Abstract
STAT3 and HIF1α are two fundamental transcription factors involved in many merging processes, like angiogenesis, metabolism, and cell differentiation. Notably, under pathological conditions, the two factors have been shown to interact genetically, but both the molecular mechanisms underlying such interactions and their relevance under physiological conditions remain unclear. In mouse embryonic stem cells (ESCs) we manage to determine the specific subset of hypoxia-induced genes that need STAT3 to be properly transcribed and, among them, fundamental genes like Vegfa, Hk1, Hk2, Pfkp and Hilpda are worth mentioning. Unexpectedly, we also demonstrated that the absence of STAT3 does not affect the expression of Hif1α mRNA nor the stabilization of HIF1α protein, but the STAT3-driven regulation of the hypoxia-dependent subset of gene could rely on the physical interaction between STAT3 and HIF1α. To further elucidate the physiological roles of this STAT3 non-canonical nuclear activity, we used a CRISPR/Cas9 zebrafish stat3 knock-out line. Notably, hypoxia-related fluorescence of the hypoxia zebrafish reporter line (HRE:mCherry) cannot be induced when Stat3 is not active and, while Stat3 Y705 phosphorylation seems to have a pivotal role in this process, S727 does not affect the Stat3-dependent hypoxia response. Hypoxia is fundamental for vascularization, angiogenesis and immune cells mobilization; all processes that, surprisingly, cannot be induced by low oxygen levels when Stat3 is genetically ablated. All in all, here we report the specific STAT3/HIF1α-dependent subset of genes in vitro and, for the first time with an in vivo model, we determined some of the physiological roles of STAT3-hypoxia crosstalk.
Similar content being viewed by others
Introduction
Hypoxia Inducible Factor (HIF) 1α, the master regulator of cell metabolism in response to low levels of available oxygen, is involved in a wide range of biological processes, such as cancer progression, angiogenesis, and differentiation of pluripotent stem cells [1, 2]. HIF1α is constitutively expressed and generally degraded by Von Hippel Lindau tumor suppressor protein (pVHL) upon hydroxylation by Prolyl Hydroxylase Domain-containing (PHD) enzymes [3, 4]. In hypoxic conditions PHD3 is unable to hydroxylate HIF1α that is therefore stable enough to migrate in the nucleus where it generates heterodimers with HIF1β, a stable and constitutively expressed protein [5]. The HIF1α/HIF1β heterodimer activates the transcription of target genes involved in angiogenesis, cell metabolism, differentiation of pluripotent stem cells, erythropoiesis, and immune cell migration [2, 6,7,8,9].
In the last decade, in vitro studies revealed connections between HIF1α and other transcription factors among which Signal Transducer and Activator of Transcription 3 (STAT3) emerged as an important modulator of hypoxia/HIF1α activity [10, 11]. STAT3 is a component of the JAK/STAT pathway that has been claimed to control cell proliferation in cancer and pluripotent stem cells, via their transcriptional and metabolic regulation [12,13,14,15,16,17].
Under pathological conditions, the crosstalk between JAK/STAT3 and hypoxia/HIF1α pathways is significant: rheumatoid arthritis is an autoimmune disease characterized by high levels of inflammation due to the crosstalk between STAT3 and hypoxia pathways [18]; glucose deprivation in brain pericytes is regulated by STAT3-mediated induction of HIF1α [19]; the self-renewal of glioma stem-like cells, which was previously considered as a hypoxia-dependent mechanism, appeared to be determined by an upregulation of STAT3 by HIF1α [20]. Notably, targeting of STAT3 cascade can block HIF1 and VEGF signaling induced by several oncogenic pathways [21].
Additionally, hypoxia has a role in differentiation and proliferation of mouse embryonic stem cells (ESCs): in particular, short exposures to low oxygen tension determine the differentiation of stem cells into definitive endodermal cells and distal lung cells [22]; it has been demonstrated that hypoxia and retinoic acid can induce differentiation of mouse ESCs in hepatic progenitor cells [7]; furthermore, HIF1α has been shown to inhibit the MEK/ERK pathway [23] and to drive the developmental progression of mouse ESCs [2]. Mouse ESCs are characterized by high levels of expression of Stat3, which is involved in the maintenance of pluripotency [24]. Indeed, mouse ESCs are cultured in presence of Leukemia Inhibitory Factor (LIF), a cytokine that activates the JAK/STAT3 pathway increasing ESCs self-renewal [25,26,27,28]. Mouse ESCs can also be maintained in a medium containing GSK3 and MEK inhibitors (2i conditions). The use of 2i allowed for the derivation and expansion of Stat3 knock-out ESCs [25], revealing a role of STAT3 also in the control of metabolism and proliferation [15, 16, 29].
Despite several studies indicating a functional interplay between HIF1α and STAT3 under pathological conditions, it is not clear whether and how the two transcription factors regulate each other. For this reason, we decided to study at the molecular level the crosstalk between HIF1α and STAT3 in the context of the physiological response to hypoxia, taking advantage of genetic models developed in ESCs and in zebrafish.
Results
STAT3 regulates the expression of a specific subset of hypoxia-dependent genes
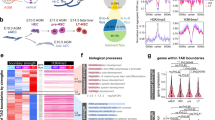
To analyze the involvement of STAT3 in hypoxia-dependent processes, we decided to perform RNAseq and compare the data of Stat3+/+ and Stat3−/− cells incubated for 24 h either in normoxia or in hypoxia (1% O2 tension). As shown in Fig. 1A and in Fig. S2, the comparison between Stat3+/+ ESCs grown in normoxia or hypoxia revealed a relevant group of differentially expressed genes. In particular, as reported in the Volcano plot (Fig. 1A), the expression of about 1’500 genes is altered in hypoxia and a large part of these genes are upregulated. As expected, genes involved in glycolysis and angiogenesis belong to the subset of upregulated genes. Among them we found Vascular Epithelial Growth Factor A (Vegfa), Hexokinase 1 (Hk1), Hexokinase 2 (Hk2), Phosphofructokinase (Pfkp), Lactate dehydrogenase A (Ldha), Aldolase A (Aldoa), Phosphoinositide-dependent kinase 1 (Pdk1) (Fig. 1A). Interestingly, when comparing the number of genes affected by hypoxia in Stat3+/+ and in Stat3−/− ESCs, we saw that the effect of hypoxia in Stat3 mutant cells was significantly attenuated: as reported in the box plots of Fig. 1B, C, only 70% of hypoxia-dependent genes are induced by hypoxia in Stat3 null cells (FC > 1 and p-value < 0.05). Hence, 30% of hypoxia-responsive genes are not significantly induced when Stat3 is knocked-out. Of note, the comparison between hypoxic and normoxic Stat3−/− cells revealed a significant difference in both down- and up-regulated transcripts, however, this response to low oxygen was weaker when compared to the one observed in hypoxic Stat3+/+ ESCs (Fig. 1B, C). Indeed, the comparison between hypoxic Stat3+/+ and hypoxic Stat3−/− cells allowed us to identify a subset of hypoxia-responsive genes whose induction is significantly dampened in Stat3−/− cells. This subset of genes is reported in the heatmap of Fig. 1D and contains canonical HIF1α targets, such as Vegfa, Hk1, Hk2, Pfkp and Hypoxia lipid droplet-associated (Hilpda) whose expressions were plotted in Fig. 1E and validated with RT-qPCR in Fig. 1F. Moreover, as shown in Fig. S1, the comparison between Stat3+/+ and Stat3−/− hypoxic ESCs shows that hypoxia does not affect the expression of STAT3 target genes: as reported in Fig. S1, Volcano plot and bar plots reveal that the overall number of STAT3-dependent genes does not change when cells are incubated in normoxia or in hypoxia. Box-plots reported in Fig. S1B demonstrated that hypoxia does not significantly affect the expression of STAT3-related genes. Stat3 itself and some of its target genes, such as Socs3 and Klf4, are downregulated in Stat3 knock-out cells, but they are not differentially expressed when comparing normoxic and hypoxic Stat3+/+ cells (Fig. S1D-I). Only Tet2 is significantly downregulated by hypoxia in Stat3+/+ ESCs, but this result is not confirmed by RT-qPCR (Fig. S1J, K). We can conclude that STAT3 plays an important role in the induction of hypoxia-dependent genes, including known HIF1α-targets involved in the regulation of glycolytic metabolism and in vascular remodeling (Fig. S2B), while HIF1α does not regulate STAT3 activity, at least as regards mouse ESCs in which STAT3 activity is constitutively induced by LIF in the culture media [15, 16, 30, 31]. It is worth mentioning that some HIF1α-dependent genes are affected by hypoxia either in Stat3+/+ and Stat3−/− cells. In Fig. S2C, D we show that the expression of Egln3, one of the main targets of HIF1α which encodes for PHD, determining the elastic feedback loop of the oxygen deprivation response [32,33,34], is not affected by Stat3 mutation, demonstrating that not all the hypoxia-dependent genes rely on STAT3 activity, but only a very specific subset of genes. Moreover, we measured the levels of Y705 and S727 phosphorylation in ESCs incubated with low oxygen tensions for 24 h and compared them with normoxic cells. As shown in Fig. S3A–D, hypoxia do not significantly affect the total level of STAT3 protein nor pSTAT3 Y705 and pSTAT3 S727 in ESCs.
A–D Transcriptome analysis by RNA-seq. A: Genes that were differentially expressed (log2[fold change (FC)] > + 0.75 or <−0.75; q-value < 0.01, Benjamini–Hochberg adjustment, as indicated by dashed lines) between Stat3 +/+ cells in normoxia and Stat3+/+ cells in hypoxia; n = 4 biological replicates. Expression levels of genes that were down- (B) and upregulated (C) in Stat3 +/+ in hypoxia relative to Stat3+/+ cells in normoxia. Each boxplot shows the 1st, 2nd and 3rd quartiles; the whiskers show the minimum and maximum values. D heatmap of RNAseq data reporting the expression of genes that are affected by hypoxia in Stat3+/+ cells compared to Stat3+/+ normoxic cells, but that require STAT3 to be properly altered in their expression. E expression level of Vegfa, Hk1, Hk2, Pfkp and Hilpda in normoxic and hypoxic Stat3+/+ and Stat3−/− mESCs taken from RNAseq data. F Gene expression analysis by RT–qPCR of Stat3+/+ and Stat3−/− cells treated in normoxic and hypoxic mESCs of the same genes shown for the RNAseq. Mean ± SEM of n = 4 experiments, with each replica shown as a dot.
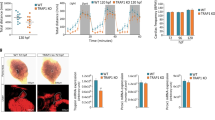
To understand how STAT3 regulates hypoxia-dependent gene expression, we decided to study the crosstalk between STAT3 and HIF1α. We first asked whether the expression levels of Hif1α mRNA are affected by the genetic ablation of Stat3. Interestingly, as reported in Fig. 2A, there are no significant differences in the expression of Hif1α in Stat3−/− cells compared to Stat3+/+ neither in normoxia or in hypoxia. Therefore, we sought to assess whether STAT3 is involved in the stabilization of HIF1α in hypoxia. Interestingly, as reported in Fig. 2B, C, western blot analysis revealed that hypoxia can stabilize HIF1α both in Stat3+/+ and in Stat3−/− cells. Once demonstrated that in mouse ESCs STAT3 is neither involved in the expression of Hif1α mRNA nor in HIF1α protein stabilization, we decided to test whether STAT3 might interact directly with HIF1α. To do so, we performed Proximity Ligation Assay (PLA) and found that PLA positive dots could be detected only in murine Stat3+/+ ESCs grown at low oxygen tensions (Fig. 2D), revealing that STAT3 and HIF1α interact with each other in hypoxic conditions. Furthermore, the cellular localization of dots indicates that the interaction between the two transcription factors mainly occurs in the nucleus.
A RT-qPCR analysis of Hif1α mRNA expression on murine Stat3+/+ and Stat3−/− murine ESCs incubated either in low and in high oxygen tensions. B, C western blot analysis of HIF1α and STAT3 from protein extracts of Stat3+/+ and Stat3−/− murine ESCs incubated either in low and in high oxygen tensions. Representative pictures (B) and quantifications (C). β-Actin was used as an internal control. 3 independent biological replicates were used. D PLA with anti-STAT3 and anti-HIF1α antibodies. Scale bar = 200 μm. Quantification of dots divided for the number of nuclei detected with DAPI (blue). n = 3 independent biological replicas were used. Scale bar = 200 μm. Mean ± SEM.
Next, we tested whether the interaction between STAT3 and HIF1α can be observed also in pseudohypoxia. Pseudohypoxia is a condition that occurs whenever HIF1α is stabilized even if the organism is exposed to atmospheric oxygen tensions. This phenomenon can be chemically triggered by some compounds like cobalt chloride (CoCl2) which substitutes with Co2+ the Fe2+ ion necessary for the catalytic activity of PHD, hence inactivating the enzyme [35,36,37]. As expected, wild type ESCs incubated for 24 hours with CoCl2 were characterized by high levels of expression of HIF1α target genes Vegfa and Hk2 compared to untreated Stat3+/+ cells (Fig. S3E, F). In contrast, Stat3−/− cells incubated with CoCl2 did not show significant upregulation of these transcripts (Fig. S3E, F), while Hif1α mRNA levels were not affected by the treatment (Fig. S3G). On the other hand, Egln3, which is a pure target gene of HIF1α involved in the elastic feedback loop of the oxygen deprivation response [32], was not affected by the absence of Stat3 (Fig. S3H). Interestingly, PLA analysis performed on ESCs treated with CoCl2 confirms the interaction between STAT3 and HIF1α (Fig. S3I). These results suggest that the formation of a nuclear STAT3-HIF1α complex is associated with full activation of hypoxia-responsive genes.
Hif1α transcriptional activity is not induced in vivo when Stat3 is knocked-out or inhibited
Zebrafish is an in vivo model in which the pathophysiological roles of HIF1α and JAK/STAT signalling have been extensively studied [9, 34, 38,39,40,41,42,43,44,45,46]. Thus, it appeared as a valid platform to analyze the STAT3-HIF1α crosstalk and the physiological implications of this interplay between transcription factors. We used the Tg(4xHRE-TATA:mCherry,cmlc2:EGFP)ia22 hypoxia reporter zebrafish line (herein called HRE:mCherry), in which the mCherry red fluorescent protein is expressed in all tissues experiencing low oxygen tensions or pseudohypoxic conditions [39]. To investigate the requirement of Stat3 in the transcriptional response to hypoxia, we combined hypoxic and pseudohypoxic treatments with chemical or genetic inhibition of Stat3. To inhibit Stat3 signaling we used AG490, which blocks the Jak-mediated Y705 phosphorylation of Stat3 and abrogates its nuclear transcriptional activity [43, 44, 47]. Larvae treated with this compound were also characterized by a reduction of stat3 gene expression (Fig. S4A). To induce hypoxia in zebrafish larvae, we incubated the animals for 3 days with 5% oxygen tension, while pseudohypoxia was forced by using either dimethyloxalylglycine (DMOG), an inhibitor of Phd-dependent degradation of Hif1α [48]; CoCl2 or dexamethasone (Dex), a synthetic glucocorticoid that contributes to the stabilization of Hif1α by degrading Vhl [39]. Notably, while hypoxia and pseudohypoxia induced the fluorescence of HRE:mCherry zebrafish larvae, such induction was abolished by AG490, suggesting that, also in zebrafish, activated Stat3 is necessary for the correct induction of hypoxia-dependent transcription (Fig. 3A–E). Moreover, we tested the expression of vegfa and hk1 at different times of exposure to low oxygen tension. We could observe that hypoxia determines a boost of expression of both vegfa and hk1 after 8 h of incubation in hypoxia; after 24 h of incubation in hypoxic conditions, both transcripts return to the levels detected in normoxic larvae. Interestingly, vegfa expression increased after 72 h of hypoxia, while hk1 expression was upregulated after 48 h of hypoxia (Fig. S4B, C). AG490 abrogated this hypoxia-dependent dynamical expression of vegfa and hk1 (Fig. S4B, C), confirming the results obtained with the HRE:mCherry reporter line and with ESCs. STAT3 transcriptional activity has also been reported to be positively affected by the phosphorylation of Serine 727 [49,50,51] and this post-translational modification is triggered by MEK/ERK pathway [52]. As we recently demonstrated that the MEK/ERK inhibitor PD98059 blocks the pS727 activities of STAT3 [43], we sought to assess whether the inhibition of S727 phosphorylation can affect the responsiveness of larvae to low oxygen tensions. When HRE:mCherry reporter larvae are treated with PD98059 and incubated in low oxygen tensions, we could not observe significant differences in reporter fluorescence compared to hypoxic DMSO-treated larvae, suggesting that the chemical inhibition of S727 phosphorylation does not affect the transcriptional responsiveness of zebrafish to hypoxia (Fig. 3F, G). To further elucidate the role of Serine or Tyrosine phosphorylation in Stat3-dependent regulation of hypoxia transcription, we injected Stat3 mRNAs (that we recently used and validated in Peron et al., [43]) in double transgenic eggs obtained from the breeding between Tg(7xStat3-Hsv.Ul23:EGFP)ia28, herein called SBE:EGFP [44], and HRE:mCherry transgenic animals to monitor at the same time the Stat3- and the hypoxia-dependent transcription. Of note, wild type Stat3 mRNA injection upregulated mCherry fluorescence suggesting that the overexpression of Stat3 increases hypoxia-dependent transcription, whereas Stat3 Y705F mRNA (that encodes a STAT3 that cannot be phosphorylated in 705 position, as described in Minami et al. [53]), cannot induce the fluorescence of both reporters (Fig. S4D, E). On the other hand, surprisingly, the overexpression of Stat3 S727A (in which the substitution of the Serine with an Alanine blocks the 727 phosphorylation, as shown in Wen et al. [54]), induces the Stat3-dependent transcriptional activity as well as the hypoxia-related fluorescence (Fig. S4D, E). These results suggest that Tyrosine phosphorylation is necessary for the regulation of hypoxia by Stat3, while Serine is not.
A Representative pictures of HRE:mCherry reporter zebrafish treated with AG490 in combination with low oxygen tension, DMOG, and Dex from 3-6 dpf and CoCl2 from 5-6 dpf. Scale bar 500 μm. n = 18 DMSO normoxia, 11 DMSO hypoxia, 18 AG490, 9 AG490 hypoxia, 11 Dex, 11 Dex AG490, 12 DMOG, 12 DMOG AG490, 18 CoCl2, 12 CoCl2 AG490 (larvae used for this experiment came from three independent breeding between wild type zebrafish). B Fluorescence quantification of HRE:mCherry reporter zebrafish treated with AG490 in combination with low oxygen tension. C Fluorescence quantification of HRE:mCherry reporter zebrafish treated with AG490 in combination with 10 μM Dex. D Fluorescence quantification of HRE:mCherry reporter zebrafish treated with AG490 in combination with 50 μM DMOG from 3–6 dpf. E Fluorescence quantification of HRE:mCherry reporter zebrafish treated with AG490 in combination with 0.1 mM CoCl2 from 5-6 dpf. F, G Representative pictures and fluorescence quantification of HRE-mCHerry reporter zebrafish larvae treated with 12.5 μM PD98059 in combination with low oxygen tension. n = 31 DMSO normoxia, 29 PD98059 normoxia, 28 DMSO hypoxia, 29 PD98059 hypoxia (larvae used for this experiment came from three independent breeding between wild type zebrafish). Scale bar = 500 μm. H, I Representative pictures and fluorescence quantification of stat3+/+, stat3+/− and stat3−/− sibling larvae in HRE:mCherry transgenic background treated with low oxygen tension from 3–6 dpf. n = 7 stat3+/+ normoxia, 9 stat3+/+ hypoxia, 6 stat3+/− normoxia, 7 stat3+/− hypoxia, 8 stat3−/− normoxia, 6 stat3−/− hypoxia (larvae used for this experiment came from three independent breeding between stat3+/− zebrafish). Scale bar = 500 μm. Mean ± SEM.
Next, we crossed the stat3ia23 mutant zebrafish line (herein called stat3−/−) [44] with the HRE:mCherry reporter line. We treated the HRE:mCherry;stat3+/+, HRE:mCherry;stat3+/− and HRE:mCherry;stat3−/− sibling larvae with 5% oxygen tension and measured mCherry fluorescence. Consistent with previous results obtained in ESCs and in embryos treated with AG490, the Hif1α-dependent reporter activity increased in hypoxic stat3+/+ larvae, while no significant increase of the reporter fluorescence was detected in hypoxic stat3+/− and stat3−/− larvae when compared with normoxic siblings (Fig. 3H, I). These results indicate that the transcriptional activation of HIF1α targets requires STAT3 also in vivo.
Nuclear crosstalk between Stat3 and Hif1α determines the regulation of hypoxia-dependent genes in zebrafish
Given that Stat3 is needed for the induction of normal Hif1α transcriptional activities and that hypoxia-induced mechanisms are impaired when Stat3 is inhibited or deleted, we wanted to determine by which mechanisms Stat3 regulates Hif1α transcriptional activity.
We first tested whether Stat3 inactivation would affect hif1α expression levels, its stabilization, or the levels of HIF1α regulators vhl and egln3; we observed no significant differences in their expression levels when comparing stat3+/+ and stat3−/− 6-dpf larvae (Fig. S5A–F). We conclude that, as already observed in ESCs (Fig. 2A, B), the lack of a functional Stat3 does not impair the expression and stabilization of Hif1α in zebrafish larvae.
Given that in ESCs STAT3 and HIF1α interact in the nucleus, we focussed our attention on the nuclear activity of HIF1α in vivo. For this purpose, we injected an mRNA that encodes a dominant active (DA) form of hif1αb bearing mutations at two prolines and one asparagine (P402A, P564G, N804A), hence preventing their hydroxylation by Phd and subsequent Vhl-dependent degradation [55, 56]. This construct activates by default nuclear targets, independently from any upstream degrading cue. We treated HRE:mCherry hypoxia reporter larvae injected with hif1αb DA mRNA at 3 dpf with AG490 and analysed the reporter fluorescence at 4 dpf. Notably, hif1αb DA mRNA determines an induction of the reporter activity that is slightly decreased (p = 0.05) in injected larvae treated with AG490 (Fig. 4A, B). We further investigated the endogenous genes induced by hif1αb DA mRNA by performing RT-qPCR analysis. The expression levels of Hif1α-dependent genes, such as vegfa and hk1 were induced by hif1αb DA mRNA, however, AG490 abrogated their upregulation (Fig. 4C, D). These results confirmed in vivo that activation of HIF1α nuclear targets rely on active STAT3, as observed in mouse ESCs.
A, B Representative pictures and fluorescence quantification of 4-dpf HRE:mCherry reporter larvae treated with either DMSO or 50 μM AG490 for 24 h after injection of hif1ab DA mRNA. n = 3 independent biological replicates; 10 DMSO, 20 DMSO hif1ab DA mRNA, 19 AG490, 24 AG490 hif1ab DA mRNA (larvae used for this experiment came from three independent breeding between wild type zebrafish). Scale bar 500 μm. C RT-qPCR analysis of vegfa of 4-dpf larvae injected with hif1ab DA mRNA and treated with AG490 (n = 4 independent biological replicates composed by pool of 15 larvae, each dot represents a pool of larvae). D RT-qPCR analysis of hk1 of 4-dpf larvae injected with hif1αb DA mRNA and treated with AG490 (n = 4 independent biological replicates composed by pool of 15 larvae, each dot represents a pool of larvae). E RT-qPCR analysis of vegfa, hk1, egln3, and hif1αb on EGFP-positive and EGFP-negative cells sorted from SBE:EGFP transgenic adult intestines (three adult intestines were pooled together in two biological replicas). Mean ± SEM.
An organ in which STAT3 plays a pivotal role is the intestine [44, 57, 58]. As we recently demonstrated in zebrafish, Stat3 is fundamental for the correct folding of gut and is particularly expressed in intestinal stem cells [44]. In particular, the Stat3 zebrafish reporter line SBE:EGFP that we recently published is characterized by EGFP positive cells in the intestinal track which represent the intestinal stem cells of zebrafish.
We took advantage of this zebrafish model to ask whether HIF1α and STAT3 are active in the same cells (cell autonomously), or whether they co-operate non-cell-autonomously in distinct cells somehow interconnected. To do so, we sorted Stat3 responsive intestinal stem cells from adult wild type intestines. EGFP-positive and -negative cells were immediately processed for RNA isolation after sorting. Results of RT-qPCR revealed a strong and significant upregulation of Hif1α transcriptional activity in EGFP-positive cells. In particular, hif1αb transcript levels were 20-fold higher in EGFP-positive cells compared to EGFP-negative cells. The upregulation of the vegfa and hk1, involved in angiogenesis and glucose metabolism, as well as of the pure Hif1α target gene egln3, involved in the Hif1α degradation feedback loop [32], revealed a massive Hif1α transcriptional activity in the EGFP-positive Stat3-responsive cells (Fig. 4E). flna, a gene that is neither affected by Stat3 nor by Hif1α was used in this experiment as a negative control. We conclude that in the context of a complex tissue like the intestine, HIF1α direct targets are physiologically strongly expressed in cells with active STAT3, further indicating cooperative transcriptional activation by the two factors and the cell-autonomous regulation of the STAT3-HIF1α crosstalk.
Hypoxia-dependent processes are impaired in stat3 mutant zebrafish
Our results so far indicate a cooperative activation of STAT3 and HIF1α in the transcriptional response to hypoxia. We then asked whether such cooperation is important also for the physiological responses in an organism.
One of the most relevant processes induced by hypoxia is angiogenesis [59]. To study how this process is affected by hypoxia in stat3 mutant zebrafish larvae, we decided to use the zebrafish endothelial cell reporter line Tg(Fli1:EGFP)y1 [60]. stat3+/+, stat3+/− and stat3−/− sibling larvae in Tg(Fli1:EGFP)y1 transgenic background were treated with 5% oxygen tension from 48 hpf to 54 hpf. After the treatment, we detected a significant increase of endothelial fluorescence in stat3+/+ hypoxic larvae when compared with untreated siblings (Fig. 5A, B), indicating that angiogenesis is induced by low oxygen tensions, hence confirming what already observed by Eyries et al. [61]. A similar induction of fluorescence was also detected in hypoxic heterozygous stat3+/− larvae when compared to normoxic heterozygous siblings, but no significant differences were detected between normoxic and hypoxic null stat3−/− larvae, indicating a pivotal role of Stat3 in this process (Fig. 5A, B).
A, B Representative pictures and fluorescence quantification of the trunk of 54-hpf stat3+/+, stat3+/− and stat3−/− in Tg(Fli1:EGFP)y1 transgenic background incubated in normoxia and hypoxia for 6 h. Scale bar: 1 mm. n = 15 stat3+/+ normoxia; 17 stat3+/+ hypoxia; 21 stat3+/− normoxia; 20 stat3+/− hypoxia; 14 stat3−/− normoxia; 14 stat3−/− hypoxia (larvae used for this experiment came from three independent breeding between stat3+/− zebrafish). C, D Representative pictures and fluorescence quantification of the tail of 54-hpf stat3+/+, stat3+/− and stat3−/− larvae in Tg(gata1:dsRed)sd2 transgenic background incubated in normoxia and hypoxia. Scale bar: 1 mm. n = 18 stat3+/+ normoxia; 14 stat3+/+ hypoxia; 41 stat3+/− normoxia; 44 stat3+/− hypoxia; 17 stat3−/− normoxia; 20 stat3−/− hypoxia (larvae used for this experiment came from four independent breeding between stat3+/− zebrafish). E, F Representative pictures of 54-hpf control (ctrl inj) and hif1α morphants (MO-hif1α) in Tg(LysC:dsRed)nz50 transgenic background incubated in normoxia and hypoxia for 6 h (scale bar: 500 μm) quantification of the ratio of dsRed-positive cells in region A and in region B. Area B between dashed lines. Arrowheads point at fluorescent cells in Area A. n = 8 ctrl inj normoxia; 9 ctrl inj hypoxia; 7 MO-hif1α normoxia; 8 MO-hif1α hypoxia (larvae used for this experiment came from three independent breeding between wild type zebrafish) G, H Representative pictures of 54-hpf stat3+/+, stat3+/− and stat3−/− larvae in Tg(LysC:dsRed)nz50 transgenic background incubated in normoxia and hypoxia for 6 h (scale bar: 500 μm); quantification of the ratio of dsRed-positive cells in region A and in region B. Area B between dashed lines. Arrowheads point at fluorescent cells in Area A. n = 11 stat3+/+ normoxia; 16 stat3+/+ hypoxia; 22 stat3+/− normoxia; 23 stat3+/− hypoxia; 11 stat3−/− normoxia; 17 stat3−/− hypoxia (larvae used for this experiment came from three independent breeding between stat3+/− zebrafish). Mean ± SEM.
Hypoxia plays a central role in erythropoiesis [62,63,64,65] and in zebrafish gata1 is a specific marker for erythropoiesis and erythrocytes [66,67,68,69,70,71]. Hence, we decided to use the Tg(gata1:dsRed)sd2 transgenic line [67], in which erythroid cells display a strong red fluorescence, to see whether Stat3 has a role in hypoxia-induced erythropoiesis. Notably, 6 h long hypoxia treatments determine a significant increase of fluorescence in stat3+/+ (Fig. 5C, D). Similarly, an upregulation of fluorescence was detected also in hypoxic stat3+/− larvae (Fig. 5C, D). Of note, no significant differences were detected between normoxic and hypoxic stat3−/− larvae, demonstrating that Stat3 is involved in the hypoxia-induced erythropoiesis (Fig. 5C, D).
Since the hypoxia/HIF1 pathway has been linked to mobilization and polarization of macrophages [56, 72,73,74,75], we used the Tg(LysC:dsRed)nz50 transgenic line [76], to focus our attention on them. First, to confirm that the migration of macrophages outside the aorta-gonad-mesonephros (AGM) region is triggered by hypoxia and relies on Hif1α, we injected 1-cell stage Tg(LysC:dsRed)nz50 transgenic embryos with a solution containing both morpholinos (MO) against hif1αa and hif1αb (as previously described in Gerri et al., [9]). 48-hpf controls and hif1α morphants were subsequently incubated in normoxia or hypoxia for 6 h and the number of macrophages was counted. As reported in Fig. 5E, F and Fig. S6A–C, hypoxia determines an increase of the ratio between the number of cells observed in the trunk of larvae (named region A) and that in the AGM (named region B), suggesting that hypoxia determines the migration of macrophages away from AGM. Interestingly, we could not detect significant differences between normoxic and hypoxic morphants, demonstrating that Hif1α is required for this process (Fig. 5E, F). Subsequently, we sought to see whether Stat3 is also involved in this process. To do so, we treated 48-hpf stat3+/+, stat3+/− and stat3−/− larvae Tg(LysC:dsRed)nz50 transgenic background with 5% oxygen tension for 6 h. Treated stat3+/+ and stat3+/− showed a significant increase of Area A/Area B ratio compared to untreated siblings, while hypoxia did not have any effect in mobilizing macrophages of stat3−/− larvae, suggesting that Stat3 is involved in this Hif1α-dependent process (Fig. 5G, H and Fig. S6D–F). Moreover, we analysed the level of expression of genes involved in macrophage activity, like mfap4, tek, and lcp1 [77,78,79]. mfap4 and tek appeared to be downregulated in their expression in stat3−/− when compared to stat3+/+ siblings (Fig. S6G), highlighting a role of Stat3 in the homeostasis of these cells. Importantly, the total number of dsRed-positive cells does not seem to be significantly affected by genetic ablation of stat3, indicating a specific effect on migration rather than survival or proliferation (Fig. S6D–F).
Discussion
In the last 20 years, several research groups suggested an interplay between STAT3 and HIF1: for instance, Gray and collaborators in 2005 demonstrated that the expression of Vegfa in pancreatic and prostate carcinomas depends on HIF1α, STAT3, CBP/p300 and Ref-1/APE [80] and this result was further investigated by Oh and collaborators [81], who showed that STAT3 and HIF1α bind the Vegfa promoter [81]. However, up to now, no clear description of STAT3-HIF1α connections have been provided yet and the mechanisms through which STAT3 and HIF1α regulate each other is still controversial: some groups demonstrated that STAT3 triggers the expression of HIF1α mRNA [82, 83], others suggested that STAT3 is involved in the stabilization of HIF1α protein [21, 84]. Moreover, Pawlus and collaborators demonstrated that STAT3 increases HIF1α DNA-binding kinetics and that HIF1α interacts with STAT3 with its bHLH and PAS domains [85]. To better elucidate this intricate mechanism of regulation and to clarify the current discrepancies between aforementioned studies, we decided to analyze the crosstalk between STAT3 and HIF1α using both in vivo and in vitro physiological models. RNAseq experiments performed on mouse ESCs allowed us to determine that a large part (about 30%) of genes induced by hypoxia and HIF1α require a functional STAT3. Of note, with this experiment we provide the list of genes that are regulated in concert by STAT3 and HIF1α, demonstrating that the hypoxia-triggered control of glycolysis and angiogenesis depends on STAT3.
Taking advantage of the Stat3 zebrafish reporter line characterized in Peron et al. [44], we demonstrated that Stat3-positive cells in the zebrafish intestine have a strong Hif1α signature, hence, we decided to test whether hypoxia-dependent activities rely on Stat3. Drugs inhibiting Stat3 signaling pathway established a severe unresponsiveness of zebrafish larvae to hypoxia and pseudohypoxia, underlying a pivotal role for Stat3 in the normal induction of hypoxia-related transcriptional activity. This result was additionally confirmed by the defective responsiveness to hypoxia of stat3 mutant zebrafish larvae: many hypoxia- and Hif1α-dependent processes like angiogenesis and macrophage mobilization from AGM are significantly affected by stat3 genetic ablation, confirming our hypothesis that Stat3 is involved in the correct activation of hypoxic adaptation. Notably, we could show both in zebrafish and mouse ESCs that STAT3 is not involved in the transcription of Hif1α gene, nor in the stabilization of HIF1α protein and that HIF1α localizes in the nucleus even when Stat3 is not expressed. Remarkably, we demonstrated that STAT3 physically interacts with HIF1α and that their interaction is specifically localized in the nucleus. These results support the hypotheses that STAT3 enhances HIF1α transcriptional activities by regulating its interaction with either HIF1β, the Hypoxia Responsive Elements on target genes or the basal transcriptional machinery. Finally, it is worth mentioning that STAT3-dependent control of HIF1α transcriptional activity affects specifically genes involved in angiogenesis (Vegfa) and metabolism (Hk1, Hk2, Pfkp) and this result was observed both in mouse ESCs (Fig. 1) and in zebrafish (Figs. 4 and 5). The regulation of these processes, representing two of the most important biological functions regulated by HIF1α, highlights the pivotal role of STAT3 in the proper response to low oxygen tensions and shows that the connection between STAT3 and HIF1α is much stronger than it was previously thought. These findings, together with the observation by Russell et al. [86] that VHL promotes ubiquitin-mediated destruction of JAK2, and the fact that JAK/STAT3 and HIF/VHL play a role in many diseases, call for a deeper analysis of their crosstalk and the implications this crosstalk might have from a therapeutic point of view.
Material and methods
Animals husbandry and zebrafish lines
Animals were staged and fed as described by Kimmel et al. [87] and maintained in a large-scale aquaria system.
Embryos were obtained after natural mating, raised in Petri dishes containing fish water (50x: 5 g NaHCO3, 39.25 g CaSO4, 25 g Instant Ocean for 1 l) and kept in a 12:12 light dark cycle at 28 °C. All experimental procedures complied with European Legislation for the Protection of Animals used for Scientific Purposes (Directive 2010/63/EU).
stat3ia23 mutant line [44] is genotyped by PCR amplification and 3% agarose gel migration. The Tg(7xStat3-Hsv.Ul23:EGFP)ia28 line, the Tg(4xHRE-TATA:mCherry,cmlc2:EGFP)ia22 line, the Tg(fli1:EGFP)y1, the Tg(gata1:dsRed)sd2 line and the Tg(LysC:dsRed)nz50 lines have been respectively characterized by Peron et al. [44], Vettori et al. [39], Lawson and Weinstein [60], Galloway et al. [67] and Hall et al. [76]. All animal experiments were performed under the permission of the ethical committee of the University of Padova and the Italian Ministero della Salute (23/2015-PR).
Sample size for experiments with zebrafish larvae was calculated for an effect of 1 and assuming the same variance for each group, by using the statistical package G*power as described in Faul et al. [88]. All experiments were blind: genotypes were assessed after the treatment.
Mouse ESCs culture and treatments
ESCs were grown as described in Betto et al. [16]. In detail, wild type (Stat3+/+) or Stat3 knock-out (Stat3−/−) (described previously in Ying et al. [25], Carbognin et al. [15], Takeda et al. [89] and provided by A. Smith’s laboratory) mouse ESC lines were routinely cultured without feeders on gelatin-coated plates (0.2% gelatin, Sigma-Aldrich, cat. G1890). Media were changed every 2 days and cells were passaged when approaching confluency (every 2-3 days); to passage, cells were replated at required density following dissociation with Accutase (ThermoFisher, cat. A1110501). Cells were grown in 2iLIF culture conditions, prepared as follows: serum-free KSR (Knockout Serum Replacement) 10% (ThermoFisher, cat. 10828028) - based medium in GMEM (Sigma-Aldrich, cat. G5154) supplemented with 1% FBS (ThermoFisher, cat. 10270106), 100 mM 2-mercaptoethanol (Sigma-Aldrich, cat. M3148), 1× MEM non-essential amino acids (ThermoFisher, cat. 11140050), 2 mM L- glutamine (ThermoFisher, cat. 25030081), 1 mM sodium pyruvate (ThermoFisher, cat. 11360070), and with small-molecule inhibitors PD (1 μM, PD0325901), CH (3 μM, CHIR99021) from Axon (cat. 1386 and 1408) and LIF (100units/ml, Qkine, cat. Qk036).
All cells were maintained at 37 °C in humidified incubators with 5% CO2.
Hypoxia treatments were performed for 24 h with 1% oxygen tensions. 0.1 mM CoCl2 was added to cell culture media for 24 h.
Sample size for experiments with zebrafish larvae was calculated for an effect of 1 and assuming the same variance for each group, by using the statistical package G*power as described in Faul et al. [88].
Proximity ligation assay
Mouse ESCs were fixed for 10 min in 4% formaldehyde at −20 °C, washed in TBS and permeabilized for 10 min with TBST + 0.5% Triton X-100 at room temperature. PLA was performed following the Duolink® protocol: cells were covered with Duolink® blocking solution and incubated for 60 min at 37 °C in humidity chamber; primary antibodies anti-STAT3 mouse monoclonal (Cell Signaling, 9139) (1:100) and anti-HIF1α rabbit monoclonal (Novus Biologicals, NB100-499) (1:100) were dissolved in Duolink® antibody diluent and samples were incubated overnight at 4 °C. Primary antibodies solution was removed and samples were washed 2 times for 5 min with washing buffer A (prepared following manufacturer’s instructions) and subsequently incubated for 1 h at 37 °C with Duolink® PLA probe mix. After 2 washes with washing buffer A for 5 min each, samples were incubated at 30 min at 37 °C with Duolink® ligation mix. Samples were washed 2 times for 2 min with washing buffer A and, subsequently, incubated for 100 min at 37 °C with Duolkin® amplification mix. After three washes with washing buffer B (prepared following manufacturer’s instructions), samples were ready for imaging.
Drug and hypoxic treatments
We used the following chemical compounds: AG490 (T3434, Sigma); DMOG (D3695, Sigma), Dex (D1756, Sigma), CoCl2 (232696, Sigma). Dex was diluted in ethanol and stored at 4 °C, while the other compounds were diluted in DMSO and stored in small aliquots at −20° C. 2 mM 1-phenyl-2-thiourea (PTU) was used to inhibit pigmentation. Larvae were treated from 3 dpf to 6 dpf with 50 μM AG490, 50 μM DMOG, 10 μM Dex; from 5 dpf to 6 dpf with 0.5 mM CoCl2. 5% oxygen tension was maintained using ProCO2 (BioSpherix) device and larvae were incubated with low oxygen tension from 3 dpf to 6 dpf.
mRNA isolation and quantitative real time reverse transcription PCR (qRT-PCR)
Total RNAs were extracted from pools of 20 larvae at 3, 4, 5 and 6 dpf with TRIzol reagent (Thermo Fisher Scientific, 15596018) and incubated at 37 °C for 30 min with RQ1 RNase-Free DNase (Promega, M6101). cDNA synthesis was performed using random primers (Promega, C1181) and M-MLV Reverse Transcriptase RNase H (Solis BioDyne, 06-21-010000) according to the manufacturer’s protocol. qPCRs were performed in triplicate with CybrGreen method by means of Rotor-gene Q (Qiagen) and the 5x HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, 08-36-00001) and zgapdh and mActnb were used as internal standard in each sample. The amplification protocol consists of 95 °C for 14 min followed by 45 cycles at 95 °C for 20 s, 60 °C for 20 s and 72 °C for 25 s. Threshold cycles (Ct) and melting curves were generated automatically by Rotor-Gene Q series software and results were obtained with the method described in Livak and Schmittgen [90]. Sequences of genes of interest primers were designed with Primer3 software [91] (http://bioinfo.ut.ee/primer3-0.4.0/input.htm) and are listed in supplementary material Table 1.
Protein extraction and western blotting
Total protein extracts were obtained by homogenization of pools of 20 6-dpf larvae in ice cold RIPA buffer (ThermoFisher, 89900) and Complete EDTA-free protease inhibitor cocktail (Sigma, 11873580001). For western blot analysis 40 μg of protein extracts were loaded per well on Bolt 4–12% Bis-Tris Plus Gels (ThermoFisher, NW04120BOX) and blotted on PVDF immobilon-p membranes (Millipore, IPFL00010). Dried membranes were then washed with PBS (Sigma, P4417) with 0.1% (w/v) Tween20 and incubated overnight with primary antibodies at 4 °C: anti-HIF1α (1:500, MA1-16504 Invitrogen); anti-STAT3 (1:1000, 9139 S Cell Singaling); anti-pSTAT3 Y705 (1:1000, D3A7 Cell Signaling), anti-pSTAT3 S727 (1:1000, 9134 Cell Signaling) and anti-βActin (1:5000, MA1-744 ThermoFisher). Secondary anti-Rabbit and anti-Mouse HRP-conjugated antibodies (1:5000, 170-6515 BIORAD, and 1:5000, 170-6516 BIORAD, respectively) were incubated for 1 h at room temperature and protein bands detected by chemiluminescence on an Alliance MINI HD 9 Blot Imaging System. Quantification of the signal was performed with ImageJ.
Imaging
For in vivo imaging, transgenic larvae were anesthetized with 0.04% tricaine, embedded in 1% low-melting agarose and mounted on a depression slide. Nikon C2 confocal system was used to acquire images from Tg(Fli1:EGFP)y1 transgenic larvae. Tg(4xHRE-TATA:mCherry,cmlc2:EGFP)ia22, Tg(LysC:dsRed)nz50 and Tg(gata1:dsRed)sd2 transgenic larvae were mounted in 1% low-melting agarose and observed with a Leica M165 FC microscope equipped with a Nikon DS-Fi2 digital camera. All images were analysed with Fiji (ImageJ) software and fluorescence integrated density was calculated setting a standard threshold on non-fluorescent samples.
Fluorescence-Activated Cell Sorting (FACS)
Adult intestines of Tg(7xStat3-Hsv.Ul23:EGFP)ia28 fish were dissected and treated as reported by Peron et al. [44]. Cells from three intestines were pooled together for two independent biological replicas.
In vitro mRNA synthesis and mRNA microinjection
The DA hif1αb mRNA was obtained from pCS2-hif1abDA vector that Dr. Phil Elks kindly sent us. The pCS2-Stat3, pCS2-Stat3Y705F and pCS2-Stat3S727A vectors were previously obtained by Peron and collaborators [43]. mRNAs were in vitro transcribed using the mMESSAGE mMACHINE® SP6 Transcription Kit (Thermo Fisher Scientific) and purified using the RNA Clean and Concentrator kit (Zymo Research). A mix containing 50 ng/μl mRNA, Danieu injection Buffer and Phenol Red injection dye, was injected into 1-cell stage embryos. hif1αa and hif1αb morpholinos, that Dr. Nana Fukuda kindly sent us, were injected together in 1-cell stage embryos as described in Gerri et al., 2017 [9].
RNA-sequencing
Quant Seq 3' mRNA-seq Library Prep kit (Lexogen) is used for library construction. Library generation is initiated by oligodT priming. The primer already contains Illumina-compatible linker sequences. After first strand synthesis the RNA is removed and second strand synthesis is initiated by random priming and a DNA polymerase. The random primer also contains Illumina-compatible linker sequences. Second strand synthesis is followed by a magnetic bead-based purification step. The library is then amplified, introducing the sequences required for cluster generation. External barcodes are introduced during the PCR amplification step. Library quantification is performed by fluorometer (Qubit) and bioanalyzer (Agilent). QuantSeq Forward contains the Read 1 linker sequence in the second strand synthesis primer, hence NGS reads are generated towards the poly(A) tail and directly correspond to the mRNA sequence. QuantSeq FWD maintains strand-specificity and allows mapping of reads to their corresponding strand on the genome, enabling the discovery and quantification of antisense transcripts and overlapping genes. Sequencing is performed on NextSeq500 ILLUMINA instrument to produce 5 million of reads (75 bp SE) for sample. The reads were trimmed using BBDuk (BBMap v. 37.62), with parameters indicated in the Lexogen data analysis protocol. After trimming, reads were aligned to the mouse genome (GRCm38.p6) using STAR (v. 2.7.6a). The gene expression levels were quantified using featureCounts (v. 2.0.1). Genes were sorted removing those that had a total number of counts below 10 in at least 4 samples out of 16. After applying this filter, we identified 12,432 expressed genes that were considered for further analyses. All RNA-seq analyses were carried out in R environment (v. 4.0.0) with Bioconductor (v. 3.7). We computed differential expression analysis using the DESeq2 R package (v. 1.28.1) [92]. DESeq2 performs the estimation of size factors, the estimation of dispersion for each gene and fits a generalized linear model. Transcripts with absolute value of log2[FC] > 0.75 and an adjusted p-value < 0.01 (Benjamini–Hochberg adjustment) were considered significant and defined as differentially expressed for the comparison in the analysis. Heatmaps were made using counts-per-million (CPM) values with the pheatmap function from the pheatmap R package (v.1.0.12; distance = ‘correlation’, scale = ‘row’) on differentially expressed genes or selected markers. Volcano plots were computed with log2[FC] and −log10[q-value] from DESeq2 differential expression analysis output using the ggscatter function from the ggpubr R package (v. 0.4.0).
Statistical analysis
Statistical analysis was performed using Graph Pad Prism software V6.0. Data are presented as the means ± SEM. Comparison between different groups of samples was performed by Student’s t-test with a confidence interval of 95%.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary figures file.
References
Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408.
Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–16.
Masson N, Ratcliffe PJ. HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O2 levels. J Cell Sci. 2003;116:3041–9.
Kaelin WG Jr. The VHL Tumor Suppressor Gene: Insights into Oxygen Sensing and Cancer. Trans Am Clin Climatol Assoc. 2017;2017:298–307.
Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–71.
Rey S, Semenza GL. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res. 2010;86:236–42.
Katsuda T, Teratani T, Chowdhury MM, Ochiya T, Sakai Y. Hypoxia efficiently induces differentiation of mouse embryonic stem cells into endodermal and hepatic progenitor cells. Biochem Eng J. 2013;74:95–101.
Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27:41–53.
Gerri C, Marìn-Juez R, Marass M, Marks A, Maischein H, Stainier DYR. Hif1α regulates macrophage-endothelial interactions during blood vessel development in zebrafish. Nat Commun. 2017;8:15492.
Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–9.
Grillo M, Palmer C, Holmes N, Sang F, Larner AC, Bhosale R, et al. Stat3 oxidation-dependent regulation of gene expression impacts on developmental processes and involves cooperation with Hif-1α. PLoS One. 2020;15:e0244255.
Avalle L, Camporeale A, Campieri A, Poli V. STAT3 in cancer: a double edged sword. Cytokine. 2017;98:42–50.
Tengesdal IW, Dinarello A, Powers NE, Burchill MA, Joosten LAB, Marchetti C, et al. Tumor NLRP3-derived IL-1β drives the IL-6/STAT3 axis resulting in sustained MDSC-mediated immunosuppression. Front Immunol. 2021;12:661323.
Dinarello A, Mills TS, Tengesdal IW, Powers NE, Azam T, Dinarello CA. Dexamethasone and OLT1177 cooperate in the reduction of melanoma growth by inhibiting STAT3 functions. Cells. 2023;12:294.
Carbognin E, Betto RM, Soriano ME, Smith AG, Martello G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016;35:618–34.
Betto RM, Diamante L, Perrera V, Audano M, Rapelli S, Lauria A, et al. Metabolic control of DNA methylation in naive pluripotent cells. Nat Genet. 2021;2021:215–29.
Tesoriere A, Dinarello A, Argenton F. Role of post translational modifications in STAT3 biological activities and function. Biomedicines. 2021;9:956.
Gao W, McCormick J, Connolly M, Balogh E, Veale DJ, Fearon U. Hypoxia and STAT3 signalling interactions regulate pro-inflammatory pathways in rheumatoid arthritis. Ann Rheum Dis. 2015;74:1275–83.
Carlsson R, Özen I, Barbariga M, Gaceb A, Roth M, Paul G. STAT3 precedes HIF1α transcriptional responses to oxygen and oxygen and glucose deprivation in human brain pericytes. PLoS One. 2018;13:e0194146.
Almiron Bonnin DA, Havrda MC, Lee MC, Liu H, Zhang Z, Nguyen LN, et al. Secretion-mediated STAT3 activation promotes self-renewal of glioma stem-like cells during hypoxia. Oncogene. 2018;37:1107–18.
Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–60.
Pimton P, Lecht S, Stabler CT, Johannes G, Schulman ES, Lelkes PI. Hypoxia enhances differentiation of mouse embryonic stem cells into definitive endoderm and distal lung cells. Stem Cells Dev. 2015;24:663–76.
Kučera J, Netušilová J, Sladeček S, Lánová M, Vašíček O, Štefková K, et al. Hypoxia Downregulates MAPK/ERK but Not STAT3 Signaling in ROS-Dependent and HIF-1-Independent Manners in Mouse Embryonic Stem Cells. Oxid Med Cell Longev. 2017;2017:4386947.
Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–9.
Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23.
Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;2:2561–74.
Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–32.
Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344:1156–60.
Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32:1149–60.
Ohtsuka S, Nakai-Futatsugi Y, Niwa H. LIF signal in mouse embryonic stem cells. JAK-STAT. 2015;4:e1086520.
Wulansari N, Sulistio YA, Darsono WHW, Kim CH, Lee SH. LIF maintains mouse embryonic stem cells pluripotency by modulating TET1 and JMJD2 activity in a JAK2-dependent manner. Stem Cells. 2021;39:750–60.
Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG Jr. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol. 2009;29:5729–41.
Walmsley SR, Chilvers ER, Thompson AA, Vaughan K, Marriott HM, Parker LC, et al. Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J Clin Invest. 2011;121:1053–63.
Santhakumar K, Judson EC, Elks PM, McKee S, Elworthy S, van Rooijen E, et al. A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 2012;72:4017–27.
Triantafyllou A, Liakos P, Tsakalof A, Georgatsou E, Simos G, Bonau S. Cobalt induces hypoxia-inducible factor-1alpha (HIF-1alpha) in HeLa cells by an iron-independent but ROS-, PI-3K- and MAPK-dependent mechanism. Free Radic Res. 2006;40:847–56.
Elks PM, Renshaw SA, Meijer AH, Walmsley SR, Van Eeden FJ. Exploring the HIFs, buts and maybes of hypoxia signalling in disease: lesson from zebrafish models. Dis Model Mech. 2015;8:1349–60.
Munos-Sanchez J, Chanez-Cardenas ME. The use of cobalt chloride as a chemical hypoxia model. J Appl Toxicol. 2018;39:556–70.
Gerri C, Marass M, Rossi A, Stainier DYR. Hif-1α and Hif-2α regulate hemogenic endothelium and hematopoietic stem cell formation in zebrafish. Blood. 2018;131:963–73.
Vettori A, Greenald D, Wilson GK, Peron M, Facchinello N, Markham E, et al. Glucocorticoids promote Von Hippel Lindau degradation and Hif-1α stabilization. PNAS. 2017;114:9948–53.
Marchi D, Kirankumar S, Markham E, Li N, Storbeck K, Krone N, et al. Bidirectional crosstalk between Hypoxia-Inducible Factor and glucocorticoid signalling in zebrafish larvae. PLOS Genet. 2020;16:e1008757.
Laquatra C, Sanchez-Martin C, Dinarello A, Cannino G, Minervini G, Moroni E, et al. HIF1α-dependent induction of the mitochondrial chaperone TRAP1 regulates bioenergetic adaptations to hypoxia. Cell Death Dis. 2021;12:434.
Liu Y, Sepich DS, Solnica-Krezel L. Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017;13:e1006564.
Peron M, Dinarello A, Meneghetti G, Martorano L, Betto RM, Facchinello N, et al. Y705 and S727 are required for the mitochondrial import and transcriptional activities of STAT3, and for regulation of stem cell proliferation. Development. 2021;148:dev199477.
Peron M, Dinarello A, Meneghetti G, Martorano L, Facchinello N, Vettori A, et al. The stem-like STAT3-responsive cells of zebrafish intestine are WNT/β-catenin dependent. Development. 2020;147:dev188987.
Dinarello A, Tesoriere A, Martini P, Fontana CM, Volpato D, Badenetti L, et al. Zebrafish mutant lines reveal the interplay between nr3c1 and nr3c2 in the GC-dependent regulation of gene transcription. Int J Mol Sci. 2022;23:2678.
Risato G, Celeghin R, Brañas Casas R, Dinarello A, Zuppardo A, Vettori A, et al. Hyperactivation of Wnt/β-catenin and Jak/Stat3 pathways in human and zebrafish foetal growth restriction models: Implications for pharmacological rescue. Front Cell Dev Biol. 2022;10:943127.
Park JS, Lee J, Lim MA, Kim EK, Kim SM, Ryu JG, et al. JAK2-STAT3 blockade by AG490 suppresses autoimmune arthritis in mice via reciprocal regulation of regulatory T Cells and Th17 cells. J Immunol. 2014;192:4417–24.
Mole DR, Schlemminger I, McNeill LA, Hewitson KS, Pugh CW, Ratcliffe PJ, et al. 2-Oxoglutarate analogue inhibitors of hif prolyl hydroxylase. Bioorg Med Chem Lett. 2003;13:2677–80.
Levy DE, Lee CK. What does Stat3 do? J Clin Investig. 2002;109:1143–8.
Zhang X, Blenis J, Li H, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–4.
Wen Z, Zhong Z, Darnell JE Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50.
Gough DJ, Koetz L, Levy DE. The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and rea-mediated transformation. PLos ONE. 2013;8:e83395.
Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci. 1996;93:3963–6.
Wen Z, Darnell JE Jr. Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucl Acids Res. 1997;25:2062–7.
Elks PM, van Eeden FJ, Dixon G, Wang X, Reyes-Aldasoro CC, Ingham PW, et al. Activation of hypoxia-inducible factor-1α (Hif-1α) delays inflammation resolution by reducing neutrophil apoptosis and reverse migration in a zebrafish inflammation model. Blood. 2011;118:712–22.
Elks PM, Brizee S, van der Vaart M, Walmsley SR, van Eeden FJ, Renshaw SA, et al. Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog. 2013;9:e1003789.
Matthews JR, Sansom OJ, Clarke AR. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ. 2011;18:1934–43.
Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–72.
Fraisl P, Mazzone M, Schmidt T, Carmeliet C. Regulation of angiogenesis by oxygen and metabolism. Dev Cell. 2009;16:167–79.
Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18.
Eyries M, Siegfried G, Ciumas M, Montagne K, Agrapart M, Lebrin F, et al. Hypoxia-induced Apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ Res. 2008;103:432–40.
Haase VH. Hypoxic regulation of erythropoiesis and iron metabolism. Am J Physiol Ren Physiol. 2010;299:F1–13.
Zhang L, Shen G, Liu X, Wang F, Zhao Y, Zhang J. Hypoxia inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J Cell Mol Med. 2012;16:1889–99.
Solak Y, Cetiner M, Siriopol D, Tarim K, Afsar B, Covic A, et al. Novel Masters of Erythropoiesis: Hypoxia Inducible Factors and Recent Advances in Anemia of Renal Disease. Blood Purif. 2016;42:160–7.
Kietzmann T. Hypoxia-inducible erythropoietin expression: details matter. Haematologica. 2020;105:2704–6.
Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci . 2002;99:5454–9.
Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of Gata1 but not Gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–16.
Bresciani E, Confalonieri S, Cermenati S, Cimbro S, Foglia E, Beltrame M, et al. Zebrafish numb and numblike are involved in primitive erythrocyte differentiation. PLoS One. 2010;5:e14296.
Quintana AM, Picchione F, Klein Geltink RI, Taylor MR, Grosveld GC. Zebrafish ETV7 regulates red blood cell development through the cholesterol synthesis pathway. Dis Model Mech. 2014;7:265–70.
Li X, Lu YC, Dai K, Torregroza I, Hla T, Evans T. Elavl1a regulates zebrafish erythropoiesis via posttranscriptional control of gata1. Blood. 2014;23:1384–92.
Lenard A, Alghisi E, Daff H, Donzelli M, McGinnis C, Lengerke C. Using zebrafish to model erythroid lineage toxicity and regeneration. Haematologica. 2016;101:e164–7.
Lewis JS, Lee JA, Underwood JC, Harris AL, Lewis CE. Macrophage responses to hypoxia: relevance to disease mechanisms. J Leukoc Biol. 1999;66:889–900.
Ke X, Chen C, Song Y, Cai Q, Li J, Tang Y, et al. Hypoxia modifies the polarization of macrophages and their inflammatory microenvironment, and inhibits malignant behavior in cancer cells. Oncol Lett. 2019;18:5871–8.
Lewis A, Elks PM. Hypoxia Induces Macrophage tnfa Expression via Cyclooxygenase and Prostaglandin E2 in vivo. Front Immunol. 2019;10:2321.
Sadiku P, Walmsley SR. Hypoxia and the regulation of myeloid cell metabolic imprinting: consequences for the inflammatory response. EMBO Rep. 2019;20:e47388.
Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42.
Walton EM, Cronan MR, Beerman RW, Tobin DM. The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PLoS One. 2020;10:e0138949.
Melcher M, Unger B, Schmidt U, Rajantie IA, Alitalo K, Ellmeier W. Essential roles for the Tec family kinases Tec and Btk in M-CSF receptor signaling pathways that regulate macrophage survival. J Immunol. 2008;180:8048–56.
Kell MJ, Riccio RE, Baumgartner EA, Compton ZJ, Pecorin PJ, Mitchell TA, et al. Targeted deletion of the zebrafish actin-bundling protein L-plastin (lcp1). PLoS One. 2018;13:e0190353.
Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–20.
Oh MK, Park HJ, Kim NH, Park SJ, Park IY, Kim IS. Hypoxia-inducible factor-1alpha enhances haptoglobin gene expression by improving binding of STAT3 to the promoter. J Biol Chem. 2011;286:8857–65.
Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19:1296–8.
Cui Y, Li YY, Li J, Zhang HY, Wang F, Bai X, et al. STAT3 regulates hypoxia-induced epithelial mesenchymal transition in oesophageal squamous cell cancer. Oncol Rep. 2016;36:108–16.
Jung JE, Kim HS, Lee CS, Shin YJ, Kim YN, Kang GH, et al. STAT3 inhibits the degradation of HIF-1alpha by pVHL-mediated ubiquitination. Exp Mol Med. 2008;40:479–85.
Pawlus MR, Wang L, Murakami A, Dai G, Hu CJ. STAT3 or USF2 contributes to HIF target gene specificity. PLoS One. 2013;8:e72358.
Russell RC, Sufan RI, Zhou B, Heir P, Bunda S, Sybingco SS, et al. Loss of JAK2 regulation via a heterodimeric VHL-SOCS1 E3 ubiquitin ligase underlies Chuvash polycythemia. Nat Med. 2011;17:845–53.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310.
Faul F, Erdfelder E, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–60.
Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci. 1997;94:3801–4.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucl Acids Res. 2012;40:e115.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Acknowledgements
We are thankful to Martina Milanetto and to the personnel at the Zebrafish Centre of the University of Padova. We are thankful to Valentina Tonelotto, Davide Volpato, Lorenzo Badenetti, Giada Vanni, Lorenzo Lupi, Saverio Fortunato and Olga Perrotti for their technical support. The study has been supported by the AIRC project IG-2017-19928 to FA G.M.’s laboratory is supported by grants from the Giovanni Armenise-Harvard Foundation, the Telethon Foundation and an ERC Starting Grant (MetEpiStem).
Author information
Authors and Affiliations
Contributions
AD acquisition, analysis, interpretation of data, drafting the work, writing-original draft, writing-review & editing. RMB acquisition, analysis, interpretation of data, drafting the work, writing-review & editing. LD acquisition, analysis, interpretation of data. CC acquisition, analysis. GMe acquisition, analysis, interpretation of data. MP acquisition, analysis. AT acquisition, analysis. RG acquisition, analysis. CL acquisition, analysis. NT funding management. GMa interpretation of data, drafting the work, writing-review & editing. FA interpretation of data, drafting the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dinarello, A., Betto, R.M., Diamante, L. et al. STAT3 and HIF1α cooperatively mediate the transcriptional and physiological responses to hypoxia. Cell Death Discov. 9, 226 (2023). https://doi.org/10.1038/s41420-023-01507-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01507-w