Abstract
During the maturation of hematopoietic stem/progenitor cells (HSPCs) to fully differentiated mature B lymphocytes, developing lymphocytes may undergo malignant transformation and produce B-cell lymphomas. Emerging evidence shows that through the endothelial-hematopoietic transition, specialized endothelial cells called the hemogenic endothelium can differentiate into HSPCs. However, the contribution of genetic defects in hemogenic endothelial cells to B-cell lymphomagenesis has not yet been investigated. Here, we report that mice with endothelial cell-specific deletion of Fbw7 spontaneously developed diffuse large B-cell lymphoma (DLBCL) following Bcl6 accumulation. Using lineage tracing, we showed that B-cell lymphomas in Fbw7 knockout mice were hemogenic endothelium-derived. Mechanistically, we found that FBW7 directly interacted with Bcl6 and promoted its proteasomal degradation. FBW7 expression levels are inversely correlated with BCL6 expression. Additionally, pharmacological disruption of Bcl6 abolished Fbw7 deletion-induced B-cell lymphomagenesis. We conclude that selective deletion of E3 ubiquitin ligase FBW7 in VE-cadherin positive endothelial cells instigates diffuse large B-cell lymphoma via upregulation of BCL6 stability. In addition, the mice with endothelial cell-specific deletion of Fbw7 provide a valuable preclinical platform for in vivo development and evaluation of novel therapeutic interventions for the treatment of DLBCL.
Similar content being viewed by others
Introduction
The lymphomas remain a major public health challenge around the globe. They represent one of the most heterogeneous groups of malignancies in all of cancer medicine. Non-Hodgkin lymphoma (NHL) which accounts for 90% of all lymphomas consists of more than 60 subtypes [1]. B-cell non-Hodgkin lymphomas are also a large heterogeneous group of hematologic malignancies. Given that the vast majority of B-cell lymphomas arise from the malignant transformation of germinal-center or post-germinal-center B cells, it is remarkable that B-cell lymphomas occur in such numerous clinically and histopathologically distinct varieties. The variability of B-cell lymphomas has been attributed to the distinct stages of differentiation and various events during which tumor progenitors become transformed [2, 3]. Recent evidence regarding the diverse origins of hematopoietic stem/progenitor cells (HSPCs) [4,5,6] might also account for this high variability. Most notably, HSPCs are derived directly from a unique population of endothelial cells known as hemogenic endothelium through a process called endothelial-hematopoietic transition (EHT) [4, 5, 7]. However, the contribution of endothelial genetic defect to B-cell lymphomagenesis remains unknown.
FBW7 is an E3-ubiquitin protein ligase that is part of the SKP1-cullin-F-box complex and functions as a tumor suppressor by targeting oncoproteins for degradation. Deletion or mutation of FBW7 is strongly implicated in numerous human cancers, including T-cell and B-cell malignancies [8,9,10,11,12]. In addition, mice with hematopoietic cell-specific Fbw7 deletion develop T-cell acute lymphoblastic leukemia [13], while T-cell deletion of Fbw7 leads to thymic lymphoma [14]. Although there is emerging evidence indicating the potential role of FBW7 in diffuse large B-cell lymphoma (DLBCL) [15, 16], direct experimental evidence implicating FBW7 in DLBCL pathogenesis is still lacking.
BCL6 is the product of a proto-oncogene that is strongly implicated in the pathogenesis of DLBCL. In many DLBCL patients, deregulation of BCL6 expression is achieved through chromosomal rearrangement or somatic mutation of the 5′-noncoding regions of BCL6 [17, 18]. BCL6 has been demonstrated to be degraded by an SKP1-CUL1-F-box protein (SCF) ubiquitin ligase complex containing the F-box protein FBXO11, which is inactivated in DLBCLs. Moreover, Park et al. reported that the signal-responsive E3 ubiquitin pellino 1 promotes B-cell lymphomagenesis by deregulating BCL6 polyubiquitination [19]. Therefore, the posttranslational modifications that regulate the BCL6 stability may also play important roles in B-cell lymphomagenesis. However, whether or not BCL6 accumulation leads to DLBCL in vivo remains largely unknown.
In this present study, we found that the mice with endothelial cell-specific deletion of Fbw7 spontaneously developed DLBCL following Bcl6 accumulation. We demonstrated that B-cell lymphomas in Fbw7 knockout mice were hemogenic endothelium-derived. Mechanistically, we found that FBW7 directly interacted with BCL6 and promoted its proteasomal degradation. In addition, pharmacological disruption of Bcl6 abolished Fbw7 deletion-induced B-cell lymphomagenesis. These findings suggest that FBW7 plays an essential role in B-cell lymphomagenesis by controlling BCL6 ubiquitination and stability.
Results
Endothelial Fbw7 deficiency induces diffuse large B-cell lymphoma
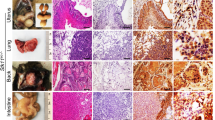
To determine the possible contribution of endothelial genetic defect to lymphomagenesis, we generated mice with endothelial cell-specific Fbw7 deletion (Fbw7ΔEC; Figure S1A–C, Supporting Information). Starting at 14 months of age, we observed that 19/36 (52.78%) Fbw7ΔEC mice spontaneously developed large neoplasms in multiple organs, including the lymph nodes (LNs), spleen, thymus, Peyer’s patches, liver, lungs, and kidneys; these neoplasms did not occur in age-matched wild-type (WT) littermates (Fig. 1A, B). After adjusting crude incidence for loss of individual mice to other causes, such as eosinophilic crystalline pneumonia, corneal plaques, and undiagnosed terminal illness, 83.69% of Fbw7ΔEC mice spontaneously developed lymphoma by 29 months of age (Fig. 1C). These neoplasms in Fbw7ΔEC mice exhibited partial or complete effacement of normal tissue architecture by diffuse infiltration of large centroblast-like or immunoblast-like cells with high-grade nuclear features, including prominent nucleoli, irregular nuclear contours, and frequent mitotic figures (Fig. 1D and Fig. S2, Supporting Information). Immunohistochemical analyses demonstrated that these lymphomas exhibited abundant expression of B-cell markers (B220 and CD20) and much weaker expression of the T-cell marker CD3e, confirming their B-cell origin (Fig. 1D and Figs. S2, S3, Supporting Information). Neoplastic cells exhibited robust proliferation and increased apoptosis, as illustrated by intense Ki67 staining, upregulation of proliferating cell nuclear antigen (PCNA), and cleaved caspase-3; these increases were accompanied by concomitant down-regulation of P27, P21, Bcl2, and Bclxl (Fig. S4, Supporting Information). Notably, Ki67 was predominantly expressed in large B cells (Fig. 1E). Together, these phenotypic characteristics strongly support the diagnosis of DLBCL in Fbw7ΔEC mice. Moreover, peripheral blood cell counts from Fbw7ΔEC and WT mice showed comparable levels of white blood cells, lymphocytes, and monocytes (Figs. S5, S6, Supporting Information), and bone marrow sections showed no obvious morphological abnormalities (Fig. 1F), further excluding the diagnosis of leukemia.
A Lymphoma incidence in Fbw7ΔEC (n = 36) and WT mice (n = 20). Statistics were determined using Fisher’s exact test. B Macroscopic lymphoma incidence in indicated organs. C Kaplan–Meier plot illustrating time to lymphoma development. Statistical analysis was performed using the log-rank test. D Macroscopic images, H&E, and immunohistochemistry analysis for B220 and CD3e in lymphoma tissue from indicated tissues. Blue and black arrows denote centroblasts and immunoblasts, respectively. Red arrows denote mitotic cells. E Immunofluorescence staining for B220 (green), Ki67 (red), and DAPI (blue) in lymphoma tissues from Fbw7ΔEC mice. F H&E staining of bone marrow from WT and Fbw7ΔEC mice.
With a panel of clinically relevant markers for B-cell lymphoma, an immuno-pathological phenotyping analysis was conducted in 11 mice with DLBCL. Based upon the expression profiles of CD10, Bcl6, and MUM1 using Han’s algorithm [20], all cases were identified as the germinal center B-cell-like subtype of DLBCL (Table S1 and Fig. S7, Supporting Information). Specifically, Bcl6 was upregulated in all DLBCL cases (Table S1 and Fig. S7, Supporting Information). Additionally, the intensity of MUM1, Bcl6, c-Myc, and CD5, or the Ki67 index, was not correlated with the clinical stage (P > 0.05 for all comparisons).
To further characterize these B-cell lymphomas, we compared gene expression profiles of mesenteric lymph nodes (mLNs) from lymphoma-bearing Fbw7ΔEC mice and normal mLNs from WT mice using an Affymetrix® GenechipTM mouse gene array. Analysis of the relative average expression of all detected genes revealed that these B-cell lymphomas had distinct patterns of gene expression (Fig. 2A–C). Gene set enrichment analysis showed that significantly altered genes are primarily cancer-associated and inflammatory immune-associated genes in mLNs from lymphoma-bearing Fbw7ΔEC mice (Fig. 2D, E), consistent with human DLBCL gene signatures [21]. Notably, numerous immunoglobulin heavy and light chain variable genes were dramatically decreased in lymphoma-bearing mLNs from Fbw7ΔEC mice compared with normal mLNs from WT mice (Fig. 2F). Consistently, Fbw7ΔEC mice showed significantly reduced levels of serum IgM and IgG (Fig. S8, Supporting Information), which is consistent with previous findings suggesting the low immunoglobulin levels in patients with non-Hodgkin lymphoma [22, 23]. Therefore, we provide an ideal animal model that resembles human DLBCL pathophysiology.
A–C Gene expression data was obtained using an Affymetrix GeneChip Mouse Gene 2.0 ST Array and mRNAs isolated from mesenteric lymph nodes (mLNs) from lymphoma-bearing Fbw7ΔEC mice and normal mLNs from WT mice (n = 3 biological replicates per group). Volcano plot (A) and heatmap of genes significantly altered (p < 0.05) by twofold (upregulated genes in B and downregulated genes in C) in lymphoma-bearing mLNs from Fbw7ΔEC mice compared with normal mLNs from WT mice. D, E Gene set enrichment analysis (GSEA) was used to identify biological terms, pathways, and processes that are coordinately up- or downregulated in lymphoma-bearing mLNs from Fbw7ΔEC mice compared with WT mLNs. GSEA analysis using KEGG (D) and hallmark (E) gene sets. F Heatmap showing some significantly downregulated immunoglobulin heavy and light chain variable genes in lymphoma-bearing mLNs from Fbw7ΔEC mice compared with normal mLNs from WT mice.
Lymphoma B cells in Fbw7 ΔEC mice are endothelium-derived
To gain insight into the mechanism underlying spontaneous B-cell lymphomagenesis in Fbw7ΔEC mice, we first assessed Fbw7 protein expression in lymphoma tissues from Fbw7ΔEC mice. Loss of Fbw7 was detected in lymphoma-bearing mLNs from Fbw7ΔEC mice, compared with normal mLNs from WT mice, where Fbw7 expression was intact (Fig. 3A). Quantitative RT-PCR analysis further confirmed decreased mRNA levels of Fbxw7 in lymphoma tissues from Fbw7ΔEC mice (Fig. 3B). Using genotyping (Fig. 3C) [24], we found that deletion of the Fbxw7 locus was detected in several lymphoid tissues, including mLNs and bone marrow from lymphoma-bearing Fbw7ΔEC mice (Fig. 3D). Furthermore, we found that B cells from the spleen, T cells from the thymus, and granulocytes and mononuclear cells from the blood also exhibited Fbxw7 locus deletion (Fig. 3E). These observations confirm that the lymphomas in Fbw7ΔEC mice were Fbxw7-deficient.
A, B Western blot and RT-PCR analysis of mLNs from WT and lymphoma-bearing Fbw7ΔEC mice. Data represent mean ± SEM (n = 3 biological replicates per group). C Exons 4–7 of the Fbxw7 locus before and after excision of the floxed exon, and primer locations. Primers B and C were used to detect WT and floxed alleles (315 and 497 bp, respectively). Primers A and C were used to detect deleted allele (662 bp). D Genotyping results for Fbw7 in mLNs and bone marrow. E Genotyping results from indicated tissues. F mTmG+/-;Fbw7flox/flox;VE-Cadherin-Cre+ mouse generation. G Immunofluorescent (IF) images and flow cytometry analysis of bone marrow cells from mTmG+/-;Fbw7flox/flox;VE-Cadherin-Cre+ and mTmG+/-;Fbw7flox/flox;VE-Cadherin-Cre- mice. Data represent mean ± SEM (n = 6 biological replicates per group). H IF of mononuclear cells and flow cytometry analysis of blood-derived B cells from mTmG+/-;Fbw7flox/flox;VE-Cadherin-Cre+ and mTmG+/-;Fbw7flox/flox;VE-Cadherin-Cre- mice. P values were determined using the student’s t-test (B, G). For all panels, *p < 0.05.
To determine the origin of lymphoma B cells in Fbw7ΔEC mice, we generated mice to perform lineage tracing: triple-transgenic Fbw7flox/flox;VE-Cadherin-Cre;ROSAmT/mG mice (Fig. 3F). Using this model, we found that 31.5% of hematopoietic cells (CD45+ cells) in the bone marrow were derived from VE-Cadherin+ cells (Fig. 3G). Furthermore, we found that over 4% of B cells are also derived from VE-Cadherin+ cells (Fig. 3H). Most importantly, we found that the lymphoma tissues developed in Fbw7flox/flox;VE-Cadherin-Cre;ROSAmT/mG mice displayed strong GFP signal (Fig. 4A–D), directly suggesting that the lymphoma B cells from Fbw7ΔEC mice are endothelium-derived.
BCL6 is a novel and specific FBW7 substrate in B cells
Next, we explored the molecular mechanism by which Fbw7 deficiency induced B-cell lymphomagenesis. Consistent with our immunohistochemistry findings (Table S1 and Fig. S7, Supporting Information), elevated Bcl6 and c-Myc expression was observed in lymphoma-bearing mLNs from Fbw7ΔEC mice, compared with normal mLNs from WT mice (Fig. 5A). As BCL6 is a proto-oncogene that is strongly implicated in the pathogenesis of human DLBCL [25,26,27], we hypothesized that BCL6 might be a novel substrate of FBW7 in B cells. As would be expected in canonical FBW7 substrates, three evolutionally conserved FBW7-recognizable degrons were found in the human BCL6 protein sequence (Fig. 5B). Although BCL6 bound to all three isoforms of FBW7 (FBW7α, β, and ɤ), only expression of FBW7β specifically and dramatically decreased BCL6 expression (Fig. 5C) in a dose-dependent manner (Fig. 5D). Cancer-associated mutant forms of FBW7β (R385H and R385C) corresponding to FBW7α (R465H and R465C) [11, 28] displayed a reduced ability to promote BCL6 degradation (Fig. S9A, B, Supporting Information). Moreover, FBW7β-mediated BCL6 elimination was abolished following treatment with the 26 S proteasome inhibitor MG-132 (Fig. 5E). These results suggest that FBW7β is the major isoform of FBW7 that regulates BCL6 protein level through the ubiquitin-proteasome pathway.
A Western blot analysis for the indicated proteins in normal lymph nodes from WT mice and lymphoma-bearing lymph nodes from Fbw7ΔEC mice. LE long exposure, SE short exposure. Data represent mean ± SEM (n = 3 for WT and n = 5 for Fbw7ΔEC mice). B FBW7 consensus sequence (CPD) and alignment of BCL6 phosphodegron sequences recognized by FBW7 in different species to CPD motifs in indicated FBW7 substrates. C Pull-down analysis of exogenous Flag-BCL6 and OFP-FBW7 (α, β, or ɤ) in HEK-293T cells. D Western blot and quantification illustrating levels of GFP-BCL6 following the introduction of increasing amounts of Myc-FBW7β in HEK-293T cells. Data represent mean ± SEM (n = 4 biological replicates per group). E Western blot and quantification showing levels of indicated proteins after treatment with MG-132 (10 µM) in HEK-293T cells for 8 h. Data represent mean ± SEM (n = 4 biological replicates per group). F Western blot showing levels of indicated proteins in HEK-293T cells expressing Myc-FBW7α, Myc-FBW7β, or PCDNA following cycloheximide (CHX, 20 µg/ml) treatment for indicated times. G Pull-down analysis illustrating Flag-BCL6 ubiquitination in the presence of GFP-FBW7 in HEK-293T cells. H Western blot analysis of indicated proteins in HEK-293T cells in the presence of Myc-tagged Cullin proteins (Cul1, 2, 3, 4A, or 5). I Western blot analysis of indicated proteins in the presence of Flag-FBW7β and indicated mutants in HEK-293T cells: Flag-BCL6/T190A/S194A (CPD1m), Flag-BCL6/S333A/S337A (CPD2m), or Flag-BCL6/S343A/S347A (CPD3m). All western blots were normalized to β-actin. P values were determined using student’s t-test (A, E) or one-way ANOVA with Tukey’s multiple comparisons test (D). For all panels, *p < 0.05; NS indicates not significant.
To further explore the role of FBW7β in modulating BCL6 stability, we examined BCL6 half-life in HEK-293T cells treated with the protein synthesis inhibitor cycloheximide. Co-expression of FBW7β with BCL6 significantly shortened the half-life of BCL6 (Fig. 5F). Consistent with a posttranslational mode of regulation, no alterations in BCL6 mRNA levels were detected after overexpression of FBW7 in HEK-293T cells (Fig. S10, Supporting Information).
Since FBW7 directly binds to its substrate to promote its ubiquitination and degradation [29], we next determined whether FBW7 could directly stimulate BCL6 ubiquitination in vitro. As expected, overexpression of FBW7β triggered polyubiquitination of BCL6 (Fig. 5G). Since FBW7 is the substrate recognition component of the E3 ubiquitin ligase SKP1-cullin-1-FBW7 complex, we next assayed if overexpression of Cullin affects BCL6 stability. Overexpression of Cullin 1, but no other Cullin family members, significantly decreased BCL6 protein abundance (Fig. 5H). When we mutated the three FBW7-recognizable degrons in BCL6, we found that the S343A/S347A mutant form of BCL6 was resistant to FBW7-mediated BCL6 degradation (Fig. 5I). These collective data suggest that the E3 ubiquitin ligase SKP1-cullin-1-FBW7 complex targets BCL6 for ubiquitination and degradation in a degron-dependent manner.
FBW7 expression levels inversely correlate with BCL6 expression
To validate the role of FBW7 in human B-cell lymphoma, we investigated the relationship of BCL6 and FBW7 in human B-cell lymphoma cell lines and found that expression of FBW7 inversely correlated with BCL6 expression (Fig. 6A). We next overexpressed FBW7β in the human B-cell lymphoma cell lines SU-DHL-4 and FARAGE, and found that BCL6 expression dramatically decreased following overexpression of FBW7β in both cell lines; levels of c-MYC and c-JUN, two well-characterized canonical FBW7 substrates, were used as positive controls (Fig. 6B). Strikingly, consistent with B-cell lymphomas in Fbw7ΔEC mice, FBW7 expression was markedly decreased in clinical samples from patients with DLBCL (Fig. 6C). Taken together, these data further support that BCL6 is a novel substrate of FBW7 in the pathogenesis of lymphoid malignancy.
A Western blot analysis of BCL6, FBW7, P21, P27, BCL2, and β-actin in three different human diffuse large B-cell lymphoma cell lines. B Western blot analysis for the indicated proteins in cell lysates from SU-DHL-4 and Farage cells expressing AAV-GFP or AAV-GFP-FBW7β. C H&E staining and immunohistochemical analysis of FBW7 in a human DLBCL tissue array (US Biomax, LY121b). The pheochromocytoma samples were used as a control.
B-cell lymphomagenesis in Fbw7 ΔEC mice is Bcl6-dependent
To validate that B-cell lymphoma development in Fbw7ΔEC mice was BCL6-dependent, we performed adoptive transfer experiments using freshly isolated DLBCL cells from Fbw7ΔEC mice. We found that fresh DLBCL cells from lymphoma-bearing Fbw7ΔEC mice rapidly induced tumor growth in young healthy recipient Fbw7ΔEC mice. Lymphoma cells showed clear and systemic metastatic capacity, and, in addition to subcutaneous tumors on the back, tumor deposits were found in different organs, including mesenteric and retroperitoneal LNs, thymus, liver, and kidney (Fig. S11, Supporting Information). Predominant expression of the B-cell marker B220 and weaker expression of the T-cell marker CD3e confirmed their B-cell origin, similar to that of the original transferred lymphoma (Fig. S11, Supporting Information). Next, we sought to determine whether lymphoma growth upon loss of Fbw7 is biologically dependent on Bcl6. To do so, we used the selective BCL6 inhibitor FX-1 [30] in in vitro and in vivo model systems (Fig. 7A). FX-1 significantly suppressed lymphoma cell growth in vitro (Fig. 7B, C), and, more importantly, mice treated with FX-1 had reduced tumor volume, tumor burden, and tumor weight compared with mice treated with vehicle control (Fig. 7D–G). Tumors from mice treated with FX-1 also displayed reduced cell proliferation and metastasis (Fig. 7H, I), suggesting that targeting BCL6 reduces this tumor growth mainly via induction of cell cycle arrest, which is consistent with the previous findings [31, 32].
A Schematic of tumor single-cell suspension preparation, primary DLBCL cell culture and treatment, and timeline for adoptive transfer experiments and FX-1 treatment. B Proliferation of primary DLBCL cells isolated from aged lymphoma-bearing Fbw7ΔEC mice following FX-1 treatment for indicated doses and times. Data represent mean ± SEM (n = 4 biological replicates per group). C Western blot analysis for indicated proteins in primary Fbw7ΔEC DLBCL cells following treatment with 10 µM FX-1 for 48 h. D Tumor growth by tumor volume during FX-1 treatment (n = 5 mice per group). E Tumor burden (area under the curve, AUC) for mice in D, calculated between initial and final volumes at day 10. F, G Tumor weight (F) and tumor volume (G) in mice upon sacrifice. Data represent mean ± SEM (n = 5 per group). H Left and middle, H&E staining of tissue sections from back tumors and kidneys of Fbw7ΔEC mice following indicated treatments. Red arrows denote mitotic cells. Scale bars: 20 µm; 100 µm in inserts. Right, representative gross images of Fbw7ΔEC mice treated with FX-1 or vehicle. Black arrows show organs infiltrated with lymphomas. I Quantification of mitotic figures in back tumors of indicated mice. Data represent mean ± SEM (n = 5 per group). P values were determined using ANOVA with Tukey’s post hoc test (B, D) and student’s t-test (E–G, and I). For all panels, *p < 0.05.
DLBCL cells isolated from aged lymphoma-bearing Fbw7ΔEC mice were next subcutaneously injected on the backs of immunocompromised NOD scid gamma (NSG) mice, which were subsequently treated with FX-1. Although subcutaneous back tumors did not develop in immunocompromised NSG mice, tumor deposits were found in several different organs, including the liver, kidney, spleen, and perirenal space (Fig. S12A, Supporting Information). Additionally, NSG mice treated with FX-1 showed significant attenuation of lymphoma growth and metastasis (Fig. S12A, B, Supporting Information). Together, these results suggest that B-cell lymphomagenesis in Fbw7ΔEC mice is Bcl6-dependent.
Discussion
In this present study, we discovered that conditional deletion of Fbw7 in VE-cadherin-positive endothelial cells promotes B-cell lymphomagenesis via upregulation of Bcl6 stability. Consistently, we demonstrated for the first time that Bcl6 is a novel substrate of FBW7, which directly interacts with Bcl6 and promotes its proteasomal degradation. Pharmacological disruption of Bcl6 abolishes Fbw7 deletion-induced B-cell lymphomagenesis. In addition, FBW7 expression is dramatically decreased in human DLBCL samples. Moreover, in human lymphoma cell lines, FBW7 expression levels are inversely correlated with BCL6 expression. Our work has provided new insights into cellular and molecular mechanisms underlying B-cell lymphomagenesis and suggests that genetic defects in endothelial cells can contribute to B-cell malignancies.
Emerging evidence indicates that HSCs maintain a unique relationship with endothelial cells and are derived from a unique population of endothelial cells via EHT [4, 5, 7]. Endothelial cells are also an important microenvironmental component of the HSCs niche [33]. Therefore, we hypothesized that the spontaneous development of DLBCL in mice following the loss of Fbw7 could be due to either EHT or microenvironmental changes in the endothelium. Given that BCL6 is a transcriptional repressor that is relatively specific for germinal center B cells [27, 32, 34,35,36], our data suggest that Fbw7 deficiency-driven BCL6 accumulation in B cells, which are derived from endothelial cells, is the major mechanism behind B-cell lymphomagenesis in Fbw7ΔEC mice (Fig. 8). Although Fbw7-deficient B cells compose only about 4% of the total B cells in young Fbw7ΔEC mice, they are the “cancer seeds” that grow and prosper as age increases. Therefore, it takes a long duration of time for these few transformed B cells with Bcl6 accumulation to ultimately outnumber and take over normal B cells; this may explain why lymphomagenesis starts at 14 months of age in Fbw7ΔEC mice. It is also worth noting that no other types of tumors or cancers were found in Fbw7ΔEC mice, likely due to the fact that Bcl6 is exclusively expressed in germinal center B cells, but not in erythroid-myeloid progenitors and other hematopoietic cell types [27, 32, 34,35,36].
Endothelial cells produce all blood cells, including HSCs, during embryonic development [37, 38]. VE-cadherin is expressed in endothelial cells. The VE-cadherin Cre we used in this study is constitutive, i.e., non-inducible. The Fbw7flox/flox;VE-cadherin-Cre mice induce FBW7 deletion in all endothelial cells and blood cells, including HSC-independent and HSC-dependent hematopoietic cells. Therefore, whether the origin of this B-cell lymphoma is HSC-independent endothelial cells (directly from endothelial cells) or HSC-derived B cells remains unknown. Both inducible CreERT2 mice are warranted to validate the endothelial cell origins of DLBCL seen in Fbw7ΔEC mice. Finally, complemental mouse models such as Fgd5-CreERT2, Vav1-Cre, CD19-Cre, or Mb1-Cre will be critical to determine which progenitor stage(s) are critical for lymphoma initiation by FBW7 deletion.
The ubiquitin ligase FBW7 targets a number of proto-oncoproteins for proteasomal degradation. Mutation or deletion of FBW7 has been strongly implicated in numerous T-cell and B-cell malignancies [11, 15, 28]. FBW7 regulates cellular apoptosis by targeting Mcl1 for ubiquitination and destruction and plays important roles in T-cell acute lymphoblastic leukemia [28]. FBXW7 mutations reduce the binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in chronic lymphocytic leukemia [28]. A low expression level of FBW7 was associated with a poor outcome in non-GCB DLBC [15]. In the present study, we observed that mice with endothelial Fbw7 deletion spontaneously developed DLBCL, with FBW7 deficiency and Bcl6 accumulation in lymphoma tissue. Moreover, FBW7 reversely correlated with BCL6 in human B-cell lymphoma. Therefore, FBW7 acts as a tumor suppressor in DLBCL.
As BCL6 is a frequently activated oncogene in the pathogenesis of DLBCL [27], it has become a critical therapeutic target in DLBCL [30, 39]. Our findings demonstrate that BCL6 inhibition had anti-lymphoma activity in our DLBCL mouse model. This further supports our experimental evidence demonstrating a role for FBW7 in B-cell lymphomagenesis through regulation of BCL6 ubiquitination and stability, suggesting that FBW7/BCL6 could be a potential target in DLBCL therapy. Importantly, our animal model also provides a valuable preclinical platform for in vivo development and evaluation of novel therapeutic interventions for the treatment of DLBCL.
In summary, we provided the first evidence that loss of E3 ubiquitin ligase FBW7 in VE-cadherin positive endothelial cells instigates diffuse large B-cell lymphoma via upregulation of BCL6 stability.
Experimental section
Mice
Vascular endothelial cadherin (VE-cadherin)-Cre transgenic mice (Stock No. 006137) [40] and Fbw7-floxed (Fbw7flox/flox) mice (Stock No. 017563) [24] were purchased from The Jackson Laboratory. VE-cadherin-Cre mice were intercrossed with Fbw7flox/flox mice to generate Fbw7flox/flox;VE-cadherin-Cre mice. Fbw7flox/flox;VE-cadherin-Cre mice were further bred with ROSAmT/mG mice (The Jackson Laboratory, Stock No. 007576) to generate triple-transgenic Fbw7flox/flox;VE-Cadherin-Cre;ROSAmT/mG mice. NSG mice (NOD.Cg-Prkdcscid;Il2rgtm1Wjl/SzJ, Stock No. 005557) were purchased from The Jackson Laboratory. Mice were housed in a controlled environment (22 °C, 12-h/12-h light/dark cycle) with free access to food and water. All mouse experimental protocols were approved by the Institutional Animal Care and Use Committee at Georgia State University and were in compliance with relevant ethical regulations. Genotyping for each strain was performed as described on The Jackson Laboratory website.
Cell lines
HEK-293A and HEK-293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM). SU-DHL-4 (ATCC® CRL-2957TM), Toledo (ATCC® CRL-2631TM), and Farage (ATCC® CRL-2630TM) cells were grown in RPMI-1640 medium. All media were supplemented with 10% FCS, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine.
Tissue collection and processing
Tissue sections were stored at −80 °C for RNA extraction or western blotting. Tissue sections for pathological diagnosis or immunohistochemistry/immunofluorescence were fixed in 10% neutral buffered formalin, and embedded in paraffin or optimal cutting temperature (OCT) embedding material. Paraffin-embedded sections or OCT-embedded sections were cut at 5 or 8 μm thickness, respectively.
Histology and immunohistochemistry
Paraffin-embedded tissue sections were stained with H & E, according to standard protocols. All images were recorded using an Olympus digital camera (Tokyo, Japan). For immunohistochemical staining, paraffin-embedded tissue sections were deparaffinized, rehydrated, and subjected to antigen retrieval. Endogenous peroxidase activity was blocked using 0.3% H2O2 for 20 min. Tissue sections were blocked with normal goat serum (Biogenex, HK112-9K) and incubated with primary antibodies against B220, CD3e, CD20, Bcl6, CD10, MUM1, and CD5. Anti-rabbit/mouse immunoglobulin G (DAKO, Cat# K5007) was used as a secondary antibody. The reaction was visualized using DAB (DAKO, Cat# K3468) and sections were counterstained with hematoxylin. Results were scored as 0 (0–25%, negative staining); 1 (25–50%, weakly positive staining); 2 (50–75%, moderately positive staining); or 3 (≥75%, strongly positive staining) according to the percentage of positive cells.
Immunofluorescence
Immunofluorescence staining was performed as previously described [41,42,43]. Briefly, OCT-embedded sections were washed with phosphate-buffered saline (PBS), fixed with acetone at 4 °C for 15 min, and permeabilized with 0.2% Triton X-100 (Dow Chemical, Midland, MI, USA) for 10 min. After blocking with goat serum for 30 min, the sections were incubated with primary antibodies against Ki67 and B220 overnight. Secondary antibodies (Alexa Flour® 555 goat anti-rabbit and Alexa Flour® 488 goat anti-rat) were added for 1 h at 37 °C, followed by nuclear DNA staining using DAPI for 10 min. Fluorescence signals were evaluated using confocal microscopy (LSM 810, Zeiss, Oberkochen, Germany) or immunofluorescent microscopy.
Isolation of B and T cells from spleen or thymus
The spleen or the thymus were removed from the mouse and transferred to a dish containing isolation buffer (PBS with 2% FBS + 2 mM EDTA). The tissues were further transferred to a cell strainer on top of a 50 ml tube and mashed into fine parts using the back of a syringe plunger. The filter was rinsed at regular intervals with an isolation buffer. The cell suspensions were filtered through a 70 µm nylon mesh and centrifuged at 300×g for 10 min at 4 °C. The cell pellets were collected, suspended with 5 ml/spleen of RBC lysis buffer (Qiagen, Cat# NC9670839), and incubated at room temperature for 5–10 min with occasional shaking. The cell suspensions were centrifuged, and the cell pellets were suspended in the appropriate buffer for use in the next step of the experimental procedure. B and T cells were further isolated using DynabeadsTM Mouse CD43 (Thermo Fisher, Cat# 11422D) and DynabeadsTM CD4 Positive Isolation Kit (Thermo Fisher, Cat# 11331D), respectively, according to the manufacturer’s instructions.
Flow cytometry
Samples were obtained from blood, bone marrow, and spleen. Erythrocytes were lysed with RBC lysis buffer (Qiagen, Cat# NC9670839) for 10 min at room temperature. Single-cell suspensions were prepared in isolation buffer (PBS, 2% fetal bovine serum, 2 mM EDTA) and stained with anti-mouse CD19 (BD biosciences, Cat# 561738) or anti-mouse CD45 (BD biosciences, Cat# 559864) antibodies. Data were obtained using the LSRFortessaTM X-20 cell analyzer (BD Biosciences).
Preparation of tumor single-cell suspension
Tumor tissues from Fbw7ΔEC mice were transferred into a 60 mm petri dish and washed with PBS. Transferred tissues were then cut into small pieces and digested in PBS containing 2 mg/ml collagenase D (Roche, Cat# 11088866001) and 100 µg/ml DNase I (Roche, Cat# 10104159001) for 1.5 h at 37 °C to obtain single-cell suspensions. Tumor cells were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine, or resuspended in an isotonic solution of saline for injection.
Adoptive transfer experiments
Freshly isolated tumor cells (1 × 107) were injected subcutaneously on the backs of 6- to 8-week-old male Fbw7ΔEC mice or immunocompromised NSG mice. For Fbw7ΔEC mice, when back tumors reached a palpable size, mice were randomly assigned into an FX-1 (Targetmol, Cat# T4089) treatment group or vehicle control group. Mice were intraperitoneally injected with FX-1 (1 mg/day, reconstituted in PEG-400) or vehicle for 10 consecutive days. Tumor size was measured using an electronic digital caliper during the 10-day treatment. Tumor volume was calculated with the following formula: tumor volume (mm3) = (smallest diameter2 × largest diameter)/2.
Constructs and mutagenesis
Plasmid information is presented in the Major Resources Tables in the Supplementary Data. Human FBW7β was cloned into an Adeno-associated viral vector modified from plasmid pAAV.CMV.PI.EGFP.WPRE.bGH. BCL6 mutant constructs were made by site-directed mutagenesis based on the pCXN2- BCL6 (Flag-tagged BCL6) wild-type construct using KOD Hot Start DNA polymerase (Xtreme, Millipore).
Adeno-associated virus (AAV) packaging
AAV-2 was packaged using triple plasmid co-transfection in HEK-293T cells. Briefly, HEK-293T cells were transfected with AAV-FBW7β-acGFP (7 µg plasmid per 10 cm dish), an AAV helper plasmid (15 µg plasmid per 10 cm dish), and AAV serotype 2 (AAV-2, 7 µg plasmid per 10 cm dish). Around 48 h after transfection, cells were harvested.
Western blotting
Total proteins were prepared from cultured cells or mouse tissues, and western blotting was performed, as briefly mentioned below. Proteins were quantified with a BCA assay (Pierce), separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat milk dissolved in TBS-T at 37 °C for 1 h, and incubated with primary antibodies at 4 °C overnight. After incubation with horseradish peroxidase-conjugated secondary antibodies at 37 °C for 1 h, the membranes were washed 3 times with TBS-T and then visualized with chemiluminescent peroxidase reagents.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from mouse tissues using Trizol reagent (Invitrogen), according to the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed into first-strand cDNA, and RT-PCR amplification was performed using SYBR Green dye and the CFX96 TouchTM Real-time PCR Detection System (BIO-RAD). The primer sequences used for the detection of Fbx7, Bcl6, and 18S are presented as follows: Fbxw7 forward: 5’-GTGATAGAGCCCCAGTTCCA-3’; Fbxw7 reverse: 5’-CCTCAGCCAAAATTCTCCAG-3’; Bcl6 forward: 5’- TGATCGCTGCCAGGCCTCCTT-3’; Bcl6 reverse: 5’- CTGGCCGATTGAACTGCGCTCC-3’; 18S forward: 5’-GTCTGTGATGCCCTTAGATG-3’; 18S reverse: 5’-AGCTTATGACCCGCACTTAC-3’. Relative mRNA expression was calculated using the comparative ΔΔCT method, and the resulting values were normalized to 18S expression. PCR was performed in triplicate for each experiment. The results presented represent three independent experiments.
Pull-down assay
Cells were washed with ice-cold PBS and lysed with IP lysis buffer (Pierce, Cat# 87787) supplemented with a proteinase inhibitor cocktail (Pierce, Cat# A32965) and phosphatase inhibitor (Thermo Fisher, Cat# 78428). Anti-FLAG M2 Affinity Gel (Sigma, Cat# A2220) was added to cell lysates and incubated overnight at 4 °C. The beads were then washed four times in a lysis buffer. After the last wash, FLAG-tagged proteins were eluted in an elution buffer and subjected to western blotting.
ELISA
For the detection of IgM and IgG, plasma samples were collected from mice and immediately stored at −80 °C in a freezer until the day of the assay. An IgG/IgM mouse uncoated ELISA kit with plates was purchased from Invitrogen (Cat# 88-50400-22 & 88-50470-22). All reagents were provided in the ELISA kit and all procedures were performed according to the manufacturer’s instructions. Plates were read at 450 nm using a TECAN Infinite M1000 Pro microplate reader.
Microarray analysis
RNA was extracted from sections of frozen biopsies using an RNeasy Mini Kit (Qiagen, CA, USA). Microarray-based gene expression profiling was performed using the Boston University Microarray and Sequencing Resource Core Facility. Mouse Gene 2.0 ST CEL files were normalized to produce gene-level expression values using the implementation of the Robust Multiarray Average (RMA) in the affy package included in the Bioconductor software suite (version 2.11) and an Entrez Gene-specific probeset mapping from the Molecular and Behavioral Neuroscience Institute (Brainarray) at the University of Michigan. Array quality was assessed by computing Relative Log Expression and Normalized Unscaled Standard Error using the affyPLM package. Principal Component Analysis was performed using the prcomp R function with expression values that had been normalized across all samples to a mean of zero and a standard deviation of one. Differential expression was assessed using the moderated (empirical Bayesian) t-test implemented using the limma package (i.e., creating simple linear models with lmFit, followed by empirical Bayesian adjustment with eBayes). Correction for multiple hypothesis testing was accomplished using the Benjamini-Hochberg false discovery rate. Human homologs of mouse genes were identified using HomoloGene. All microarray analyses were performed using the R environment for statistical computing.
Gene set enrichment analysis (GSEA)
GSEA was used to identify biological terms, pathways, and processes that were coordinately up- or downregulated within each pairwise comparison. The Entrez Gene identifiers of human homologs of the genes interrogated by the array were ranked according to the t statistic computed using a knockout vs WT comparison. Any mouse genes with multiple human homologs (or vice versa) were removed prior to ranking, so that the ranked list represented only those human genes that match exactly one mouse gene. This ranked list was then used to perform pre-ranked GSEA analyses (default parameters with random seed 1234) using the Entrez Gene versions of the Hallmark, Biocarta, KEGG, Reactome, Gene Ontology, and transcription factor and microRNA motif gene sets obtained from the Molecular Signatures Database (MSigDB), version 6.0.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 software. Unpaired two-tailed Student’s t-tests were used to calculate significant differences between the two groups. Multiple comparison correction analysis was performed using one-way ANOVA with Tukey’s post hoc HSD test. A log-rank (Mantel–Cox) test was used to analyze Kaplan–Meier curves. Fisher’s exact test was applied to test for association between categorical variables. P values were considered significant when P < 0.05.
Data availability
All data is available in the main text or the supplementary files.
References
Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017;390:298–310.
Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–9.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11.
Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–11.
Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–20.
Ciau-Uitz A, Walmsley M, Patient R. Distinct origins of adult and embryonic blood in Xenopus. Cell. 2000;102:787–96.
Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–5.
Yoshimoto N, Yanagi T, Matsumura W, Ujiie I, Izumi K, Ando S, et al. Primary cutaneous diffuse large B-cell lymphoma presenting as a solitary subcutaneous nodule with TP53 and FBXW7 mutations. Int J Dermatol. 2017;56:1459–61.
Mihashi Y, Mizoguchi M, Takamatsu Y, Ishitsuka K, Iwasaki H, Koga M, et al. C-MYC and its main ubiquitin ligase, FBXW7, influence cell proliferation and prognosis in adult T-cell leukemia/lymphoma. Am J Surg Pathol. 2017;41:1139–49.
Vazquez-Dominguez I, Gonzalez-Sanchez L, Lopez-Nieva P, Fernandez-Navarro P, Villa-Morales M, Cobos-Fernandez MA, et al. Downregulation of specific FBXW7 isoforms with differential effects in T-cell lymphoblastic lymphoma. Oncogene. 2019;38:4620–36.
Close V, Close W, Kugler SJ, Reichenzeller M, Yosifov DY, Bloehdorn J, et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood. 2019;133:830–9.
Sakhdari A, Ok CY, Patel KP, Kanagal-Shamanna R, Yin CC, Zuo Z, et al. TP53 mutations are common in mantle cell lymphoma, including the indolent leukemic non-nodal variant. Ann Diagn Pathol. 2019;41:38–42.
Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Gene Dev. 2008;22:986–91.
Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–88.
Yao S, Xu F, Chen Y, Ge Y, Zhang F, Huang H, et al. Fbw7 regulates apoptosis in activated B-cell like diffuse large B-cell lymphoma by targeting Stat3 for ubiquitylation and degradation. J Exp Clin Cancer Res. 2017;36:10.
Yao S, Guo T, Zhang F, Chen Y, Xu F, Luo D, et al. Fbw7 inhibits the progression of activated B-cell like diffuse large B-cell lymphoma by targeting the positive feedback loop of the LDHA/lactate/miR-223 axis. Front Oncol. 2022;12:842356.
Offit K, Lo Coco F, Louie DC, Parsa NZ, Leung D, et al. Rearrangement of the bcl-6 gene as a prognostic marker in diffuse large-cell lymphoma. N. Engl J Med. 1994;331:74–80.
Pasqualucci L, Bereshchenko O, Niu H, Klein U, Basso K, Guglielmino R, et al. Molecular pathogenesis of non-Hodgkin’s lymphoma: the role of Bcl-6. Leuk Lymphoma. 2003;44:S5–12.
Park HY, Go H, Song HR, Kim S, Ha GH, Jeon YK, et al. Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J Clin Invest. 2014;124:4976–88.
Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82.
Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–61.
Biggar RJ, Christiansen M, Rostgaard K, Smedby KE, Adami HO, Glimelius B, et al. Immunoglobulin subclass levels in patients with non-Hodgkin lymphoma. Int J Cancer. 2009;124:2616–20.
Ellison-Loschmann L, Benavente Y, Douwes J, Buendia E, Font R, Alvaro T, et al. Immunoglobulin E levels and risk of lymphoma in a case-control study in Spain. Cancer Epidemiol Biomark Prev. 2007;16:1492–8.
Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–408.
Ci W, Polo JM, Melnick A. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr Opin Hematol. 2008;15:381–90.
Baron BW, Anastasi J, Montag A, Huo D, Baron RM, Karrison T, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci USA. 2004;101:14198–203.
Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–55.
Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–9.
Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93.
Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–11.
Schlager S, Salomon C, Olt S, Albrecht C, Ebert A, Bergner O, et al. Inducible knock-out of BCL6 in lymphoma cells results in tumor stasis. Oncotarget. 2020;11:875–90.
Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–60.
Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–34.
Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9.
Polo JM, Juszczynski P, Monti S, Cerchietti L, Ye K, Greally JM, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci USA. 2007;104:3207–12.
Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–14.
Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–36.
Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–91.
Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, et al. Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J Clin Invest. 2016;126:3351–62.
Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, et al. VE-cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235:759–67.
Nie P, Li DD, Hu LH, Jin SX, Yu Y, Cai ZH, et al. Atorvastatin improves plaque stability in ApoE-knockout mice by regulating chemokines and chemokine receptors. PLoS ONE. 2014;9:e97009.
Cui M, Cai Z, Chu S, Sun Z, Wang X, Hu L, et al. Orphan nuclear receptor Nur77 inhibits angiotensin II-induced vascular remodeling via downregulation of β-catenin. Hypertension. 2016;67:153–62.
Cai Z, Wang Z, Yuan R, Cui M, Lao Y, Wang Y, et al. Redox-sensitive enzyme SENP3 mediates vascular remodeling via de-SUMOylation of β-catenin and regulation of its stability. EBioMedicine. 2021;67:103386.
Funding
This work was supported by the national institute grants to M-HZ and the Nurture projects for basic research of Shanghai Chest Hospital (Grant Number: 2022YNJCQ03 to ZC). This work was, in part, supported by the Georgia Research Alliance. M-HZ is a Georgia Research Alliance Eminent Scholar in Molecular Medicine.
Author information
Authors and Affiliations
Contributions
ZC designed and executed the experiments, analyzed the data, and wrote the manuscript. SY, ZL, and PS provided advice and technical assistance and analyzed the data. FZ, JA, and YD provided reagents and technical assistance. BH provided advice, analyzed the data, and revised the manuscript. M-HZ conceived and designed the experiments, analyzed the data, and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee at Georgia State University and were in compliance with relevant ethical regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr Georgia Atkin-Smith
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cai, Z., You, S., Liu, Z. et al. Selective deletion of E3 ubiquitin ligase FBW7 in VE-cadherin-positive cells instigates diffuse large B-cell lymphoma in mice in vivo. Cell Death Dis 15, 212 (2024). https://doi.org/10.1038/s41419-024-06597-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-024-06597-7