Abstract
Helicobacter (H.) pylori-induced gastritis is a risk factor for gastric cancer (GC). Deleted-in-liver-cancer-1 (DLC1/ARHGAP7) inhibits RHOA, a downstream mediator of virulence factor cytotoxin-A (CagA) signalling and driver of consensus-molecular-subtype-2 diffuse GC. DLC1 located to enterochromaffin-like and MIST1+ stem/chief cells in the stomach. DLC1+ cells were reduced in H. pylori gastritis and GC, and in mice infected with H. pylori. DLC1 positivity inversely correlated with tumour progression in patients. GC cells retained an N-terminal truncation variant DLC1v4 in contrast to full-length DLC1v1 in non-neoplastic tissues. H. pylori and CagA downregulated DLC1v1/4 promoter activities. DLC1v1/4 inhibited cell migration and counteracted CagA-driven stress phenotypes enforcing focal adhesion. CagA and DLC1 interacted via their N- and C-terminal domains, proposing that DLC1 protects against H. pylori by neutralising CagA. H. pylori-induced DLC1 loss is an early molecular event, which makes it a potential marker or target for subtype-aware cancer prevention or therapy.
Similar content being viewed by others
Introduction
Chronic gastritis following infection with the gram-negative bacterium Helicobacter (H.) pylori is a risk factor for gastric cancer (GC) [1], especially by strains positive for the virulence factor cytotoxin-A (CagA) [2]. Among the four consensus-molecularsubtypes (CMS) [3], mutations in RHOA and RHO GTPase-activating proteins (GAPs) are major drivers of diffuse GC (CMS2). These mutations foster constitutive GTPase activities [4], conferring adverse prognosis and therapeutic resistance [5]. Thus, identifying proteins/drugs that inhibit RHO signalling remains an unmet medical need.
Tumour suppressor gene “deleted-in-liver-cancer-1” (DLC1) is frequently silenced in GC owing to methylation or genomic rearrangements [6]. Human [7] and murine [8] DLC1/Dlc1 genes are expressed as multiple mRNA splice variants and protein isoforms. Full-length (FL) DLC1 (DLC1v1, >170 kDa) and N-terminally truncated DLC1v4 (>90 to 130 kDa) share identical multi-domain organisations, containing a sterile-alpha motif (SAM), a serine-rich region, a nuclear localisation sequence (NLS), a caveolae/caveolin-binding motif (Cavbm), followed by RHO GTPase-activating protein (GAP) and steroidogenic acute regulatory protein-related lipid-transfer (START) domains. DLC1 acts on inter-cellular contacts and focal adhesions to alter cell migration and invasion by inducing cytoskeleton reorganisation via GTPases (RHO, RAC, CDC42) and interaction with structural/adapter proteins (catenins, cadherins, caveolins, integrins, talin, tensin, focal adhesion kinase), involved in inflammation, carcinogenesis, and metastasis [6]. DLC1 also functions as a positive predictor of survival and responsiveness to chemotherapy in patients with GC [9, 10].
Based on previous observations that the pathogen-endocytosis receptor caveolin-1 (CAV1) recruits DLC1v4 upon H. pylori infection [11], we hypothesised that DLC1 antagonises CagA to neutralise its pro-inflammatory, cell-damaging oncogenic effector functions. Herein, we found that (i) CagA physically interacted with DLC1, (ii) DLC1 inhibited intracellular CagA signalling, and (iii) CagA downregulated transcription from DLC1v1/4 promoters, proposing DLC1-mediated inhibition of CagA/RHO signalling as a potential therapeutic target.
Results
Correlation between DLC1 expression and clinical factors in patients with GC
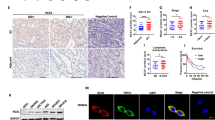
To detect in situ expression of DLC1 protein, we analysed tissue microarrays (TMAs) from patients with GC. Formalin-fixed paraffin-embedded (FFPE) tissue sections from normal/non-tumour stomach (NT: n = 30) and tumour (TU: n = 116) cases were stained by immunohistochemistry (IHC) with DLC1 antibody (Ab) against the N-terminus, including the SAM domain (S1a). Quantitative analyses of graded scores displayed prominent DLC1 staining in intestinal-type, differentiated tumours both in the cytoplasm and at membranes, compared with diffusetype, undifferentiated tumours with marked stroma positivity. Compared with epithelial cells of the non-neoplastic gastric mucosa, DLC1 protein was downregulated in tumour cells (*p < 0.05 vs. NT, Kruskal–Wallis test with Dunn post-tests). No differences were detected by Lauren histology (NT: n = 30; TU: n = 34 diffuse; n = 56 intestinal). DLC1 downregulation could be associated with the grade of tumour dedifferentiation (S1b). DLC1 was also reduced by UICC (S1c) and pTNM staging, as determined by tumour size (T) (S1d) and nodal status (N) (S1e). Conversely, DLC1 was upregulated in cases exhibiting metastases (S1f) (TU: n = 36 M0; n = 11 M1; *p < 0.05 vs. NT, Mann–Whitney test). No significant alterations were recorded in the stroma. Supportive data from independent patient cohorts are provided in Supplementary Results (S2, Tables S3–5).
Expression of DLC1 variants in human GC cell lines
To explore the expression of DLC1 variants in GC [7] (Fig. 1A), the DLC1 gene was examined for COSMIC/GISTIC entries on somatic mutations and copy number variations (Table S6). Common putative driver splice mutations were detected at identical or adjacent codons (aa 438/439/501/522) within the human FL DLC1v1 gene and located N- and C-terminal of the SAM domain where the DLC1v4 CDS (aa 512) begins (Fig. 1B). Cancer hotspot mutations and splice variants were similar in cancer cell lines (CCLE) and pan-cancer patient studies (from EC/GC).
A Scheme of human DLC1 mRNA/protein variants. FL DLC1v1 and the N-terminally truncated DLC1v4 isoform exhibit identical C-terminal multi-domain organisation: sterile-alpha motif (SAM), serine-rich region, nuclear localisation sequence (NLS), caveolin-1 (CAV1)-binding motif (Cavbm), RHO GTPase-activating protein (GAP), and steroidogenic acute regulatory protein-related lipid-transfer (START) domains. DLC1v4 (start codon: MKLEI, aa 513-1528) used in the present study lacks the SAM region compared to DLC1v1 (aa 1-1528). B Comparison of putative driver splice mutations in the human FL DLC1v1 gene located N- and C-terminal of the SAM domain where the DLC1v4 transcript starts. Point mutation data were retrieved from cBioPortal (Table S6) based on COSMIC/GISTIC entries. Cancer hotspots (red) and splice variants (yellow) are similar in human cancer cell lines (CCLE) and pan-cancer studies (from patients with EC/GC). C Detection of DLC1 mRNA variants. Total RNA from human gastrointestinal cancer and non-cancer (HEK293T) cell lines were subjected to RT-qPCR using primers directed against N- and C-terminal regions of the FL DLC1v1 cDNA. Quantitative analyses (top) and representative images from PCR-amplification products upon ethidium bromide-stained agarose gel electrophoresis (bottom). Ct-values were normalised to B2M and calculated as -fold ± S.E. (*p < 0.05 N vs. C, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). D Downregulation of DLC1 mRNA variants in human GC. Total RNA from matched TU and NT tissues of patients with GC (n = 5 cases) was subjected to RT-qPCR using primers directed against N- and C-terminal regions of FL DLC1 cDNA. Quantitative analyses (top) and representative gels (bottom). Ct-values were normalised to B2M and calculated as -fold ± S.E. (*p < 0.05 TU vs. NT, 2way-ANOVA with Bonferroni post-tests). E Detection of endogenous (left, source file: “Original Western Blots” page 2) and ectopic (right, source file: page 3) DLC1 protein variants. Cells (HEK293T, AGS) were transfected with EV or DLC1 expression plasmid for 48 h, followed by extraction of total cell lysates. FL DLC1v1 (170 kDa) was detected by an Ab specific for the N-terminus/SAM + domain of the protein (PA5-18290), ΔSAM DLC1v4 (110 kDa) by anti-FLAG Ab. Representative images from Western blots. Note the presence of endogenous DLC1 (130–170 kDa) in GC cells detected by the Ab directed against the N-terminal part of DLC1, including the SAM domain.
To measure DLC1 mRNA in human gastrointestinal cancer and non-cancer (HEK293T) cell lines, primers were designed against N- and the C-terminal parts of the FL DLC1 cDNA. Consistent with previous data [11], RT-qPCR analyses (Fig. 1C) revealed that FL DLC1v1 was present in HEK293T, whereas truncated DLC1v4 was detectable in all cancer cell lines examined (*p < 0.05 N vs. C, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line).
RT-qPCR analyses of total RNA extracted from frozen samples of matched normal NT and TU tissues from patients with GC (Fig. 1D) confirmed that DLC1v1 mRNA was reduced by ~40% compared with controls (*p < 0.05 TU vs. NT, 2way-ANOVA with Bonferroni post-tests, n = 5 cases).
Similar results were observed at the protein level (Fig. 1E). We amplified the DLC1v4 (start codon: MKLEI, aa 513-1528) CDS lacking the SAM region when compared with FL DLC1v1 (aa 1-1528) [7]. The cDNAs were inserted into FLAG-tagged expression vectors and transfected into HEK293T or AGS cells for 48 h. Western blotting confirmed overexpression of FL DLC1v1 (170 kDa) and N-terminally truncated ΔSAM DLC1v4 (110 kDa). A smaller sized, endogenous DLC1 protein (130–170 kDa) was detected in GC cell lines (AGS, MKN45, KATO3, N87) using an Ab specific for the N-terminal part, including the SAM domain, but not in HEK293T or SNU1, indicative of cell line-specific isoforms.
H. pylori and CagA downregulate DLC1 expression in vivo
As DLC1 is frequently downregulated in GC, we speculated whether H. pylori infection contributes to human or mouse DLC1/Dlc1 gene silencing. FFPE tissue sections from endoscopic gastric biopsies of naïve and infected individuals were stained by IHC using DLC1 Ab (Fig. 2A). After dichotomous grouping as negative (scores 0/1) vs. positive (scores 2/3) cases, DLC1 staining was correlated to the infection status (n = 9–10 per group). Quantitative analyses of the corpus region exhibited the disappearance and displacement of DLC1 + cells by leucocyte infiltration into inflammatory regions (Fig. 2B). DLC1+ cells accumulated in the periphery of inflammatory follicles. This distribution pattern proposed that H. pylori gastritis leads to downregulation of DLC1 positivity in the stomach.
A DLC1 positivity in uninfected and H. pylori-infected human gastric biopsies. Sections from FFPE human stomach tissue (n = 9 Hp+ vs. n = 10 Hp- cases) from endoscopic material were stained with DLC1 Ab by IHC. Positivity scores for DLC1 were correlated to the infection status. Data are absolute case numbers after dichotomous grouping of stainings as negative (scores 0/1) vs. positive (scores 2/3) (n.s., Hp+ vs. Hp-, Fisher-Exact test). B H. pylori-induced inflammatory cell infiltrates outcompete DLC1+ cells in human gastric biopsies. Representative IHC images from (A) of the corpus region (left, healthy; right, gastritis). Colour code: Brown = DLC1; Blue = nuclei (hematoxylin). Original magnifications ×200; Scale bar = 50 µm. Note the displacement of DLC1+ cells by accumulating intramucosal leucocytes in inflammatory regions. C H. pylori downregulates DLC1+ positivity in vivo. C57BL/6J mice were infected with mouse-adapted CagA+ injection-proficient H. pylori PMSS1 WT strain or CagA-injection-deficient isogenic mutant ΔCagE for 1 or 6 months. Sections from FFPE gastric tissue were stained with Abs against chief/stem cell marker MIST1 or DLC1. Quantitative analyses (top) and representative IHC images of the corpus region (bottom). Data are cell numbers per mm2 (*p < 0.05 vs. WT, Kruskal–Wallis test with Dunn or Sidak post-tests, n = 2–9 mice per group or time point). Colour code: Brown = DLC1/MIST1; Blue = nuclei (hematoxylin). Original magnifications ×100; Scale bar = 100 µm.
To explore the impact of H. pylori virulence factors on DLC1 expression, C57BL/6J mice were infected for 1 and 6 months with the mouse-adapted CagA+ injection-proficient H. pylori PMSS1 WT strain or isogenic ΔCagE mutant, and DLC1 was visualised by IHC. The CagE mutant cannot translocate CagA into epithelial cells, as CagE is an essential protein for the functionality of the type IV secretion system (T4SS) [2]. Notably, PMSS1 WT reduced the numbers of DLC1+ cells in the stomach of infected mice when compared with uninfected controls (*p < 0.05 vs. WT, Kruskal–Wallis test, n = 2–9 mice per group or time point); however, no changes in the numbers of DLC1+ cells were observed in the absence of CagA translocation (Fig. 2C).
To determine if DLC1 localises to gastric cell types other than ECL cells, the putative stem/chief cell marker MIST1 was examined. Supportive immunofluorescent (Fig. 3A) stainings revealed a scattered distribution of DLC1+ cells in the vicinity of abundant MIST1+ cells at the base of the gland, presenting single double-positive cells, suggesting partial colocalisation. Compared with DLC1+/CHGA+ ECL cells and DLC1+/MIST1+ stem/chief cells, parietal cells showed no colocalisation (Fig. 3B). Notably, after 1 and 6 months of infection, both DLC1+ and MIST1+ cell counts were reduced in PMSS1 WT infected animals vs. littermates receiving the isogenic CagE-deficient mutant (after 1 month) or were uninfected (Fig. 2C), indicating a negative impact of CagE/T4SS-mediated injection of CagA on the expression of both markers.
A Partial colocalisation of DLC1 with MIST1+ stem/chief cells. Detection of DLC1 protein in murine gastric chief/stem cells. FFPE sections from stomachs of mock (control) and WT PMSS1-infected (6 months) mice (from Fig. 2C) were co-stained using DLC1 and MIST1 Abs for immunofluorescence microscopy. Quantitative analyses (left) and representative images (right) of the corpus region. Data are numbers of double-positive cells per mm2 presented as mean (n = 2–9 uninfected control mice per time point, n.c. = no double pos. cases detectable in infected mice). Colour code: Green = DLC1; Red = MIST1; Blue = nuclei (DAPI); original magnifications ×200; Scale bar = 20 and 100 µm. White arrow/frame: Yellow = zoomed-in overlay (DLC1+/MIST1+). Insert (top left): Representative images of PCR-amplification products and murine DLC1 protein isoforms (GC: 70 kDa vs. NT: 170 kDa) from total tissue lysates detected by Western blot using Abs against the N-terminal part of DLC1 including the SAM domain (source file: “Original Western Blots” page 4). Legend: STO = Stomach (Corpus); L = Liver (pos. control); GC = gastric cancer. B Colocalisation of DLC1 with ECL cells, but not with parietal cells. FFPE sections from stomachs of uninfected mice were co-stained using DLC1, chromogranin A (CHGA), and H+K+ATPase (HK) Abs for immunofluorescence microscopy. Quantitative analyses from images of the corpus region presented in S3. Data are numbers of single or double-positive cells per field presented as mean (*p < 0.05, Kruskal–Wallis test, n = 2–9 uninfected control mice, n ≥ 5 fields per image). C Scheme of promoters identified in the human DLC1 gene according to Low et al. [7]. Note that the promoters generate four different transcripts/protein isoforms with identical C-terminus but distinct N-termini. Legend: E = Exon. D H. pylori downregulates DLC1 mRNA. AGS cells were infected with CagA+ injection-proficient H. pylori strain G27 (MOI = 100) for 3 days. Ct-values from RT-qPCRs on total RNA were normalised to B2M and calculated as % ± S.E. (*p < 0.05 vs. mock, 2way-ANOVA with Bonferroni post-tests, n = 3). E CagA is sufficient for DLC1 promoter inhibition. Cells (HEK293T, AGS, N87) were co-transfected for 72 h with EV or CagA expression plasmid and reporter vector pGL3 containing the human proximal promoter sequences of DLC1v1 or v1. Luciferase activity was normalised to protein concentration and calculated as -fold ± S.E. (*p < 0.05 vs. EV, one-sample t test, n = 3 per cell line).
Conclusively, H. pylori CagA downregulated DLC1 in the basal stem cell niche of the gastric gland, which gives rise to both chief and ECL lineages, thus explaining the presence of DLC1 in both cell types. Supportive data on the expression of DLC1 in ECL cells are provided in Supplementary Results (S3).
H. pylori and CagA downregulate DLC1 expression in vitro
An internal promoter is located upstream of exon 9 to allow transcription of N-terminally truncated DLC1v2/4 mRNAs [7]. Binding motifs for stress-sensitive transcription factors were predicted to bind this promoter (e.g. p53, STAT, HSF), factors known to be also activated by H. pylori infection (Fig. 3C).
As shown in Fig. 1, AGS cells expressed DLC1v4 but no FL DLC1v1 mRNA, making this cell line suitable to study the effect of CagA on this promoter. AGS cells were infected with the CagA+ injection-proficient H. pylori strain G27 (MOI = 100) for 3 days (Fig. 3D). RT-qPCRs revealed a reduction of DLC1v4 mRNA by ~90% when compared with controls (*p < 0.05 vs. mock, 2way-ANOVA with Bonferroni post-tests, n = 3).
To confirm whether H. pylori CagA is causal for inhibiting DLC1 mRNA transcription, we amplified the promoters [7] of DLC1v1 and v4, followed by the insertion of genomic DNA sequences into the pGL3 reporter vector. AGS, N87, and HEK293T cells were transfected for 72 h with EV or GFP-CagA-FL expression plasmid, along with pDLC1v1 or v4 reporter constructs. CagA reduced luciferase activity of both promoters when compared with control (HEK293T: >95%; AGS: >95%; N87: 10–60%; *p < 0.05 vs. EV, one-sample t test, n = 3 per cell line) (Fig. 3E).
DLC1 counteracts cell phenotypes evoked by CagA
Based on the observed CagA-mediated DLC1 downregulation, we hypothesised that, vice versa, DLC1 inhibits CagA signalling. To explore the mutual antagonism of the two proteins, we examined whether DLC1 prevents CagA-mediated cytoskeleton rearrangements, detectable as spike-like cell elongations, termed the “humming bird” phenotype [2].
AGS and HEK293T were transfected with EV, CagA, or DLC1 expression plasmids or a combination thereof for 36 h, followed by fixation and staining using DLC1 or FLAG Ab for immunofluorescent microscopy (Fig. 4A). Numbers of adherent cells normalised to total cell counts were increased (HEK293T: *p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). DLC1 was localised to the periphery of spread-out cells and in focal adhesions (DLCv1) or neurite-like extensions (DLC1v4), whereas CagA was targeted to the plasma membrane and promoted needle-like cell elongations (“humming bird”). In cells receiving both plasmids, DLC1 reduced CagA-dependent formation of stress spikes (AGS: *p = 0.0374, Fisher-Exact test; n = 3) (Fig. 4B). Similar results were obtained for AGS (S4) and HEK293T (S5) cells. Supportive data on DLC1v1/4 effects on morphologies are provided in Supplementary Results (S6–8).
Cells (AGS, HEK293T) were transfected with EV, CagA, or DLC1 expression plasmids or a combination thereof for 36 h, followed by fixation and staining using DLC1 and FLAG Abs for immunofluorescence microscopy. Quantitative analyses (top) and representative images (bottom). Numbers of adherent cells with morphologies (spread-out, round, elongated) were counted, normalised to total cell counts and calculated as % ± S.E. (A, HEK293T: *p < 0.05 vs. EV (t = 0), 2way-ANOVA Bonferroni post-tests, n = 3 per cell line; B AGS: *p = 0.0374, Fisher-Exact test; n = 3). Colour code: Red = DLC1; Green = CagA (GFP); Yellow = DLC1/CagA (overlay); Blue = nuclei (DAPI); Original magnifications ×400 (zoomed-in 630×); Scale bar = 20 µm. White frames/arrows: DLC1 promotes neurite-like extensions (abbrev. “nl”) (DLC1v4+) or localises to focal adhesions (abbrev. “fa”) (DLC1v1+); membrane-bound (abbrev. “mb”) CagA forms needle-like elongations (“humming bird”).
DLC1 inhibits CagA-mediated oncogenic signalling
Downstream signalling cascades were assessed to elucidate molecular mechanisms on how CagA and DLC1 functionally antagonise each other. AGS cells were transfected with EV or DLC1v1 for 24 h, followed by serum-deprival for 16 h, and restimulation with FCS (20% v/v) or EGF (50 ng/ml) for the indicated times. Western blotting (S9a) demonstrated that DLC1v1 augmented phosphorylation of MAPKs (p38, JNK1, ERK1/2: *p < 0.05 vs. EV, 2way-ANOVA, n = 3 per cell line) but not of AKT or FAK (not shown).
Next, we examined whether DLC1v1-mediated activation of stress/growth-related kinases is translated to the nucleus towards c-FOS transcription, a cell proliferation surrogate (Fig. 5A). HEK293T, AGS and N87 cells were co-transfected for 72 h with EV, DLC1v1, or CagA expression plasmids or combinations thereof, along with pGL3 reporter containing the SRE from the human c-FOS gene promoter. Compared with CagA-transfected controls, luciferase activity was reduced by DLC1v1 to 10% (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line).
A DLC1 inhibits c-FOS promoter activity. Cells (HEK293T, AGS, N87) were co-transfected for 72 h with EV, DLC1v1, or CagA expression plasmids or combinations thereof along with pGL3 reporter vector containing the SRE from the human c-FOS gene promoter. Luciferase activity was normalised to protein concentration and calculated as -fold ± S.E. (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). B DLC1 inhibits HMGB2 promoter activity. Cells were transfected as in (A), along with pGL3 reporter containing the HRE from the human HMGB2 gene promoter. Luciferase activity was normalised to protein concentration and calculated as -fold ± S.E. (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). C DLC1 inhibits RHO-driven promoter activity. Cells were transfected as described above, with EV, DLC1v1/4, or CagA expression plasmids, along with pSRE.L reporter plasmid containing an RHO-driven modified SRE. Luciferase activity was normalised to protein concentration and calculated as -fold ± S.E. (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). D DLC1 inhibits CagA-driven activation of RHOA. TsA201 (top, source file: “Original Western Blots” page 5–9) and AGS (bottom, source file: page 10–14) cells were co-transfected for 72 h with EV, DLC1v1/4, or CagA expression plasmids or combinations thereof. Total cell lysates were subjected to GST-RHOA pulldown assays and Western blotting to detect active and total (input) RHOA, respectively. Representative images are shown.
H. pylori triggers oxidative stress by releasing reactive oxygen species from infected host cells [12]. To determine whether DLC1v1 interferes with hypoxia-driven stress responses (Fig. 5B), cells were transfected as above with a pGL3 reporter containing the HRE from the promoter of the human HMGB2 gene [13]. DLC1v1 decreased luciferase activity to 20% when compared with CagA-transfected controls (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line). Overall, FL DLC1v1 protected host cells from CagA-mediated damaging and oncogenic effects.
DLC1 inhibits RHOA via its RHO-GAP domain [6], whereas CagA activates RHOA [14]. We thus examined whether DLC1 inhibits CagA-driven RHOA activity. Cells were transfected as before with EV, DLC1v1/4, or CagA plasmids, along with a modified pSRE.L reporter [15] containing an RHO-driven SRE deficient for the TCF-binding site in the human c-FOS promoter (Fig. 5C). Both DLC1 isoforms reduced RHO-driven luciferase activity compared with CagA-transfected controls. Moreover, DLC1 counteracted RHO-driven reporter gene expression in presence of CagA overexpression (*p < 0.05 vs. EV, 2way-ANOVA with Bonferroni post-tests, n = 3 per cell line).
Specificity was verified by employing an RHOA-activating plasmid expressing a constitutively active α subunit of heterotrimeric G-protein α13 (G13qL) [16] and an RHO-inhibiting plasmid encoding for C. botulinum C3 toxin (C3T) [17] targeting RHO. C3T abolished CagA-mediated activation of the RHO-driven SRE, whereas DLC1-mediated inhibition of the SRE pertained in presence of G13qL (S9b).
GST-pulldown assays confirmed these findings using recombinant RHO-binding domain of rhotekin as bait for binding active RHOA (Fig. 5D). TsA201 cells were co-transfected with EV, DLC1v1/4, or CagA plasmid or combinations for 72 h. Total cell lysates were subjected to pulldown assays and Western blotting to detect active and total (input) RHOA. CagA increased G-protein-coupled RHOA activation, an event inhibited by both DLC1 isoforms when compared with input controls. Consistently, RHOA specificity was verified by C3T and G13qL for RHOA pulldown. C3T inhibited CagA- and G13qL-mediated RHOA activation (S9c).
DLC1 interacts with and neutralises CagA
We inferred that DLC1 neutralises oncogenic and cell-damaging effects of CagA by physically interacting with the two proteins. Several CagA deletion constructs were generated as described previously (Fig. 6A) [18]. CagA-FL (1-1216 aa) plasmid was complemented by two C-terminal fragments, CagA-C (838-1216 aa) and CagA-CT (1029-1216 aa), and the N-terminal fragment, CagA-NT (1-877 aa). CagA-CT and CagA-NT did not contain EPIYA motifs and multimerisation domains. DLC1 plasmids included FL DLC1v1 and truncated DLC1v4 without SAM domain [7]. DLC1 constructs were FLAG-tagged, CagA mutants fused to GFP.
A Scheme of DLC1 and CagA expression constructs. Legend: SAM = sterile-alpha motif, RHO-GAP = RHO GTPase-activating protein (catalytic domain for RHO inhibition); START = StAR-related lipid-transfer (domain), Cav1bm = caveolin-1 binding motif, EPIYA = Src kinase tyrosine phosphorylation motif; grey = multimerisation domain. B, C CagA interacts with DLC1. HEK293T cells were transfected with CagA-FL (aa 1-1216), CagA-NT (aa 1-877), or CagA-C (aa 838-1216), combined with DLC1v1 or v4 expression plasmids, respectively. CoIP was performed on cell lysates using Abs against CagA, DLC1 (v1) and FLAG (v4). Quantitative analyses (B) and representative images (C) from Western blots (Source file: “Original Western Blots” page 15–17). OD values from gels were normalised to ERK1/2 (as loading control) and presented as -fold ± S.E. (*p < 0.05 vs. bead control, 2way-ANOVA with Bonferroni post-tests, n = 3 per plasmid). D HEK293T cells were transfected with CagA-FL (aa 1-1216) in combination with DLC1v1 or v4 plasmids. Representative images from PLA microscopy. Colour code: Red dots = DLC1/CagA complex; Green = CagA (GFP); Blue = nuclei (DAPI); original magnifications ×630; Scale bar = 10 µm. Note accumulation of DLC1+ PLA complexes in CagA-GFP+ cell areas (yellow = overlay). E Model of DLC1 and CagA cross-talk in GC. Legend: NT = non-tumour healthy epithelial (“stem”) cell; GC = gastric cancer cell. Normal cells express FL DLC1v1 protein in a “closed” conformation mediated by intra-molecular interaction between the N- and the C-terminus, tethering it to focal adhesion complexes and cellular junctions. Tumour cells express N-terminally truncated DLC1v4 protein lacking this intra-molecular interaction, resulting in an “open” conformation recognised by CagA and promoting stress and malignancy-related cell phenotypes. H. pylori infection may evoke loss (LOF) of DLC1v1 and gain (GOF) of DLC1v4 isoforms over time (Δt) during chronic infection.
CoIP experiments were performed to verify complex formation between the two proteins. HEK293T cells were transfected with CagA-FL combined with DLC1v1 or v4 plasmids. IP was performed on cell lysates using Abs against CagA or DLC1 (Fig. 6B, C). Western blotting corroborated the interaction between CagA-FL and both DLC1 isoforms. We observed that DLC1v4 was more efficiently precipitated than DLC1v1 (*p < 0.05 vs. bead control, 2way-ANOVA with Bonferroni post-tests, n = 3 per plasmid). To determine which CagA domains are necessary for the interaction, cells were transfected with CagA-NT or CagA-C in combination with DLC1v1 or v4 plasmids, respectively. Both N- and C-terminal parts of FL CagA interacted with the two DLC1 isoforms. Again, DLC1v4 was more efficiently precipitated than DLC1v1 (*p < 0.05 vs. bead control, 2way-ANOVA with Bonferroni post-tests, n = 3 per plasmid). PLA microscopy confirmed the proximity of the two proteins. Oligonucleotide-labelled secondary Abs generated a red fluorescent signal on colocalisation of GFP-CagA and DLC1 (Fig. 6D). All examined CagA constructs interacted with both DLC1 isoforms (S10).
Collectively, these findings suggested that DLC1 binds and neutralises CagA in vitro [11]. Supportive data on phenotypes of Dlc1-deficient mice are shown in Supplementary Results (S11-15), confirming that DLC1 also prevents gastric inflammation in vivo.
Discussion
Herein, we demonstrated that the RHO GTPase inhibitor DLC1 prevented gastric inflammation in vivo and counteracted the oncogenic actions of CagA, a major virulence factor of H. pylori and a risk factor for GC [14]. Mechanistically, we provided evidence indicating mutual functional antagonism of the two proteins, wherein DLC1 inhibits CagA-dependent RHO signalling and, vice versa, CagA prevents transcription from the DLC1 promoter, thereby lowering DLC1 protein in living cells.
This cross-talk (Fig. 6E) was driven by protein-protein interaction mediated via the N- and C-terminal CagA domains and the internal/C-terminal regions of FL DLC1v1 and ΔSAM DLC1v4. The N-terminal membrane domain of CagA inhibits the signalling activity of the C-terminal EPIYA motifs [18], and the N-terminal SAM domain in DLC1 inhibits the enzymatic activity of the RHO-GAP domain [19]. Hence, steric competition between these mutual intra- and inter-molecular regulatory protein interactions determines the outcome of signalling activities for both proteins. Notably, N-terminally truncated DLC1v4 protein, retaining the C-terminal RHO-GAP and START domains such as the caveolin-binding motif (Cavbm), was recruited to caveolin-rich areas of the plasma membrane by H. pylori [11]. Moreover, this truncation variant was present after transformation, whereas FL DLC1v1 was lost in GC cells. Thus, DLCv4, lacking the auto-inhibitory N-terminal domain and/or harbouring other unidentified mutations [20], may gain aberrant oncogenic function in gastric cells, a feature which could explain the observed enrichment of DLC1 mRNA/protein in diffuse-type (CMS2) and advanced stages of GC with distant metastases [6]. Consistently, therapeutic response in patients with GC has been correlated to DLC1 expression [9, 10].
Genomic alterations in DLC1 contribute to drug sensitivity and confer growth advantage upon activation of RHO-ROCK signalling, rendering tumour cells more susceptible to ROCK-inhibitors (e.g., Y-27632). Low DLC1 expression also predicts poor therapeutic efficiency of fluoropyrimidine/oxaliplatin in adjuvant chemotherapy and was associated with advanced, metastatic disease and shortened time to recurrence and overall survival. Likewise, ROCK-inhibitor fasudil [21] attenuated murine GC growth in vivo. Joshi et al. have designed cell-permeable peptides derived from the SAM domain of DLC1 [22], which destroy the intra-molecular auto-inhibition between the SAM and the RHO-GAP domains. Thereby, the peptides trigger a conformational reactivation of DLC1, from a “closed” towards an “open” state (Fig. 6E), allowing recruitment and interaction with potent tumour suppressors like PTEN, culminating in decreased RHOA activation, tumour cell growth and migration in vitro, proposing DLC1 as a druggable target.
To date, DLC1 has not been functionally connected with H. pylori in patients or animal models, although gene hypermethylation was detected upon infection [23]. Similar to humans [7], at least four murine splice/protein variants have been identified [8]. Gene trapped Dlc1gt/+ mice harbour an insertion between exon 1 and 2 of the 6.1 kb transcript and exhibit developmental defects and disease-prone phenotypes [8, 24, 25]. However, Dlc1gt/+ mice do not per se progress to cancer, suggesting that cooperative damage cues or oncogenic mutations are required for transformation. Consistently, we showed that Dlc1-deficient animals exhibit gastritis and disturbed expression of neuro-entero-endocrine hormones, characterised by moderate leucocyte infiltration and enhanced proliferation of epithelial cells, indicating an early step in the transition from inflammation to malignancy (S15). The highest in situ expression of DLC1 protein both in mouse and human gastric tissue was found in ECL cells [26], known to produce a majority of the gut peptide and histidine/tryptophan-derived hormones [27]. Therefore, DLC1 may play an additional role in stomach homeostasis via auto/paracrine regulation of secretory factors, in addition to its well-established function in epithelial cells as a cytoskeleton regulator. H. pylori infection correlates with deterioration of the integrity and secretory activity of ECL cells (e.g. somatostatin producing D cells [28]) and hypergastrinemia (by gastrin producing G cells [26]), resulting in altered histamine release and subsequent gastric acid secretion from adjacent parietal cells in the oxyntic mucosa and pre-neoplastic alterations in humans and animal models. Consistent with this sequence of events, Dlc1gt/+ mice exhibited reduced levels of several ECL hormones, primarily somatostatin [29], which counteracts the pro-angiogenic and trophic effects of gastrin [27]. However, further in-depth studies need to clarify the causality, considering the unidentified stem cell origin of gastric adenocarcinoma and neuroendocrine tumours (carcinoids).
According to Lauren’s classification, H. pylori can be associated with intestinal GC, via the “Correa” sequence of histomorphological changes, and with diffuse GC [2], in addition to sporadic mutational events causative for this subtype. The current CMS classification [3] highlights the function of RHO signalling in diffuse (CMS2) GC. This is underscored by the fact that several mutations in RHOA and RHO GAP (e.g. CLDN18-ARHGAP6 or CTNND1-ARHGAP26 fusions) genes, to which DLC1 (also termed ARHGAP7) also belongs, were identified as oncogenic drivers for diffuse GC [3, 4]. Therefore, we propose that loss of the RHO-inhibitor DLC1 correlates with H. pylori infection in certain GC subtypes. Despite the declining incidence of intestinal GC, that of diffuse GC continues growing and is characterised by poor prognosis and rapid metastasis. Thus, there is an urgent medical need to specifically address this GC subtype. Consistently, our data propose that novel subtype-specific treatment strategies could be developed for RHOA inhibition.
Materials and methods
Subjects
All patients provided written informed consent. Studies were performed in accordance with the principles of the Declaration of Helsinki and approved by the Medical Ethics Committees of the Universities of Heidelberg, TU München and Kiel. Patients’ tissue specimens were collected after surgical resection, stored and classified by Lauren’s histological classification. Tissue microarrays (TMAs) were custom-made [30] or purchased from US Biomax (Rockville, MD).
Animals
Tissue biobanks from C57BL/6J mice infected with H. pylori strains SS1 [11] or PMSS1 [31] were generated previously. Dlc1gt/+ mice [8] were bred in a pathogen-free mouse facility. Animal studies were conducted in agreement with animal welfare guidelines and approved by the governments of Bavaria and Baden-Württemberg (55.2-1-54-2531-74-08/-99-10; -2532-155-12; 35-9185.81/G-146/15; 55.2-1-55-2532-196-2016).
DNA-constructs
The ~1 kb fragment of the proximal human DLC1v1 (NC_018919.2; position 135.59.304–135.60.451) promoter [7] was amplified by PCR from genomic DNA of normal human liver and inserted into the KpnI/SacI sites of pGL3-luc luciferase reporter plasmid (Promega, Mannheim, Germany). The upstream ~0.6 kb region of the proximal human DLC1v4 (NG_015998.1; position 242.923-243.520) promoter [7] was amplified as detailed above and inserted into the KpnI/HindIII sites of pGL3-luc. Isoform 1 of FL human DLC1 cDNA (NM_182643, start aa 1 MSVAI, DLC1v1) was obtained in pCMV-SPORT6.1 (Open Biosystems, Thermo Fisher Scientific, Waltham, MA) and subcloned into the BamHI/NotI sites of mammalian expression vector pTarget (Promega). Isoform 4 of human DLC1 cDNA (NM_001164271.1; ΔSAM, start aa 513 MKLEI, DLC1v4) [7] was amplified from total RNA isolated from human hepatoma HepG2 cells and inserted into pTarget, as described above. Both DLC1 cDNA variants were FLAG-tagged on their respective N-termini. The pEGFP fusion constructs with CagA-FL, CagA-N (aa 1-877) and CagA-C (aa 838-1216), and pTRE-Tight with GFP-tagged CagA-CT (aa 1029-1216) were described previously (both from BD Biosciences, San Jose, CA) [18]. Firefly luciferase reporter gene plasmids were pHRE-luc [13] driven by the human HMGB2 promotor in pGL3 (Promega), pSRE-luc (Stratagene/Agilent, La Jolla, CA) and pSRE.L-luc containing a mutated TCF-binding-deficient SRE from the human c-FOS promotor [15], and renilla luciferase control plasmid pR.L-TK-luc (Promega). Expression plasmids for RHO pulldown assays were GST-tagged RHO-binding domain (aa 7-89) of rhotekin in pGEX-2T (Merck/Sigma), C3 toxin of Clostridium botulinum (C3T) [17], and G-protein α subunit 13 (G13qL) [16] in pcDNA3 (Thermofisher).
Reagents
Chemicals were from Merck/Sigma (Darmstadt, Germany). Primary antibodies (Abs) are listed in Table S1. Secondary Abs were anti-mouse or rabbit IgG conjugated to Alexa Fluor® 488/594 (Thermofisher) or ECL™ HRP (GE Healthcare, Stafford, UK).
Cell culture
Human TsA201 cells were purchased from the European Collection of Authenticated Cell Cultures (ECACC, Salisbury, UK); MKN45 from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany); HEK293T and other human cancer cell lines (Gastric: AGS, N87, MKN45, SNU1, KATO3; Colorectal: SW480; Liver: HepG2) from the American Type Culture Collection (ATCC, Rockville, MD). All lines were maintained as suggested by the distributors. Transient transfection, reporter gene and cellular assays are described in Supplementary Methods.
Bacterial culture and infection of mammalian cells
H. pylori wildtype (WT) SS1 (Sydney strain, CagA+ injection-deficient, mouse-adapted), PMSS1 (pre-mouse Sydney strain, CagA+ injection-proficient, mouse-adapted), and its CagA-injection-deficient isogenic mutant (ΔCagE) or G27 (clinical isolate, CagA+ injection-proficient, cell culture-adapted) were recovered from −80 °C glycerol stocks and grown on blood agar plates under microaerobic conditions as detailed in Supplementary Methods [11, 31]. Bacteria were harvested from agar plates, propagated on AGS cell monolayers for in vitro experiments or animal inoculations.
Reverse transcription PCR (RT-PCR) and quantitative PCR (qPCR)
Total RNA was extracted from cells or frozen tissues with RNeasy™ Mini Kit (Qiagen, Hilden, Germany) followed by cDNA synthesis (Verso™ cDNA Kit, Thermofisher). Two-step PCR (95 °C 15 s, 60 °C 1 min, 40 x cycles) was performed using Power SYBR® Green Master Mix and ABI PRISM Real Time 7900HT device (Thermofisher). Relative mRNA expression was calculated with the ∆∆Ct method [32]. Oligonucleotides are listed in (Table S2) [33].
RHO pulldown, co-immunoprecipitation (CoIP) and Western blotting
GTPase activity (pull-down) assays were performed as detailed by the manufacturer (Biozol/Cell Biolabs Inc., San Diego, CA) or custom-made [16]. Procedures are presented in Supplementary Methods [11].
Immunofluorescence
Proximity ligation assay (PLA) was conducted as recommended by the manufacturer (Duolink® In Situ Red Starter Kit Mouse/Rabbit, Merck/Sigma). Staining protocols for cell lines and formalin-fixed and paraffin-embedded (FFPE) tissue sections are given in Supplementary Methods. Slides were visualised in triple-colour mode using a digital camera-connected (Axiovision, release 4.4) fluorescence microscope (Axiovert 200M, Carl Zeiss MicroImaging, Hallbergmoos, Germany). Single or double-positive cells (n > 5 per field; n = 5 fields per image) were quantified with ImageJ.
Immunohistochemistry (IHC)
Procedures for Ab (Table S1) staining on FFPE tissue sections were performed as described by the Vectastain ABC kit (Vector Labs, Burlingame, CA) using 3,3-diaminobenzidine (DAB), followed by hematoxylin counterstaining. Frequency and intensity of positivity [30] were determined in epithelial and lamina propria (stroma) cells [Scores: 0+= negative (0–25%), 1+= weak (25–50%), 2+= moderate (50–75%), and 3+= strong (75–100%)]. Percentages were calculated as positive cells divided by the total number of cells per field. Signals were quantified by a blinded observer using a bright-field microscope (n > 10 cells per field; n = 5 fields per image) or BX61VS slide scanner (Olympus, Hamburg, Germany). Field areas were defined by the morphology of a given anatomical structure, such as the foveolar-neck-base unit, and measured in mm2.
Statistics
Sample size estimates (POWER 0.8) for animal experiments were calculated with SAS (Cary, NC). Results are displayed as means ± standard error (S.E.) from at least three independent experiments (n ≥ 3) from different cell passages or individuals (mice, patients). Optical densities (ODs) of bands in gels from Western blots and PCRs were measured using automated imaging devices and quantified with ImageJ (Washington, DC). Data were normalised to house-keeping genes or proteins and calculated as -fold change or % when compared with control. Statistical analyses were performed with GraphPad Prism (La Jolla, CA). Data were tested for non- vs. parametric distribution and equal vs. unequal variance, followed by the recommended test with correction for multiple comparisons. Tests were 2-sided and unpaired if not stated otherwise in the legends to figures. P-values < 0.05 were considered significant (*).
Data availability
For all data requests, please contact the corresponding author.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249.
Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–14.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–7.
Yoon C, Cho SJ, Aksoy BA, Park DJ, Schultz N, Ryeom SW, et al. Chemotherapy resistance in diffuse-type gastric adenocarcinoma is mediated by RhoA activation in cancer stem-like cells. Clin Cancer Res. 2016;22:971–83.
Zhang Y, Li G. A tumor suppressor DLC1: the functions and signal pathways. J Cell Physiol. 2020;235:4999–5007.
Low JS, Tao Q, Ng KM, Goh HK, Shu XS, Woo WL, et al. A novel isoform of the 8p22 tumor suppressor gene DLC1 suppresses tumor growth and is frequently silenced in multiple common tumors. Oncogene. 2011;30:1923–35.
Sabbir MG, Wigle N, Loewen S, Gu Y, Buse C, Hicks GG, et al. Identification and characterization of Dlc1 isoforms in the mouse and study of the biological function of a single gene trapped isoform. BMC Biol. 2010;8:17.
Su Y, Lin L, Zhang J, Jiang Y, Pan C, Sun L, et al. Low expression of DLC1 is predictive of poor therapeutic efficiency of fluoropyrimidine and oxaliplatin as adjuvant chemotherapy in gastric cancer. Mol Med Rep. 2015;12:5771–9.
Park H, Cho SY, Kim H, Na D, Han JY, Chae J, et al. Genomic alterations in BCL2L1 and DLC1 contribute to drug sensitivity in gastric cancer. Proc Natl Acad Sci USA. 2015;112:12492–7.
Hitkova I, Yuan G, Anderl F, Gerhard M, Kirchner T, Reu S, et al. Caveolin-1 protects B6129 mice against Helicobacter pylori gastritis. PLoS Pathog. 2013;9:e1003251.
Salama NR, Hartung ML, Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385–99.
Regel I, Merkl L, Friedrich T, Burgermeister E, Zimmermann W, Einwachter H, et al. Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology. 2012;143:99–109 e110.
Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–94.
Lutz S, Freichel-Blomquist A, Rumenapp U, Schmidt M, Jakobs KH, Wieland T. p63RhoGEF and GEFT are Rho-specific guanine nucleotide exchange factors encoded by the same gene. Naunyn Schmiedebergs Arch Pharm. 2004;369:540–6.
Rumenapp U, Asmus M, Schablowski H, Woznicki M, Han L, Jakobs KH, et al. The M3 muscarinic acetylcholine receptor expressed in HEK-293 cells signals to phospholipase D via G12 but not Gq-type G proteins: regulators of G proteins as tools to dissect pertussis toxin-resistant G proteins in receptor-effector coupling. J Biol Chem. 2001;276:2474–9.
Wilde C, Genth H, Aktories K, Just I. Recognition of RhoA by Clostridium botulinum C3 exoenzyme. J Biol Chem. 2000;275:16478–83.
Pelz C, Steininger S, Weiss C, Coscia F, Vogelmann R. A novel inhibitory domain of Helicobacter pylori protein CagA reduces CagA effects on host cell biology. J Biol Chem. 2011;286:8999–9008.
Kim TY, Healy KD, Der CJ, Sciaky N, Bang YJ, Juliano RL. Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J Biol Chem. 2008;283:32762–70.
Wang D, Qian X, Sanchez-Solana B, Tripathi BK, Durkin ME, Lowy DR. Cancer-associated point mutations in the DLC1 Tumor suppressor and other Rho-GAPs occur frequently and are associated with decreased function. Cancer Res. 2020;80:3568–79.
Hinsenkamp I, Schulz S, Roscher M, Suhr AM, Meyer B, Munteanu B, et al. Inhibition of Rho-associated kinase 1/2 attenuates tumor growth in murine gastric cancer. Neoplasia. 2016;18:500–11.
Joshi R, Qin L, Cao X, Zhong S, Voss C, Min W, et al. DLC1 SAM domain-binding peptides inhibit cancer cell growth and migration by inactivating RhoA. J Biol Chem. 2020;295:645–56.
Liu JB, Wu XM, Cai J, Zhang JY, Zhang JL, Zhou SH, et al. CpG island methylator phenotype and Helicobacter pylori infection associated with gastric cancer. World J Gastroenterol. 2012;18:5129–34.
Sabbir MG, Prieditis H, Ravinsky E, Mowat MR. The role of Dlc1 isoform 2 in K-Ras2(G12D) induced thymic cancer. PLoS ONE. 2012;7:e40302.
Basak P, Dillon R, Leslie H, Raouf A, Mowat MR. The Deleted in Liver Cancer 1 (Dlc1) tumor suppressor is haploinsufficient for mammary gland development and epithelial cell polarity. BMC Cancer. 2015;15:630.
Waldum HL, Rehfeld JF. Gastric cancer and gastrin: on the interaction of Helicobacter pylori gastritis and acid inhibitory induced hypergastrinemia. Scand J Gastroenterol. 2019;54:1118–23.
Schubert ML, Rehfeld JF. Gastric peptides-gastrin and somatostatin. Compr Physiol. 2019;10:197–228.
Junior MF, Batista SA, Barbuto RC, Gomes AD, Queiroz DM, Araujo ID, et al. CagA-positive Helicobacter pylori strain containing three EPIYA C phosphorylation sites produces increase of G cell and decrease of D cell in experimentally infected gerbils (Meriones unguiculatus). Adv Med Sci. 2016;61:231–6.
Sundaresan S, Kang AJ, Hayes MM, Choi EK, Merchant JL. Deletion of Men1 and somatostatin induces hypergastrinemia and gastric carcinoids. Gut. 2017;66:1012–21.
Boger C, Kruger S, Behrens HM, Bock S, Haag J, Kalthoff H, et al. Epstein-Barr virus-associated gastric cancer reveals intratumoral heterogeneity of PIK3CA mutations. Ann Oncol. 2017;28:1005–14.
Mejias-Luque R, Zoller J, Anderl F, Loew-Gil E, Vieth M, Adler T, et al. Lymphotoxin beta receptor signalling executes Helicobacter pylori-driven gastric inflammation in a T4SS-dependent manner. Gut. 2017;66:1369–81.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Ebert MP, Tanzer M, Balluff B, Burgermeister E, Kretzschmar AK, Hughes DJ, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N. Engl J Med. 2012;366:44–53.
Acknowledgements
The authors want to thank Frank Herweck, Friedrich Behne, and Alexandra Kerner for their excellent technical assistance. Editing of the main text for language and grammar accuracy was supported by https://asco.editage.com.
Funding
EB received funding from the Deutsche Forschungsgemeinschaft (DFG, BU2285). NMB received funds from the DFG (#410981386) and the DFG Clinician Scientist-Programme “Interfaces and interventions in complex chronic conditions” (ICON). EB and ME are supported by GRK 2727 [DFG Graduate School: Innate Immune Checkpoints in Cancer and Tissue Damage (InCheck)], and ME obtained a grant provided by Merck Heidelberg Innovation Call (Darmstadt, Germany). MM had grant support from CancerCare Manitoba Foundation. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors cooperated and contributed to, critically reviewed and approved the manuscript. EB wrote the paper. EB and ME defined the research theme. CF, EB, IsH, IvH, JPK, SEM and TW designed methods, carried out the experiments, analysed the data, and interpreted the results. CR, MG and TG collected patient samples and performed and analysed tissue stainings. ME, NMB and RV collected biopsies and conducted clinical studies. KT, MG, MM, MQ and RML provided transgenic mice and tissue stainings. CW performed statistical calculations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest. Parts of the cell line data have been deposited under https://doi.org/10.11588/heidok.00025333 and URLs: http://www.ub.uni- heidelberg.de/archiv/24484; http://mediatum.ub.tum.de/?id=1140675.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hinsenkamp, I., Köhler, J.P., Flächsenhaar, C. et al. Functional antagonism between CagA and DLC1 in gastric cancer. Cell Death Discov. 8, 358 (2022). https://doi.org/10.1038/s41420-022-01134-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-022-01134-x