methylcytosine (m5C), a posttranscriptional RNA modification. Aberrant m5C modification has been implicated in the development of multiple malignancies. However, its function in pancreatic cancer (PC) needs to be elucidated. Herein, we determined that NSUN2 was overexpressed in PC tissues and related to aggressive clinical features. Silence of NSUN2 by lentivirus weakened the capability of proliferation, migration and invasion of PC cells in vitro and inhibited the growth and metastasis of xenograft tumors in vivo. Contrarily, overexpression of NSUN2 stimulated PC growth and metastasis. Mechanistically, m5C-sequencing (m5C-seq) and RNA-sequencing (RNA-seq) were carried out to identify downstream targets of NSUN2 and results showed that loss of NSUN2 led to decreased m5C modification level concomitant with reduced TIAM2 mRNA expression. Further validation experiments proved that NSUN2 silence accelerated the decay of TIAM2 mRNA in a YBX1-dependent manner. Additionally, NSUN2 exerted its oncogenic function partially through enhancing TIAM2 transcription. More importantly, disruption of the NSUN2/TIAM2 axis repressed the malignant phenotype of PC cells through blocking epithelial-mesenchymal transition (EMT). Collectively, our study highlighted the critical function of NSUN2 in PC and provided novel mechanistic insights into NSUN2/TIAM2 axis as promising therapeutic targets against PC.
Similar content being viewed by others
Introduction
Pancreatic cancer (PC), characterized by highly metastatic courses, is a fatal malignancy of the digestive system. The incidence of PC is stably increasing over the past decades, ranking the 12th most prevalent cancer worldwide [1, 2]. Despite improvements in diagnosis and treatment have been made, the prognosis of PC turns out to be disappointing, with an overall 5-year relative survival rate of 6% and the 7th leading cause of cancer-related mortality [2, 3]. The high incidence and mortality impose a great threat on human health and has become an enormous burden globally. Hence, it is imperative to figure out the molecular mechanisms underlying progression of PC and further identify promising targets for effective treatments.
Recently, RNA modification such as N6-methyladenine (m6A), 5-methylcytosine (m5C) and pseudouridine, has received considerable attention due to their fundamental role in cancer development [4, 5]. These aberrant epigenetic regulations in PC were also widely explored [6,7,8,9]. It has been elucidated that m6A modification of PIK3CB could promote PTEN-deficient PC progression by activating AKT signaling pathway [8]. Guo’s research revealed that m6A demethylase ALKBH5 serves as a tumor-suppressor in PC via activating PER1 in a posttranscriptional manner [9]. m5C, methylation of carbon 5 in cytosine, is another prevalent modification of RNA in eukaryotic cells. Similar with m6A, m5C modification is a dynamic reversible process, which can be catalyzed by the m5C methyltransferases (also called “writers”: NSUN, DNMT and TRDMT family members) and removed by the m5C demethylases (namely “erasers”: TET families) [10,11,12]. Moreover, m5C can be recognized by its binding proteins (“Readers”: ALYREF, YBX1) to affect RNA’s processing, including RNA export and stability [13, 14].
As the key m5C methyltransferase, NSUN2 (NOP2/Sun domain family, member 2) has been demonstrated to be overexpressed in a wide range of malignancies, including cancers of bladder, prostate, kidney, cervix, esophagus, stomach, liver, thyroid and breast [15]. Emerging data suggested that NSUN2 exerts a nonnegligible function in diverse biological process, such as cellular proliferation, differentiation, migration and involves in tumorigenesis in an m5C-dependent manner [16,17,18,19]. As elucidated by Su et al., NSUN2-mediated RNA m5C modification promoted esophageal squamous cell carcinoma (ESCC) progression through upregulating GRB2 by stabilizing its mRNA in an LIN28B-dependent manner [19]. Another study conducted by Lin et al. demonstrated that NSUN2 contributes to gastric cancer development by suppressing p57Kip2 [17]. However, the functional role of NSUN2 in PC is still obscure.
In this study, we identified that elevated expression of NSUN2 was correlated with unfavorable survival outcome in PC patients. Subsequent functional experiments revealed that the overexpression of NSUN2 could facilitate the proliferation, migration and invasion of PC. Mechanistically, TIAM2 was identified as the potential downstream target of NSUN2 using m5C-seq and RNA-seq. Further functional experiments confirmed that decreased TIAM2 could partially reverse the promotion effect of NSUN2 on the malignant phenotypes of PC. Additionally, NSUN2/TIAM2 axis enhanced epithelial-mesenchymal transition (EMT) process. Collectively, our study suggested that NSUN2 may be a promising prognostic marker and therapeutic target of PC.
Results
Differential expression profiles and prognostic value of m5C regulators in PC
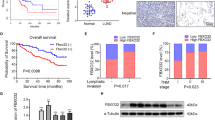
To determine the expression profiles of m5C-related regulators in PC, the sequencing data of 179 PC samples and 171 control normal samples extracted from TCGA and GTEx was analyzed. Results demonstrated that m5C-related regulators were generally differentially expressed in PC tissues compared with normal tissues. Of these, 13 genes (NOP2, NSUN2, NSUN3, NSUN4, NSUN5, DNMT1, DNMT2, DNMT3A, TET1, TET2, TET3, ALYREF and YBX1) were aberrantly upregulated and DNMT3B was downregulated (Fig. 1A). Moreover, NSUN2, NSUN4, DNMT3A and YBX1 were significantly correlated with the overall survival (OS) of PC patients (Fig. 1B, Fig. S1A). Subsequently, the aberrant expression of these four regulators was verified in 9 independent PC cohorts from the GEO database (Fig. 1C). However, only the overexpression of NSUN2 and YBX1 predicted a poor survival status of PC patients (Fig. 1D). Previous studies have demonstrated that overexpression of YBX1 could accelerate PC growth and recognize m5C modification catalyzed by NSUN2, thereby promoting pathogenesis of bladder cancer [20, 21]. However, the role of NSUN2 in PC remains little known. Hence, we selected NSUN2 as our target gene for the subsequent experiment and determined whether NSUN2 exerts an oncogenic role in PC progression.
A TCGA and GTEx databases were used to analyze the mRNA expression of m5C-related genes in PC tissues and normal pancreas tissues. ***P < 0.001. B Kaplan–Meier survival analysis was performed based TCGA database. C Heatmap showed the altered mRNA expression of m5C related genes in GEO datasets. Red indicates the related gene is upregulated and blue indicates the related gene is downregulated; White indicates that the related gene is absent in the dataset. D GSE62452 dataset was used for survival analysis in patients with low or high expression of NSUN2. NS, not significant.
NSUN2 is overexpressed in PC and correlated with an unfavorable prognosis
Next, the protein expression of NSUN2 was further validated by IHC staining on a microarray containing 90 paired PC tissues and adjacent normal tissues. In accordance with the aforementioned data, aberrant overexpression of NSUN2 was observed in PC tissues (Fig. 2A, B). Moreover, correlations between NSUN2 expression and clinical characteristics of patients with PC were explored (Table 1). We observed that elevated NSUN2 expression indicated an advanced TNM stage (Fig. 2C, D) and distant metastasis (Fig. 2E, F) in PC. Consistently, Kaplan–Meier analysis revealed that patients with NSUN2 overexpression were prone to have a shorter OS time than those with low expression (median: 15.60 months vs. 21.78 months, Fig. 2G). However, NSUN2 expression was not identified as an independent risk factor for OS in PC by the multivariate Cox regression analysis in this cohort (Supplementary Table 1, Fig. S1B). Altogether, these results provide further strong evidence that NSUN2 upregulation is an unfavorable prognosticator for PC.
A Representative IHC staining images and B IHC scores of NSUN2 expression in paired PC tissues and adjacent non-tumor tissues. Scale bar = 50 μm. C–F NSUN2 overexpression in PC was associated with C, D advanced TNM stage and E, F metastasis. G Upregulation of NSUN2 was significantly associated with shorter OS in PC patients.
NSUN2 accelerates the proliferation of PC cells both in vitro and in vivo
Then we explored the function of NSUN2 in PC cells. Firstly, NSUN2 protein expression was validated in the human pancreatic ductal epithelial cell line (HPDE6C7) and several PC cell lines (SW1990, PANC-1, CFPAC-I and MIA PaCa-2). With the exception of CFPAC-I cells, other PC cell lines all exhibited higher NSUN2 expression than HPDE6C7 (Fig. S2). Subsequently, we established cell lines with NSUN2 silence or overexpression by lentiviral transduction, and Western blot analysis proved the successful transduction (Fig. 3A, B). Functional experiments revealed that knockdown of NSUN2 substantially retarded PC cells proliferation as evidenced by CCK-8, EdU and colony formation assays (Fig. 3C, E, G), while opposite effects were observed in cells with NSUN2 upregulation (Fig. 3D, F, H).
A, B Western blot confirmed the knockdown and overexpression of NSUN2 by lentiviral constructs in PC cell lines. C, D Cell viability was determined by CCK-8 in cells with stable NSUN2 knockdown or overexpression. E, F EdU staining assays were performed to evaluate cell proliferation. G, H Colony formation assays. I, K Tumor growth and weight were monitored in mice. J, L IHC staining of NSUN2 and Ki-67 expression in tumor sections. Scale bar = 100 μm. Data are presented as the mean ± SD of at least 3 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001.
To further determine the impact of NSUN2 on tumor growth in vivo, we established a subcutaneous xenograft model. It was verified that NSUN2-silenced cells formed smaller tumor nodules than those control cells formed (Fig. 3I), while upregulation of NSUN2 in PC cells significantly stimulated tumor growth as evidenced by an accelerated growth curve and increased tumor weight (Fig. 3K), which was consistent with our previous observation in vitro. Additionally, IHC staining of sections from the xenograft tumors showed a decreased portion of Ki-67-positive cells in NSUN2 silence group (Fig. 3J), but an increased portion in NSUN2 overexpression group (Fig. 3L). Taken together, these findings indicate that NSUN2 could promote the growth capacity of PC.
NSUN2 modulates PC migration and invasion in vitro and metastasis in vivo
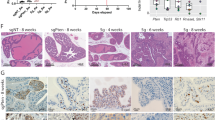
Subsequently, we further explored whether NSUN2 regulates the motility of PC cells via Transwell migration and invasion assays. Results displayed that attenuation of NSUN2 expression markedly impaired the migratory and invasive ability of SW1990 and MIA PaCa-2 cells (Fig. 4A, C), whereas overexpression of NSUN2 was stimulative (Fig. 4B, D). Next, a peritoneal dissemination model and a hematogenous metastasis model were established to validate the role of NSUN2 on metastasis in vivo. Remarkably, extensive intestinal and mesenteric metastases, liver metastases, and ascites formation were observed in the control group, whereas only one mouse in the shNSUN2 group suffered intestinal metastasis (Fig. 4E). Although peritoneal dissemination occurred in all mice, the number of intestinal metastatic nodules formed in the NSUN2 overexpression group were statistically higher than the control vector group (Fig. 4F). Similarly, SW1990/shCtrl cells formed remarkably large and excessive lung and liver metastatic foci than NSUN2-silenced SW1990 cells (Fig. 4G), and more extensive liver metastases were observed in the NSUN2 overexpression group (Fig. 4H). Taken together, these findings suggest that NSUN2 could facilitate migration and invasion of PC cells in vitro and metastasis in vivo.
A, B Migration ability and C, D invasion ability of PC cells with NSUN2 silence or overexpression were evaluated by Transwell assays. Data are shown as the mean ± SD of 3 replicates. *P < 0.05, **P < 0.01, ***P < 0.001. E, F Representative images of tumor peritoneal dissemination and statistical data were indicated. G, H Representative images of hematogenous metastasis model established by injecting PC cells through the tail vein. Scale bar = 100 μm. I, J Altered expression of EMT-associated genes in cells with NSUN2 silence or overexpression. Scale bar = 100 μm.
Considering the crucial role of EMT in cancer metastasis, we further investigated whether NSUN2 expression affected EMT. Interestingly, NSUN2-silenced SW1990 and MIA PaCa-2 cells exhibited a lower expression of N-cadherin, MMP9, MMP2 and vimentin than control cells (Fig. 4I). Conversely, enforced NSUN2 expression had the opposite effects (Fig. 4J). We also observed that silencing NSUN2 resulted in the formation of cobblestone-like PC cells (Fig. S3A), whereas cells with overexpressed NSUN2 exhibited a spindle-like shape (Fig. S3B). Based on these findings, it is plausible that NSUN2 may play a metastasis-prompting role in PC by inducing EMT.
Characterization of mRNA m5C profile by m5C-seq in PC cells
Given that NSUN2 is one of the m5C RNA methyltransferases, we explored whether NSUN2 affected the global RNA m5C modification level of PC cells. As anticipated, NSUN2-silenced groups showed decreased methylation levels, and increased levels were observed in NSUN2-overexpressing cells (Fig. 5A), indicating the m5C methyltransferase activity of NSUN2 in PC cells. Next, m5C-seq assays were performed to investigate the effect of NSUN2 silencing on the m5C profile of PC cells. Results showed that m5C sites were distributed in all regions of the mRNA, and the distribution patterns in two groups were similar (Fig. 5B). Of note, in addition to about half of m5C sites distributed within CDS, the region near the start codon is also an important source of m5C peaks, suggesting that m5C may be involved in translation regulation. These observations accord with previously published reports [14, 22]. Moreover, we also investigated the sequence context of methylated regions using DREME software. “AGSCDGG” (S “Strong” = C/G, D “Not C” = A/G/U) was identified as the preferred motif in PC cells with the lowest E-value (4.1e−197) (Fig. 5C). Although this sequence context was not exactly consistent with the preferred motif previously measured by bisulfite sequencing, it also demonstrated that m5C sites are embedded in CG-rich environments [22, 23].
A Relative m5C levels were assessed in cells with or without NSUN2 silence or overexpression, respectively. B The distribution of m5C sites within distinct mRNA regions. C The preferred sequence context of m5C sites in PC cells. D, E KEGG analysis for genes with downregulated m5C modification after knockdown of NSUN2. The ten most enriched pathways are shown (for full list see Supplementary Table 6).
To get insights into the regulatory role of m5C, we performed KEGG analysis on mRNAs with differentially downregulated m5C sites after NSUN2 silencing and observed that they were mainly enriched in pathways in cancer, MAPK pathways, focal adhesion and other oncogenic pathways (Fig. 5D, E), which suggests that NSUN2 may be involved in cell proliferation and migration in an m5C-dependent manner.
TIAM2 was identified as a target of NSUN2 by m5C-seq and RNA-seq in PC
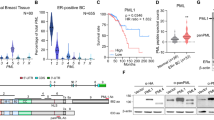
It has been confirmed that enhanced mRNA transcription is positively correlated with NSUN2 mediated-m5C modification and consequently achieved by m5C reader YBX1 through maintaining mRNA stability [21]. Considering the aberrant expression and prognostic role of NSUN2 and YBX1 in PC patients (Fig. 1A–D), we hypothesized that similar regulatory mechanisms may exist in PC. Therefore, we analyzed m5C-seq and RNA-seq data in PC cells to excavate the potential targets of NSUN2 in PC. Results demonstrated that among 2099 annotated genes with 4138 differential hypomethylated m5C sites, 1387 (66.08%) genes were downregulated, and 712 (33.92%) genes were upregulated in mRNA level (Supplementary Table 2). Twelve potential targets (LURAP1L, ZNF221, RASSF2, MPP2, ZNF574, LYSMD2, LRRC39, TIAM2, SMPDL3A, CPQ, KRCC1, TOGARAM2), which were downregulated in mRNA level with top hypomethylated sites, were screened out (Fig. 6A and Table 2). Of these, TIAM2, LURAP1L, ZNF574 and KRCC1 were upregulated in PC and positively correlated with NSUN2 (Fig. 6B, C, Fig. S4A, B), but only TIAM2 had a statistically significant impact on both OS and relapse-free survival (RFS) in PC patients (Fig. S5A, B).
A Venn diagram showed the selection for the downstream target of NSUN2 according to RNA-seq and m5C-seq. B, C TIAM2 expression was positively correlated with NSUN2 expression in PC based on TCGA data. D–G TIAM2 expression upon NSUN2 knockdown or overexpression were determined by D, E qRT-PCR and F, G Western blot. H, I The relative TIAM2 mRNA expression upon NSUN2 knockdown in SW1990 and MIA PaCa-2 cells treated with 5 μg/ml actinomycin D for indicated times. Data are presented as the mean ± SD of 2 replicates. J A positive correlation between TIAM2 and YBX1 mRNA expression in PC based on TCGA data. K, L TIAM2 mRNA(K) and protein expression(L) in SW1990 and MIA PaCa-2 cells transfected with YBX1 shRNA and the corresponding control lentivirus. Data are presented as the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
TIAM2, namely T-cell lymphoma invasion and metastasis 2, is a Rac1-Guanine nucleotide exchange factor (Rac1-GEF), which facilitates the exchange of GDP for GTP, thereby activating Rac1 [24]. It has been demonstrated that TIAM2 could facilitate cancer cells proliferation and migration, thereby facilitating cancer progression [25,26,27]. Although very limited evidence suggested that TIAM2 is correlated with poor prognosis in PC [28], its regulatory mechanism remains unclear. Therefore, we selected TIAM2 as a candidate target of NSUN2 for further investigation. Strikingly altered TIAM2 mRNA (Fig. 6D, E) and protein (Fig. 6F, G) expression upon NSUN2 silencing or overexpression further supported our postulation.
It has been validated that NSUN2 could affect metabolism of mRNA such as degradation and nuclear export [14, 18, 21]. We further investigated whether NSUN2 regulates TIAM2 expression by affecting its mRNA stability. Actinomycin D assay showed that the stability of TIAM2 mRNA was significantly reduced upon NSUN2 silence (Fig. 6H, I). Although there was no significant difference in the stability of TIAM2 mRNA compared to the control group, possibly due to the high level of endogenous NSUN2 expression in SW1990 cells, we still observed enhanced stability of TIAM2 mRNA in PANC-1 cells with NSUN2 overexpression (Fig. S6A, B).
The stability of m5C-modified mRNAs was preferentially positively regulated by the YBX1(m5C reader) [13, 21]. Therefore, we further explored whether YBX1 influences the expression of TIAM2 in PC. Based on the TCGA database, a positive correlation between YBX1 and TIAM2 mRNA expression in PC was identified (Fig. 6J). In line with this data, we observed that the transduction of shRNAs targeting YBX1 led to the reduction of TIAM2 mRNA stability (Fig. S6C, D) and reduced the expression of TIAM2 (Fig. 6K, L). Moreover, YBX1 plasmids were transfected into the control cells and NSUN2 silenced cells, respectively. We observed that overexpression of YBX1 significantly upregulated the expression of TIAM2 in the control cells. However, forced expression of YBX1 in NSUN2 silenced cells only slightly restored the downregulation of TIAM2 induced by NSUN2 knockdown (Fig. S6E, F, G), suggesting that YBX1 may regulate TIAM2 expression through NSUN2-mediated m5C modification.
TIAM2 is responsible for NSUN2-mediated PC progression
To further investigate the impact of TIAM2 on NSUN2-induced cancer progression, we co-transfected siRNA (siTIAM2 or siCtrl) into vector control or NSUN2-overexpressing PC cells. The transfection efficiency was confirmed by Western blot (Fig. 7A). Intriguingly, compared with the vector+siCtrl group, cells in vector+siTIAM2 group exhibited lower proliferation capacity and formed less and smaller colonies (Fig. 7B–G). Additionally, impaired migratory and invasive abilities were observed in TIAM2-silencing cells (Fig. 7H–K). Most notably, TIAM2 downregulation largely abolished the promoting effects of NSUN2 overexpression on abovementioned malignant phenotypes (Fig. 7B–K). Subsequently, we performed KEGG analysis and GSEA based on TCGA dataset. Results indicated that proliferation- and metastasis-related pathways such as EMT were significantly enriched in patients with high TIAM2 expression (Fig. 7L, Fig. S7). Further study displayed that TIAM2 silencing substantially decreased the expression of EMT-related molecules (N-cadherin, MMP9, MMP2, Vimentin) in PC cells, and partly reversed their expression in the presence of overexpressed-NSUN2 (Fig. 7M). These data provide compelling evidence that upregulation of TIAM2 by NSUN2-mediated m5C modification is significant for promoting PC progression.
A Western blot was conducted to examine TIAM2 expression in the Vector+siCtrl, NSUN2+siCtrl, Vector+siTIAM2, NSUN2+siTIAM2 cells. B Cell viability of indicated cells was determined by CCK-8 assays. C–E EdU staining assays for indicated cells. F, G The ability of colonies formation for indicated cells. H-I The migration ability and J, K invasion ability of indicated cells was evaluated by Transwell assays. L GSEA indicated that EMT pathway was enriched in the patients with high TIAM2 expression. M TIAM2 silence could suppress EMT-related genes expression and rescue these genes expression upregulated by NSUN2. Scale bar = 100 μm. Data are shown as the mean ± SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Clinical relevance of the NSUN2/TIAM2 axis in PC patients
Eventually, we performed IHC assays to determine the expression of TIAM2 on PC tissues. IHC staining confirmed that TIAM2 can be mainly detected in the cell nuclei and also in the cytoplasm of PC cells. Additionally, a positive correlation between TIAM2 and NSUN2 was observed in PC tissues (Fig. 8A). Moreover, survival analysis based on TCGA dataset demonstrated that patients with high NSUN2 and high TIAM2 mRNA expression showed decreased overall survival than others (Fig. 8B, Fig. S8). Taken together, these results further underscore the notion that NSUN2 promotes PC tumorigenesis partially by enhancing TIAM2 expression and consequently activating EMT process. The suggested regulatory mechanism is illustrated in Fig. 8C.
A IHC staining of NSUN2 and TIAM2 in PC tissues and the positive correlation between them. (n = 31). Scale bar =100 μm. B Patients with high NSUN2 and high TIAM2 expression had poor overall survival. C The schematic figure shows that NSUN2 upregulates m5C level of TIAM2 mRNA, and may sustain the stability of TIAM2 mRNA; TIAM2 overexpression could promote cell proliferation, and activate EMT process in PC.
Discussion
Pancreatic cancer is a leading cause of cancer death worldwide. Despite advances in treatment, the prognosis of PC remains deadly poor. Thus, it’s urgently needed to investigate the underlying mechanisms related to PC progression and search for novel therapeutic strategies. m5C methylation is an abundant modification on eukaryotic RNA and emerging studies have demonstrated the m5C dysregulation are implicated in human diseases including cancers [29,30,31,32]. However, thus far, studies on m5C modification and its functions in PC are limited.
In this study, we determined that NSUN2 was overexpressed in PC tissues compared with normal tissues and its elevated expression indicated an unfavorable prognosis, which accords with the previous work [15, 23]. Functional experiments showed that NSUN2 could accelerate PC proliferation and promote migration and invasion in vitro, thereby enhancing tumor growth and metastasis in vivo. Consistent with our findings, it has been reported that NSUN2 is involved in the development of gastric cancer, esophageal squamous cell carcinoma (ESCC), gallbladder carcinoma and other human cancers [16, 17, 19, 33,34,35]. Lin et al.’s research showed that NSUN2 could be modulated by SUMO-2/3 and promote gastric cancer cell proliferation and motility [17]. Besides, NSUN2 has been proven to enhance the initiation and progression of ESCC by upregulating GRB2 expression [19]. Our findings further support the oncogenic role of NSUN2 in malignancies.
To further investigate the regulatory role of NSUN2 during PC progression, we conducted m5C quantification assays and found that NSUN2 silence in PC cells resulted in the significantly decreased m5C level. m5C-seq revealed the distribution patterns of m5C along mRNAs in PC. Complementary to the prior studies in which bisulfite sequencing was utilized, the most common motif “AGSCDGG” was identified by m5C-antibody based m5C-seq. KEGG analysis showed that differentially methylated sites-harboring genes were mainly enriched in several oncogenic pathways, which are closely related with tumor growth and metastasis [36, 37]. Subsequently, we identified TIAM2 as a downstream target of NSUN2 by sequencing and bioinformatics analysis. Accumulating data showed that TIAM2 was involved in some tumor-associated biological processes, including immune response, cell proliferation, apoptosis, motility [38,39,40,41,42,43]. Recent studies showed that TIAM2 was frequently upregulated in various human malignancies such as liver cancer, lung cancer [25, 27, 44]. Importantly, Jiang and coworkers found that TIAM2 could exert a tumor-promoting effect in PC [28]. Likewise, we found a positive correlation between TIAM2 and NSUN2, and TIAM2 silence could partially reverse the oncogenic effects of NSUN2 overexpression. Moreover, TIAM2 may mediate NSUN2-induced metastasis via regulating EMT-related proteins expression. These findings revealed a novel regulatory mechanism that NSUN2 exerts its oncogenic effect through, at least in part, upregulating TIAM2 expression in PC.
Previous studies demonstrated that m5C modification plays an important role in regulating mRNA metabolic process, including pre-mRNA splicing, mRNA export, stability and translation [29]. Mechanistic studies have determined that m5C methylation written by NSUN2 can be recognized by “m5C reader” YBX1 or ALYREF, thereby impeding degradation or enhancing nuclear export of mRNA, respectively [13, 14, 18, 21]. Chen et al. verified that YBX1 can bind the m5C methylation site in the HDGF 3’ untranslated region catalyzed by NSUN2 and stabilize HDGF mRNA to drive bladder cancer progression [21]. It was also reported that YBX1 could enhance mRNA stabilization by recognizing m5C-modified sites through π-π interactions to facilitate the maternal-to-zygotic transition of zebrafish [13]. Herein, we found that NSUN2 and YBX1 coregulate the stability of TIAM2 mRNA. After NUN2 silencing, the ability of YBX1 to regulate TIAM2 expression was greatly weakened, suggesting that YBX1 might be implicated in NSUN2-mediated overexpression of TIAM2. However, a clearer detailed mechanism needs to be further studied. Besides, very recently, Su and coworkers identified LIN28B as a novel m5C reader to inhibit m5C-modified GRB2 mRNA decay in ESCC [19]. Therefore, whether other readers or m5C-related enzymes contribute to the TIAM2 expression requires further investigation.
In summary, our study revealed that NSUN2 is elevated in PC and predicts an unfavorable prognosis. We reported for the first time that NSUN2 facilitates PC progression by upregulating TIAM2 expression in an m5C-dependent manner. NSUN2 may represent a promising therapeutic target for PC treatment.
Materials and methods
Bioinformatics analysis
Data retrieved from The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx) and Gene Expression Omnibus (GEO) dataset was used for analysis. Gene set enrichment analysis (GSEA)was conducted with normalized data using the R language tools. Two gene sets were utilized for GSEA (HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION,GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP).
Human tissue specimens and cell lines
A tissue microarray (TMA) containing 90 paired pancreatic cancer and adjacent normal tissues was provided by Shanghai Outdo Biotech Co., Ltd. and another TMA containing 32 pancreatic cancer tissues was purchased from Expect laboratory (Tsingtao, China). Human pancreatic ductal epithelial cell line HPDE6C7 and PC cell lines MIA PaCa-2 and CFPAC-1 were preserved by our laboratory. SW1990, PANC-1 were kindly provided by Dr. Qiankun Luo at Zhengzhou University. HPDE6C7, SW1990, PANC-1and CFPAC-1 were cultivated in the DMEM (VivaCell, Shanghai, China) with 10% fetal bovine serum (FBS, VivaCell, Shanghai, China), whereas MIA PaCa-2 cells were maintained in DMEM with 10% FBS and 2.5% super horse serum (Sangon Biotech, Shanghai, China). All cells were cultured in a humidified 5% CO2 atmosphere at 37 °C.
Lentiviral transduction and cell transfection
Lentivirus constructs were purchased from HanBio (Shanghai, China). 2 × 105 cells were seeded into a six-well plate and then transfected with NSUN2 overexpression (namely NSUN2) or knockdown (shNSUN2, shYBX1) recombinant lentivirus after 24 h. Subsequently, PC cells were maintained in medium containing 3ug/ml puromycin after 72 h to select stably transfected cells for further studies. For TIAM2 silence experiments, the lentivirus-infected PC cells were transfected with small-interfering RNAs (siRNAs, RiboBio, Guangzhou, China) targeting TIAM2 (si-TIAM2) or negative control RNAs (si-Ctrl) following the manufacturer’s instructions. Nucleotide sequences for the siRNAs and shRNAs were listed in Supplementary Table 3. PCMV-YBX1WT plasmids were kind gifts from Prof. Yun-Gui Yang (Beijing Institute of Genomics, Beijing, China). Transient transfection was performed using EZ Trans cell transfection reagent (Life-iLab, Shanghai, China) according to the manufacturer’s instructions. After 48 h of culture, cells were harvested for western blotting.
Western blot
Cells were lysed in RIPA buffer containing PMSF (Solarbio, Beijing, China). After quality control, protein samples were separated by 8% or 10% Bis-Tris SDS-PAGE gel and then transferred to the polyvinylidene fluoride membranes. Membranes were incubated with primary antibodies at 4 °C overnight, followed by incubation with the corresponding horseradish peroxidase-conjugated or fluorescent-labeled secondary antibody. Signals were detected by the enhanced chemiluminescence (ECL) (SH-Focus 523, Hangzhou, China) or fluorescence imaging system (Odyssey, Nebraska, USA). anti-GAPDH or anti-β-actin antibody was used as the internal control. The information of antibodies and all original western blot images were presented in Supplementary Table 4.
Quantitative real-time PCR (qRT-PCR)
Total RNA isolated by TRIzol (Ambion, Texas, USA) was utilized for synthesizing complementary DNA (cDNA) by using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, Massachusetts, Scientific, USA). Subsequently, the complementary DNA was amplified to test TIAM2 by real-time PCR using a SYBR Green Master Mix (Servicebio, Wuhan, China). GAPDH was used as the internal control. The relative mRNA expression was quantified using 2-ΔΔCt method. The PCR primers were listed in Supplementary Table 5.
Cell viability assay
Cell viability was assessed by CCK-8 assay. Briefly, cells were seeded on 96-well plates ((3–5) × 103 cells/well). The culture medium and CCK-8 solution (Dojindo, Kyushu, Japan) were mixed with a 9:1 ratio and then incubate cells for 1.5 h. The optical density (OD) values were measured at 450 nm using a Microplate reader (BioTek, Vermont, USA) at 24th, 48th, 72nd, 96th and 120th h.
Colony formation assay
For this assay, 1000 cells/well were cultured in 6-well plates (or 500 cells/well in 12-well plates) for 12–14 days. The colonies were stained with 0.1% crystal violet (Solarbio, Beijing, China) for 10 min after fixation using paraformaldehyde for 15 min. Finally, the colonies were counted.
5-Ethynyl-2e′-deoxyuridine (EdU) assay
EdU staining assay was performed by EdU assay kit (RiboBio, Guanghzou, China). Briefly, cells were incubated with 25 μM EdU for 5 h and then fixed with 4% paraformaldehyde. Subsequently, cells were dyed by Apollo stain mixture and Hoechst 33342 for 30 min, respectively. A fluorescence microscope (Olympus, Tokyo, Japan) was utilized to take images to calculate the proportion of EdU-positive cells.
Transwell migration and invasion assays
Transwell chambers (Corning, New York, USA) were used for these assays. For invasion assays, the Matrigel (Corning, New York, USA) was thawed at 4 °C overnight. 100 µL Matrigel diluted by serum-free medium was added to the upper chamber and incubated for 30 min for solidification at 37 °C. Next, 3 × 104 cells suspended in 200 µL serum-free medium were placed to the upper chamber, and 600 µL 10% FBS-supplemented medium to the lower chamber. After 36-h incubation at 37 °C, both chambers were immersed in 10% formaldehyde and dyed with 0.1% crystal violet for 20 min, respectively, and then cells in the upper chamber were wiped off with cotton swabs. Finally, stained cells on the lower membrane surface were counted under a microscope. Transwell migration experiment shared the same protocols with invasion assays but without 100 µL Matrigel in the upper chambers.
m5C quantification
The global m5C level was assessed using the MethylFlash™ 5-mC RNA Methylation ELISA Easy Kit (Fluorometric) (EpiGentek, New York, USA). Briefly, adding 100 µl binding solution and 200 ng sample total RNA into each well, followed by incubation at 37 °C for 90 min for RNA binding. Then, 50 µl of m5C Detection Complex Solution containing m5C antibody was applied into each well after washing. Subsequently, diluted m5C antibody were removed after 50-min incubation at room temperature. Finally, wells were incubated with Fluorescence Development Solution at room temperature for 2–4 min away from direct light. The signal was determined by a fluorescence microplate reader within 2 to 10 min at 530ex/590em nm.
RNA-seq and m5C-methylated RNA immunoprecipitation sequencing(m5C-seq)
Cloudseq Biotech Inc. (Shanghai, China) provided RNA-seq and m5C-seq service. RNA-seq was conducted as previously described [45]. For m5C-seq, NanoDrop ND-1000 (Thermo Fisher Scientific, Massachusetts, USA) was firstly used to evaluate the quality and quantity of total RNA, followed by integrity evaluation using denaturing agarose. If OD260/OD280 values range from 1.8 to 2.1, the RNA purity is qualified and the RNA extracted from all samples met this standard. Then, RNA was randomly fragmented into fragments of about 200 nt and subsequently incubated with m5C antibody (NEB, Massachusetts, USA) for immunoprecipitation. Immunoprecipitated RNA was analyzed by high-throughput sequencing on the NovaSeq 6000 sequencer (Illumina, California, USA) to generate raw reads. Then, paired-end reads harvested from Novaseq 6000 sequencer were quality controlled by Q30 and a Q30 > 80% indicates good sequencing quality. Following removal of 3′ adaptor-trimming and inferior quality reads, clean reads were aligned to the reference genome (UCSC HG38) by Hisat2 software (v2.0.4) [46]. Methylated sites on RNAs were identified using MACS software [47]. Differentially methylated sites (|fold change | ≥2 and p < 0.000001) were selected by diffReps [48]. The pathway enrichment analysis was conducted on the differentially methylated sites-harboring genes based on Kyoto Encyclopedia of Genes and Genomes (KEGG) database. P < 0.05 was statistically significant.
mRNA stability assays
5 μg/ml actinomycin D (ActD, GlpBio, California, USA) were added to treat PC cells for 0, 4,8 h. Then, samples were harvested for total RNA extraction. qRT-PCR was performed to analyze the remaining TIAM2 mRNA expression. GAPDH was used as the internal control.
Animal experiment
The 4–6-week-old male BALB/c nude mice provided by Ziyuan Eperimental Animal Technology Co., Ltd. (Hangzhou China) were housed in specific pathogen-free units. Mice were humanely sacrificed by cervical dislocation after anesthesia at the endpoint of the experiment. Animal studies were approved by the ethics committee of the First Affiliated Hospital of Zheng Zhou University.
For the subcutaneous model, mice were randomly assigned into each group according to the random table method (n = 5 per group). 5 × 106 SW1990(transfected with shCtrl or shNSUN2) or PANC-1 cells (transfected with Vector or NSUN2) were subcutaneously injected into the lower flanks of mice to establish the xenograft tumors. Tumor weight and volume were recorded subsequently and tumor tissues were collected for Hematoxylin and eosin (HE)-staining and immunohistochemistry (IHC). Tumor volume (mm3) = (width)2 × length/2.
To evaluate the peritoneal dissemination ability, 1 × 106 transfected PC cells were injected into the peritoneal cavity of mice (n = 5/6 per group). Mice were carefully monitored and sacrificed at the 30th day after injection. Tissue with metastatic nodules was examined histopathologically.
For construction of hematogenous metastasis model, 1 × 106 transfected PC cells were injected into the tail veins of mice (n = 5/6 per group). Mice were sacrificed at the endpoint. Lungs and livers were collected for histopathological examination.
Immunohistochemistry (IHC) and evaluation
IHC and evaluation were performed as previously described [49]. The score of 11 and 12 was defined as high expression while the others as low expression for statistical analysis. Detailed information of antibodies for IHC was shown in Supplementary Table 4.
Statistical analysis
Statistical analysis was performed by SPSS Statistics 23.0 (IBM, New York, USA) and GraphPad Prism software8.0.1 (GraphPad, California, USA). Differences between two groups were evaluated by a two-tailed Student’s t test, χ2 test or Fisher’s exact test. Spearman’s correlation analysis was utilized to estimate the correlation between two genes. Survival analysis was conducted by Kaplan–Meier and log-rank test. Cox’s proportional hazard regression model was used for identifying significant independent prognostic factors. P < 0.05 (two-tailed) was statistically significant.
Data availability
The authors declared that all the data and materials are available on reasonable request.
References
Pourshams A, Sepanlou SG, Ikuta KS, Bisignano C, Safiri S, Roshandel G, et al. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4:934–47.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846–61.
Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22.
Nombela P, Miguel-Lopez B, Blanco S. The role of m(6)A, m(5)C and Psi RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20:18.
Li J, Wang F, Liu Y, Wang H, Ni BN. (6)-methyladenosine (m(6)A) in pancreatic cancer: Regulatory mechanisms and future direction. Int J Biol Sci. 2021;17:2323–35.
Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, et al. N(6)-methyladenosine-mediated upregulation of WTAPP1 promotes WTAP translation and wnt signaling to facilitate pancreatic cancer progression. Cancer Res. 2021;81:5268–83.
Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D, et al. N(6)-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Gut. 2020;69:2180–92.
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020;19:91.
Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM, et al. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J Biol Chem. 2021;296:100087.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639.
Yang Y, Wang L, Han X, Yang W-L, Zhang M, Ma H-L, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–1202.e1111.
Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–25.
Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–71.
Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin Y, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019;110:3510–9.
Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020;11:270.
Xu X, Zhang Y, Zhang J, Zhang X. NSun2 promotes cell migration through methylating autotaxin mRNA. J Biol Chem. 2020;295:18134–47.
Su J, Wu G, Ye Y, Zhang J, Zeng L, Huang X, et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene. 2021;40:5814–28.
Liu Z, Li Y, Li X, Zhao J, Wu S, Wu H, et al. Overexpression of YBX1 promotes pancreatic ductal adenocarcinoma growth via the GSK3B/cyclin D1/cyclin E1 pathway. Mol Ther Oncolytics. 2020;17:21–30.
Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90.
Schumann U, Zhang HN, Sibbritt T, Pan A, Horvath A, Gross S, et al. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020;18:40.
Chen SY, Chen KL, Ding LY, Yu CH, Wu HY, Chou YY, et al. RNA bisulfite sequencing reveals NSUN2-mediated suppression of epithelial differentiation in pancreatic cancer. Oncogene. 2022;41:3162–76.
Chiu CY, Leng S, Martin KA, Kim E, Gorman S, Duhl DM. Cloning and characterization of T-cell lymphoma invasion and metastasis 2 (TIAM2), a novel guanine nucleotide exchange factor related to TIAM1. Genomics. 1999;61:66–73.
Chen JS, Su IJ, Leu YW, Young KC, Sun HS. Expression of T-cell lymphoma invasion and metastasis 2 (TIAM2) promotes proliferation and invasion of liver cancer. Int J Cancer. 2012;130:1302–13.
Zhao ZY, Han CG, Liu JT, Wang CL, Wang Y, Cheng LY. TIAM2 enhances non-small cell lung cancer cell invasion and motility. Asian Pac J Cancer Prev. 2013;14:6305–9.
Cooke M, Kreider-Letterman G, Baker MJ, Zhang S, Sullivan NT, Eruslanov E, et al. FARP1, ARHGEF39, and TIAM2 are essential receptor tyrosine kinase effectors for Rac1-dependent cell motility in human lung adenocarcinoma. Cell Rep. 2021;37:109905.
Jiang B, Zhou L, Lu J, Wang Y, Liu C, Zhou W, et al. Elevated TIAM2 expression promotes tumor progression and is associated with unfavorable prognosis in pancreatic cancer. Scand J Gastroenterol. 2021;56:59–67.
Bohnsack K, Höbartner C, Bohnsack M. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019;10:102.
Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842.
Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906–19.
Cui M, Qu F, Wang L, Liu X, Yu J, Tang Z, et al. m5C RNA methyltransferase-related gene NSUN4 stimulates malignant progression of hepatocellular carcinoma and can be a prognostic marker. Cancer Biomark. 2022;33:389–400.
Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–81.
Luo G, Xu W, Chen X, Wang S, Wang J, Dong F, et al. NSUN2-mediated RNA m(5)C modification modulates uveal melanoma cell proliferation and migration. Epigenetics. 2022;17:922–33.
Sun Z, Xue S, Xu H, Hu X, Chen S, Yang Z, et al. Effects of NSUN2 deficiency on the mRNA 5-methylcytosine modification and gene expression profile in HEK293 cells. Epigenomics. 2019;11:439–53.
Yamaguchi N, Knaut H. Focal adhesion-mediated cell anchoring and migration: from in vitro to in vivo. Development. 2022;149:dev200647.
Lin X, Zhuang S, Chen X, Du J, Zhong L, Ding J, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol Ther. 2022;30:688–702.
Rooney C, White G, Nazgiewicz A, Woodcock SA, Anderson KI, Ballestrem C, et al. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010;11:292–8.
Chan YL, Lai WC, Chen JS, Tseng JT, Chuang PC, Jou J, et al. TIAM2S mediates serotonin homeostasis and provokes a pro-inflammatory immune microenvironment permissive for colorectal tumorigenesis. Cancers (Basel). 2020;12:1844.
Woroniuk A, Porter A, White G, Newman DT, Diamantopoulou Z, Waring T, et al. STEF/TIAM2-mediated Rac1 activity at the nuclear envelope regulates the perinuclear actin cap. Nat Commun. 2018;9:2124.
Wong RWJ, Ngoc PCT, Leong WZ, Yam AWY, Zhang T, Asamitsu K, et al. Enhancer profiling identifies critical cancer genes and characterizes cell identity in adult T-cell leukemia. Blood. 2017;130:2326–38.
Gaitanos TN, Koerner J, Klein R. Tiam-Rac signaling mediates trans-endocytosis of ephrin receptor EphB2 and is important for cell repulsion. J Cell Biol. 2016;214:735–52.
Boespflug ND, Kumar S, McAlees JW, Phelan JD, Grimes HL, Hoebe K, et al. ATF3 is a novel regulator of mouse neutrophil migration. Blood. 2014;123:2084–93.
Cooke M. Novel insights into the RTK-dependent metastatic phenotype of KRAS-mutant lung adenocarcinoma. Mol Cell Oncol. 2021;8:2013723.
Wang Y, Zeng L, Liang C, Zan R, Ji W, Zhang Z, et al. Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics. 2019;11:1693–715.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8:e65598.
Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10:5447–59.
Acknowledgements
The authors are grateful to National Natural Science Foundation of China for its support.
Funding
The study was supported by the National Natural Science Foundation of China (82070643, U1904164, and U2004121).
Author information
Authors and Affiliations
Contributions
GYC, RRS, and WHX performed study concept and design; GZZ and LWL performed the experiment; JHL, YC, and YW performed tissue specimen-related experiment; JHL, YZZ, and ZHD interpretated the data and analyses; GZZ drafted the manuscript. GYC, RRS and WHX revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, G., Liu, L., Li, J. et al. NSUN2 stimulates tumor progression via enhancing TIAM2 mRNA stability in pancreatic cancer. Cell Death Discov. 9, 219 (2023). https://doi.org/10.1038/s41420-023-01521-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01521-y