Abstract
Alzheimer’s disease (AD), an age-related progressive neurodegenerative disorder, exhibits reduced cognitive function with no cure to date. One of the reasons for AD is the accumulation of Amyloid-beta 42 (Aβ42) plaque(s) that trigger aberrant gene expression and signaling, which results in neuronal cell death by an unknown mechanism(s). Misexpression of human Aβ42 in the developing retina of Drosophila exhibits AD-like neuropathology. Small non-coding RNAs, microRNAs (miRNAs), post-transcriptionally regulate the expression of their target genes and thereby regulate different signaling pathways. In a forward genetic screen, we identified miR-277 (human ortholog is hsa-miR-3660) as a genetic modifier of Aβ42-mediated neurodegeneration. Loss-of-function of miR-277 enhances the Aβ42-mediated neurodegeneration. Whereas gain-of-function of miR-277 in the GMR > Aβ42 background downregulates cell death to maintain the number of neurons and thereby restores the retinal axonal targeting defects indicating the functional rescue. In addition, gain-of-function of miR-277 rescues the eclosion- and climbing assays defects observed in GMR > Aβ42 background. Thus, gain-of-function of miR-277 rescues both structurally as well as functionally the Aβ42-mediated neurodegeneration. Furthermore, we identified head involution defective (hid), an evolutionarily conserved proapoptotic gene, as one of the targets of miR-277 and validated these results using luciferase- and qPCR -assays. In the GMR > Aβ42 background, the gain-of-function of miR-277 results in the reduction of hid transcript levels to one-third of its levels as compared to GMR > Aβ42 background alone. Here, we provide a novel molecular mechanism where miR-277 targets and downregulates proapoptotic gene, hid, transcript levels to rescue Aβ42-mediated neurodegeneration by blocking cell death. These studies shed light on molecular mechanism(s) that mediate cell death response following Aβ42 accumulation seen in neurodegenerative disorders in humans and provide new therapeutic targets for neurodegeneration.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD), a fatal, progressive neurodegenerative disorder, is highly prevalent in the elderly aged 65 or above. It is characterized by progressive neuronal loss, cognitive decline, and memory defects [1, 2]. Furthermore, AD has been reported to be a leading cause of death worldwide with no available cure to date. The presence of amyloid plaques and intracellular tau neurofibrillary tangles are hallmarks of AD. Normally, the amyloid precursor protein (APP) is processed by α-secretase and γ-secretase to form amyloid-beta 40 (Aβ40) peptide. However, if APP is sequentially cleaved by β-secretase and γ-secretase, hydrophobic amyloid-beta 42 (Aβ42) peptides are formed. These Aβ42 monomers aggregate to form amyloid plaques and trigger the aberrant activation of signaling pathways and oxidative stress resulting in neuronal cell death [3,4,5]. Recently the FDA has approved lecanemab, an anti-amyloid antibody that slows down the cognitive and functional decline in early-stage AD patients, according to a phase 3 clinical trial [6]. Thus, amyloid plaques target(s) are being pursued as potential therapeutic targets [6].
Since the genetic machinery is conserved, several vertebrate and invertebrate model systems are being developed and used to study AD pathology and underlying mechanism(s) [7,8,9,10,11]. Invertebrate model system like Drosophila melanogaster can be easily genetically manipulated and offers a unique advantage to study the molecular mechanism(s) and pathogenesis of AD [11,12,13,14]. The Drosophila eye has been extensively used for modeling neurodegenerative models like AD [13, 15,16,17]. The adult compound eye, which develops from larval eye-antennal imaginal disc, is comprised of approximately 800 units called ommatidia. Each ommatidium consists of 8 photoreceptors and several support cells [17,18,19,20,21]. The undifferentiated epithelial cells in the eye imaginal disc begin to differentiate into retinal neurons and other cell types at the late larval stage onto pupal development [18, 19]. Between 24h and 40 h after pupa formation (APF), any extra cells are eliminated through programmed cell death (PCD) to refine the hexagonal lattice [18, 19]. The precise organization of Drosophila eye makes it highly sensitive to genetic manipulations and allows quick screening of large sample size [21,22,23,24]. Therefore, the Drosophila eye is employed to mimic many neurodegenerative disorders including AD and study the different signaling pathways as well as screen genetic modifiers and therapeutic targets for AD [13, 15, 25,26,27].
Using the Gal4/UAS transgenic target system [28], human Aβ42 is spatiotemporally misexpressed in the differentiating photoreceptor neurons of the developing eye to mimic AD [15,16,17]. This results in the accumulation of amyloid plaques exhibiting a progressive neurodegenerative phenotype in the Drosophila eye as compared to the wild-type eye, and thus phenocopies AD-like neuropathology [17, 26]. Therefore, the Drosophila eye model, due to repertoire of genetic tools, can be exploited for genome-wide screening to identify genetic modifiers of AD-like neuropathology. Accumulation of Aβ42 plaques triggers cell death, which is the primary cause of neurodegeneration in AD [29]. The proapoptotic factor head involution defective (hid) along with other caspases like Dronc, Drice, and Dark are involved in regulating PCD during pupal eye development [19]. In Drosophila, upon apoptotic stimuli, three proapoptotic genes: head involution defective (hid), reaper (rpr) and grim (grim) are expressed and thereby trigger cell death by inhibiting Drosophila inhibitor of apoptosis (DIAP1) [30,31,32]. Upon DIAP1 degradation, initiator caspase: Dronc (caspase 9) as well as effector caspase: Drice (caspase 3) are activated. This activation of the caspase cascade results in cell death. Caspase-dependent cell death can be prevented by high levels of baculovirus protein P35 [33]. Cell death observed in various human diseases is an outcome of aberrant signaling due to abnormal gene expression.
Complex gene regulatory mechanisms control the expression of genes during development. MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression of different signaling pathways [34]. Recent findings indicate that miRNAs play a crucial role in modulating multiple signaling pathways associated with various diseases [35]. These miRNAs confer specificity to the RNA-induced silencing complex (RISC) through partial sequence complementarity with specific mRNA targets. Recruitment of miRNA and the RISC complex mostly results in repression of the target mRNA by an increase in turnover and/or translational inhibition. miRNAs regulate many biological events, including growth, development, differentiation, and neurodegenerative processes [36, 37]. Therefore, we hypothesized that miRNAs could regulate the expression of genes, which are members of signaling pathways that are involved in AD.
In a forward genetic screen using candidate miRNAs, we screened for potential genetic modifiers of Aβ42-mediated neurodegeneration phenotype. The rationale of the screen was to individually co-express a microRNA transgene along with GMR > Aβ42 in the developing retina, and to screen for modifiers of the neurodegenerative phenotype. In this screen, we identified miR-277 as a potential miRNAs genetic modifier using the Drosophila eye model. Here, we report that upregulation of miR-277 rescues Aβ42-mediated neurodegeneration phenotype whereas downregulation of miR-277 enhances Aβ42-mediated neurodegenerative phenotype of reduced eye with glazed surface. Furthermore, we have identified hid mRNA as a target of miR-277 using bioinformatics, genetic and molecular approaches. Here, we present an insight on the underlying mechanism of how miR-277 ameliorates Aβ42-mediated neurodegeneration by regulating proapoptotic gene, hid, transcript levels and demonstrate the novel neuroprotective role of miR-277 in AD neuropathology.
Materials and methods
Stocks
The fly stocks used in this study are listed in Flybase (http://flybase.bio.indiana.edu). Stocks used in this study are GMR-Gal4 [38], Elav-Gal4 [39], OK107-Gal4 [40, 41], UAS-Aβ42 [42], UAS miR-277, miR-277 mutant generated by TALEN editing, Df(3 R)miR-277-34-KO (BL#58908), GMR-hid; GMR-Gal4, GMR>rpr, GMR>grim, hid5’FWT-GFP [43]. The UAS-Aβ42 transgenic flies were generated by microinjecting a UAS-construct where two tandem copies of human amyloid—β1-42 (Aβ42) fused to signal peptide for secretion were cloned [44, 45]. The rationale of bi-cistronic construct was to mimic APP duplications associated with early onset of familial AD and to express high levels of Aβ42 to induce strong eye phenotype.
Generation of miR-277 mutant by TALEN gene editing
We employed the transcription activator-like effector nucleases (TALEN) system for gene editing [46] to generate miR-277 mutant lines. A 150 base pairs of flanking sequences that surrounded miR-277 mature sequences were selected and added to the TALE-NT website (https://tale-nt.cac.cornell.edu/about) for designing target sites pairs for TALENs binding and editing [47, 48]. One pair of TALEN target sites, TALEN-miR-277-L_5′-GAAACTATCTGAAGCAT-3′ and TALEN-miR-277-R_5′-TCTGGAATGTCGTACC-3′, was chosen for the TALEN plasmids generation. The TALEN plasmids were synthesized by ZGENEBIO Biotech Inc. The midi-scale of TALEN plasmids was prepared by following the QIAGEN Plasmid Midi Kit (QIAGEN, #12145) and then transferred to BestGene Inc for Drosophila embryo injection services.
After embryo microinjection, all the G0 adults were crossed with stubble (TM3) balancer strains by combining one male and one female to generate the individual family. From each family, 15 G1 offspring were randomly picked up to establish the homozygous subfamily strains for mutant line screening. Genomic DNA isolations and PCR reactions were performed using the published protocol [49]. The primer pairs used for PCR reaction and Sanger sequencing are mentioned in Table 1.
pTub-miR-277 plasmid
A 500 base pairs of DNA fragment that included miR-277 stem-loop precursor with each 200 base pairs of up and downstream flanking sequences were amplified from genomic DNA using the miR-277 specific primer set (Table 2). The miR-277 fragment was cloned into NotI/XhoI sites of the pTub-miR plasmid (gift from Dr. Stephen M. Cohen) to generate the pTub-miR-277 plasmid.
Genetic crosses
Gal4/ UAS targeted misexpression system was used in our study [28]. The genetic crosses were maintained at 25 °C, while the egg-lays were transferred to 29 °C for further growth. Misexpression of Aβ42 in the differentiating retina (GMR-Gal4 > UAS-Aβ42) exhibits a stronger neurodegenerative phenotype at 29 °C with no penetrance [38]. GMR-Gal4 directs the expression of transgenes in the differentiating retinal precursor cells of the developing eye imaginal disc and pupal retina [38]. All crosses with Df(3 R)miR-277-34-KO (BL#58908) stock are mentioned as Df(3 R)miR-277KO.
Adult eye imaging
The adult flies of similar age from both sexes were stored at −20 °C for approximately 2 h for imaging. After the incubation, the flies were mounted on a dissection needle. It was placed horizontally over a glass slide using clay putty. The adult eye was imaged on the Axiomager.Z1 Zeiss Apotome and the Z-stacks were obtained [42, 50]. The final images were generated by compiling individual stacks from the Z section using the extended depth of focus function of Axiovision software version 4.6.3.
Frequency of eye phenotype
For each genetic cross, three independent sets of two hundred flies were screened (200 × 3 = 600) and the frequency of eye phenotype(s) was calculated [51]. The eye phenotypes were categorized as severely reduced eye, reduced eye, and rescue of neurodegenerative phenotype. Graphs were plotted in GraphPad Prism.
Quantitative analyses of severity score of eye degenerative phenotype
We examined the eye phenotypes from 200 flies per genotype and these flies were selected for scoring according to the following criteria: “No-eye” was assigned to category 6, 80% eye degeneration was assigned to category 5, 60–80% eye degeneration was assigned to category 4, 40–60% eye degeneration was assigned to category 3, 20–40% eye degeneration was assigned to category 2, 0–20% eye degeneration was assigned to category 1 and wild-type was assigned to category 0 [51]. Comparisons were made using non-parametric: Mann–Whitney t test and graphs were plotted in GraphPad Prism.
Quantitative analyses of relative surface area of the eye
The adult eye images were opened in Image J software, and region of interest (ROI) was drawn and represented as yellow dotted line. We measured the surface area of the eye of five flies per genotype by using Image J software and plotted graph in GraphPad Prism.
Immunohistochemistry
Eye-antennal imaginal discs were dissected from the third instar larvae in cold 1× phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in 1× PBS for 20 min, and then quickly washed once in 1× PBS. It was followed by three washes in 1× PBST. The tissues were stained with a combination of antibodies following a previously published protocol [42, 52]. The primary antibodies used were rat anti-Embryonic Lethal Abnormal Vision (ELAV) (1:100; Developmental Studies Hybridoma Bank, DSHB), mouse anti-Discs-large (Dlg) (1:100; DSHB), and mouse anti-Chaoptin (24B10) (1:100; DSHB) [53]. Secondary antibodies (Jackson Laboratory) used were goat anti-rat IgG conjugated with Cy5 (1:250), and donkey anti-mouse IgG conjugated with Cy3 (1:250). The tissues were mounted in the antifading agent: Vectashield (Vector Laboratories). The immunofluorescent images were captured at 20× magnification by using Olympus Fluoview 3000 Laser Scanning Confocal Microscope [54]. All final figures were prepared using Adobe Photoshop software.
Detection of cell death
Apoptosis was detected by using terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay detection kit from Roche Diagnostics. TUNEL assay labels the DNA fragments in dying cells. This protocol involves labeling DNA breakage by adding fluorescently labeled nucleotides to free 3′-OH DNA ends in a template-independent manner using terminal deoxynucleotidyl transferase (TdT). The fluorescein labels (TMR red) incorporated in nucleotide polymers can be detected by fluorescence microscopy [32, 55]. After secondary-antibody staining, eye-antennal discs were blocked in 10% normal goat serum in phosphate-buffered saline with 0.2% Triton X-100 (PBST) and labeled for TUNEL assays using a cell-death detection kit from Roche Diagnostics. The TUNEL positive cells were counted from five sets of imaginal discs per genotype and were used for the statistical analysis using Microsoft Excel 2013 [56]. The graphs were plotted in GraphPad Prism, the p values were calculated using student’s t test, and the error bars represent standard error of mean (SEM). The symbols above the error bar signify *p value < 0.05, **p value < 0.01, ***p value < 0.001.
Pupal retina staining
Early white pre-pupae per genotype were selected and kept on a moist kim-wipe in a petri plate. After 48 h of collection of pre-pupae, pupal retina was dissected in cold 1× PBS carefully from the pupa. The retinae were then fixed in 4% paraformaldehyde in 1× PBS for 20 min, and then washed once in 1× PBS. It was followed by three washes in 1× PBST. The dissection, washing, and antibody staining was done in nine well plate. The tissues were stained with a combination of antibodies following a previously published protocol [42, 52]. The primary antibodies used were rat anti-Embryonic Lethal Abnormal Vision (ELAV) (1:100; Developmental Studies Hybridoma Bank, DSHB), and mouse anti-Discs-large (Dlg) (1:100; DSHB). Secondary antibodies (Jackson Laboratory) used were goat anti-rat IgG conjugated with Cy5 (1:250), and donkey anti-mouse IgG conjugated with Cy3 (1:250). The tissues were mounted in Vectashield (Vector Laboratories). The immunofluorescent images were captured at ×60 magnification by using Olympus Fluoview 3000 Laser Scanning Confocal Microscope [54]. All final figures were prepared using Adobe Photoshop software. The pupal retina was opened in Image J and ROI of 500px × 500px was drawn and the number of photoreceptor cells and the number of pigment cells were counted within the ROI, respectively. The five sets of pupal retinae per genotype were used for statistical analysis using Microsoft Excel. The graphs were plotted in GraphPad Prism. The p values were calculated using student’s t test, and the error bars represent standard error of mean (SEM) *p value < 0.05, **p value < 0.01, ***p value < 0.001.
DHE
The third instar larval eye-antennal imaginal discs were dissected in cold 1× Schneider’s Drosophila medium (Gibco, Cat. #21720024). The samples were incubated in Dihydroethidium (DHE, Life Technologies Cat. # D11347) dye solution [(1:300) in 1× PBS] [57, 58] for 5 min and were washed three times with cold 1× PBS. The eye discs were then mounted on a slide and were immediately imaged on a Laser Scanning Confocal microscope (Olympus Fluoview 3000) [54]. All final figures were prepared using Adobe Photoshop software. The number of ROS puncta were quantified from five sets of imaginal discs per genotype by using automated quantification method [58]. The Interactive H watershed plugin of Image J/Fiji free software was used for automated quantification and the statistical analysis was performed using Microsoft Excel [58]. The p values were calculated using student’s t test, and the error bars represent the standard error of mean (SEM) *p value < 0.05, **p value < 0.01, ***p value < 0.001.
Luciferase reporter plasmid
The 3′UTRs of three apoptosis-related genes, hid, dark, and drice, were amplified from genomic DNA of W[1118] strain with the following gene-specific 3′UTR primer pairs (Table 3).
The 3′UTR DNA fragments were individually cloned into the XbaI/StuI sites of the pTub-ffluc reporter plasmid (gift from Dr. Stephen M. Cohen) to generate three luciferase reporter plasmids, pTub-ffuc-hid, pTub-ffuc-dark and pTub-ffuc-drice, by In-Fusion HD Cloning Kit (Clontech, 639646).
Luciferase assay
The luciferase reporter gene assay has recently been adapted to test whether a certain mRNA is the target for a specific miRNA [59]. The day prior to transfection, Drosophila Schneider S2 (S2) cells were seeded at a density of 1–1.5 × 106 cells per well in 12-well plate in Schneider’s Drosophila medium (SDM) (Gibco, Cat. #11720034) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Cat. #A3160402) and 1× penn/strep (Invitrogen, Cat. #15070-063). Before transfection, cells were washed with SDM only and changed the cultured medium into SDM (300 μl/well) only without serum and penn/strep for 1 h. The diluted plasmid reagent was prepared by mixing the following plasmids, 100 ng of pTub-rLuc (gift from Dr. Stephen M. Cohen), 100 ng of pTub-ffLuc luciferase reporter (containing 3′UTR sequence of the target gene), and 1 μg of pTub-miR-277 with 100 μl SMD for each well. The diluted Cellfectin (Invitrogen, 10362-010) reagent was prepared by adding 5 μl Cellfectin in 100 μl SDM for each well and wait for 5 min. The diluted Cellfectin was mixed with diluted plasmids and waited for 30 min. The mixture was added to cells for 4–6 h transfection reaction, and then medium was changed into SDM with 10% FBS and 1× penn/strep. The pTub-miR-1 (gift from Dr. Stephen M. Cohen) was performed at the same time in each transfection for negative control [60]. After 48-hour transfection, cells were lysed to detect the luciferase activities by using the Dual-Luciferase Reporter Assay System (Promega, E1910). Renilla luciferase activity provided normalization for firefly luciferase activity. The value of relative luciferase activities of the different pTub-ffLuc-3′UTR constructs was calculated from the normalization of the luciferase activities of miR-1. The targeting effect of miR-277 on the luciferase gene expression can be shown as the relative luciferase activity.
Real-time quantitative polymerase chain reaction
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed according to the standardized protocol [61, 62]. Total RNA was extracted in 500 μl of TRIzol Reagent (Thermo Fisher, Cat. No. # 15596926) from twenty pairs of third instar larvae eye-antennal imaginal discs (n = 40), which were dissected from GMR-Gal4, GMR > Aβ42, GMR>miR-277, GMR > Aβ42+ miR-277, GMR>miR-277 mutant, GMR > Aβ42+ miR-277 mutant, GMR > Df(3 R)miR-277KO and GMR > Aβ42 + Df(3 R)miR-277KO. The quality of isolated RNA was determined by using the Nanodrop 2000 spectrophotometer (Thermo Scientific). The quality of samples was checked by A260/A280 ratio which were >2. cDNA was produced from total RNA through RT-PCR using the first-strand cDNA synthesis kit (GE healthcare, Cat# 27926101). RT-qPCR was performed using iQ™ SYBR Green Supermix (Bio-Rad) and Bio-Rad iCycler (Bio-Rad) following the kit’s protocol for 25 μl. Primers used for hid were: (fwd: CCACCGACCAAGTGCTATAC; rev: CGGCGGATACTGGAAGATTT). Primers used for miR-277 were: (fwd: GCGTGTCAGGAGTGCATTT; rev: GTACGTTCTGGAATGTCGTACC). The expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (fwd: CAATGGATTTGGTCGCATCG; rev: CCGTTGACCACCAGG AAACC) was used as an internal control to normalize the results. The fold change was calculated relative to the expression level of the respective control using delta deltaCT (ΔΔCT) method [61].
Eclosion assay
Eclosion assays are used for screening the effects of various genetic backgrounds on eclosion of flies. We used Elav-Gal4 line to drive the expression of Aβ42 and other transgene in the central nervous system [39]. We collected eggs on a grape plate from Elav-Gal4 (control), Elav > Aβ42, Elav > Aβ42 + miR-277, Elav > Aβ42 + miR-277 mutant and Elav > Aβ42 + Df(3 R)miR-277KO. We seeded the first instar larvae (30 in each set) from a synchronous culture in each vial. 270 larvae (9 sets of 30 larvae) were counted for each cross. The larvae were allowed to develop to adulthood, and the eclosion rate [42] was counted. All unhatched pupae were also counted. The graph was plotted in GraphPad Prism.
Climbing assay
Climbing assays were performed to characterize the locomotor dysfunction. We used OK107-Gal4 line to drive the expression of Aβ42 and other transgene in the larval mushroom body as well as in adult mushroom body lobes [40, 41]. Since, Elav > Aβ42 flies were lethal, we used OK > Aβ42 flies for detecting if miR-277 can affect the locomotor defects observed in AD flies. Flies were aged from 1 to 30 days in the regular food. Groups of 10 flies per genotype were transferred into cylindrical glass tube after anesthetization and left for 5-10 min for the revival and acclimatization at room temperature. Tubes were marked at 10 cm above the bottom of the vial. After acclimatization, the flies were gently tapped down to the bottom of vial, and the number of flies that crossed the 10 cm mark were recorded after 10 sec. Three trials were performed, and numbers were then averaged, and the resulting mean was used as the overall value for each single group of flies. For all genotypes, three replicates were carried out.
Results
miR-277 is a genetic modifier of Aβ42-mediated neurodegeneration
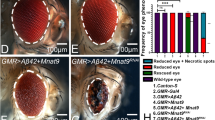
Drosophila larval eye imaginal disc (Fig. 1A) develops into an adult compound eye comprising around 800-unit eyes called ommatidia (Fig. 1B). Note that the eye imaginal disc is stained for membrane-specific marker- discs large (Dlg, green) and pan neural marker- embryonic lethal abnormal vision (Elav, red) (Fig. 1A, C, E, G, I, K, M, O, Q). The GMR-Gal4 driver flies have wild-type adult eye (Fig. 1D) and eye imaginal discs (Fig. 1C). Misexpression of human Aβ42 in the differentiating retinal neurons of the developing eye by using GMR-Gal4 driver (GMR > Aβ42), results in a progressive neurodegenerative phenotype with increased spaces in the photoreceptors mostly at the posterior margin of the eye disc (Fig. 1E), which further worsens in the adult eye as evident from the reduced eye size, disorganized and fused ommatidia in the adult eye (Fig. 1F). The progressive neurodegenerative phenotype of highly reduced adult eye has 100% penetrance (n = 600, 600/600 = 100%) (Fig. 1F, S, T, U) [17]. In a forward genetic screen (Supplementary Figure 1), we identified miRNA−277 (miR-277) as the modifier of the neurodegenerative phenotype of GMR > Aβ42 (Fig. 1M, N, S, T, U). Misexpression of miR-277 alone (GMR > miR-277) serves as a control and exhibits near-normal eye phenotype in the eye imaginal disc and adult eye (Fig. 1G, H). Whereas gain-of-function of miR-277 in GMR > Aβ42 (GMR > Aβ42+miR-277) background (Fig. 1M, N, S, T, U) significantly rescues the Aβ42-mediated neurodegeneration (Fig. 1E, F, S, T, U) as seen in eye imaginal disc as well as the adult eye (n = 600, 600/600 = 100%).
A Wild-type larval eye imaginal disc develops in to B an adult eye comprises of ~800 organized ommatidia. Note that the eye imaginal disc is stained with a membrane-specific marker, discs large (Dlg; green), and a pan neural marker ELAV (red) that marks the photoreceptors (C, D) GMR-Gal4, drives the expression of target transgenes in the developing eye, serves as a control. E, F Misexpression of human Aβ42 in the developing (E) eye imaginal disc (GMR > Aβ42) leads to highly reduced (F) adult eye phenotype with fused ommatidia. G, H GMR>miR-277, I, J GMR>miR-277 mutant, and K, L GMR > Df(3 R)miR-277KO eye imaginal discs and adult eye also served as controls. M, N Gain-of-function of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277) results in significant rescue in eye disc and adult eye as compared to the GMR > Aβ42. O–R However, reducing miR-277 levels in GMR > Aβ42 flies using (O, P) miR-277 mutant (GMR > Aβ42+ miR-277 mutant), (Q, R) Df(3 R)miR-277KO [GMR > Aβ42 + Df(3 R)miR-277KO] enhances GMR > Aβ42 neurodegenerative phenotype. S Bar graph shows frequency as average between 3 repetitions. Two hundred flies were counted per repetition (200 × 3 = 600) to calculate the frequency for each genotype (1. GMR-Gal4, 2. GMR > Aβ42, 3. GMR > Aβ42+miR-277, 4. GMR > Aβ42+miR-277 mutant, 5. GMR > Aβ42 + Df(3 R)miR-277KO). Statistical analysis was performed using the student’s t test for independent samples. T Quantitative analyses of severity score of neurodegenerative phenotypes(s) in eye. Flies from each genotype were selected for scoring according to the criteria described in methods section. Comparisons were made using non-Parametric: Mann–Whitney t test. U Quantitative analyses of area of the eye. The surface area of the eye (within white dotted line) was calculated using Image J. Statistical analysis was performed using the student’s t test for independent samples. The surface area is significantly rescued in GMR > Aβ42+miR-277 (n = 5; p = 0.0000000146) whereas significantly reduced in GMR > Aβ42+miR-277 mutant (n = 5; p = 0.017) and GMR > Aβ42 + Df(3 R)miR-277KO (n = 5; p = 0.02) as compared to GMR > Aβ42. V Relative expression of miR-277 at the transcriptional level using quantitative PCR (qPCR) in genotypes (1. GMR-Gal4 2. GMR > Aβ42, 3. GMR > Aβ42+ miR-277). Triplicate was used for the calculation. Statistical analysis was performed using student’s t test for independent samples. The data plotted shows mean ± SEM (Standard Error of Mean), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05 respectively. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. Scale bar = 100 μm.
To further validate our hypothesis, we investigated the loss-of-function of miR-277 by using miR-277 mutant (Fig. 1I, J) generated by TALEN system (Supplementary Figure 2A–C) or a deficiency, Df(3 R) miR-277KO that knocks out miR-277 function [GMR > Df(3 R)miR-277KO; Fig. 1K, L] in GMR-Gal4 background. Interestingly, loss-of-function of miR-277 in GMR > Aβ42 background (GMR > Aβ42+ miR-277 mutant) enhances the neuronal loss and hence show reduced eye phenotype (Fig. 1O, P, S, T, U) (Supplementary Figure 2) as compared to the GMR > Aβ42 alone (Fig. 1F). The severity of the phenotype is more in frequency with Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO) (n = 600, 333/600 = 55.5%) (Fig. 1S) than with the molecular mutant (n = 600, 306/600 = 51%) (Fig. 1S). Moreover, gain-of-function of miR-277 in the GMR > Aβ42 flies significantly suppressed the neurodegenerative phenotype (Fig. 1T) and increased the surface area of the eye (Fig. 1U). This further validates our findings from the forward genetic screen that miR-277 is a genetic modifier of Aβ42-mediated neurodegeneration in the Drosophila eye.
We further quantitated the miR-277 transcript levels by qPCR and found that miR-277 levels were reduced to half in GMR > Aβ42 background as compared to the control, GMR-Gal4. Furthermore, miR-277 levels were increased by 2.5 folds in GMR > Aβ42+ miR-277 background as compared to the control (Fig. 1V). Thus, our data suggest that a reduction in miR-277 levels triggers neurodegenerative response.
Gain-of-function of miR-277 can restore axonal targeting defects
In AD, neurons die due to the deposition of amyloid plaques. Since miR-277 can rescue the reduced eye phenotype of GMR > Aβ42, we investigated if miR-277 can rescue the functionality of the neurons. One of the facets of the neurodegenerative phenotypes in retinal neurons is the disruption of axonal transport due to impaired axonal targeting and guidance from retinal neurons to the brain. We used Chaoptin (24B10, DSHB), a reliable marker for axonal targeting that marks the photoreceptor neurons and their axons [53]. During the late third instar eye imaginal disc, R1-R6 axons of each ommatidium project to the lamina, whereas R7 and R8 axons project to the medulla, a separate layer of the optic lobes [16, 63]. We counted a total of n = 50 eye discs per genotype, and frequency of eye discs indicating rescue of axonal targeting from GMR > Aβ42 was recorded (Fig. 2A–J). The wild-type (n = 50) 100% (Fig. 2A, J) and GMR-Gal4 (n = 50) 100% (Fig. 2B, J) eye imaginal discs showed similar axonal projections in the brain. In comparison to the controls, GMR > Aβ42 imaginal discs exhibit severe defects in axonal targeting, which contributes to the neurodegenerative phenotype in AD (Fig. 2C, J) [16, 64]. Misexpression of miR-277 in GMR > Aβ42 (GMR > Aβ42+ miR-277; Fig. 2G, J) background, significantly restored the axonal targeting in comparison to the GMR > Aβ42 (Fig. 2C, J). However, GMR> miR-277 (Fig. 2D), or GMR>miR-277 mutant (Fig. 2E), or GMR > Df(3 R)miR-277KO (Fig. 2F) alone did not show any axonal targeting defects. The axonal targeting was further impaired or worsened when miR-277 levels were downregulated in the GMR > Aβ42 background by either miR-277 mutant (GMR > Aβ42 + miR277 mutant, Fig. 2H, J) or by Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO, Fig. 2I, J). This evidence suggests that upregulating the levels of miR-277 restores the axonal targeting defects, as seen in the GMR > Aβ42 background. It has been shown that impaired axonal targeting is associated with neuronal cell death. Therefore, we tested the effects of modulation of miR-277 in cell death in GMR > Aβ42 background.
Chaoptin (24B10) marks the photoreceptors and their neurons. The photoreceptor neurons bundle up in the optic stalk and innervate the medulla and lamina of the brain. A In the wild-type eye imaginal disc, the retinal axons marked by 24B10 innervates the two layers of the brain marked with yellow arrows. B GMR-Gal4 serves as the control. C Misexpression of Aβ42 (GMR > Aβ42) results in impaired axonal targeting from retina to the brain. The retinal axons fail to innervate the two layers of the brain and end abruptly. Controls (D) GMR>miR-277 (E) GMR>miR-277 mutant, and (F) GMR > Df(3 R)miR-277KO do not impact axonal targeting. G Gain-of-function of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277) significantly restores the axonal targeting to near wild-type. Loss-of-function of miR-277 in GMR > Aβ42 flies using (H) miR-277 mutant (GMR > Aβ42+ miR-277 mutant), and (I) Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO) disrupt the axonal targeting. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. The magnification of all eye imaginal discs is ×20. J Bar graph shows the frequency of restoration of axonal targeting phenotype. Sample size was 5 for each genotype 1. GMR-Gal4, 2. GMR > Aβ42, 3. GMR > Aβ42+miR-277, 4. GMR > Aβ42+miR-277 mutant, 5. GMR > Aβ42 + Df(3 R)miR-277KO. GMR > Aβ42+miR-277 significantly restores the axonal targeting (n = 5; p = 0.000029) whereas GMR > Aβ42+ miR-277 mutant (n = 5; p = 0.37) and GMR > Aβ42 + Df(3 R)miR-277KO (n = 5; p = 0.38) disrupt the axonal targeting as compared to GMR > Aβ42. The data plotted shows mean ± SEM (Standard Error of Mean), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05 respectively. Scale bar = 100 μm.
Gain-of-function of miR-277 can block Aβ42-mediated cell death
We employed terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining to mark the nuclei of dying cells. Fluorescein-dUTP is transferred by the enzyme terminal deoxynucleotides transferase (TdT) at 3’OH where single or double-strand breaks occur during apoptosis [55, 56]. The TUNEL-positive cells were counted within yellow dotted ROI from five imaginal discs of each genotype and were used for statistical analysis. A few cells undergo cell death in the wild-type eye imaginal disc (Fig. 3A, J), whereas the GMR > Aβ42 eye imaginal disc shows significantly increased TUNEL positive nuclei (Fig. 3C, J). Gain-of-function of miR-277 in the GMR > Aβ42 background (GMR > Aβ42+miR-277; Fig. 3G, J) results in a significant reduction (~4 times) in the number of dying cells as compared to the GMR > Aβ42 (Fig. 3C, J). Loss-of-function of miR-277 in GMR > Aβ42 background either by miR-277 mutant (GMR > Aβ42+ miR-277 mutant; Fig. 3H, J) or by Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO; Fig. 3I, J), results in significant increase in number of dying nuclei as compared to the wild-type (Fig. 3A, J). There is a slight increase in TUNEL-positive nuclei in case of deficiency of miR-277 (GMR > Aβ42 + Df(3 R)miR-277KO; Fig. 3I, J) as compared to the miR-277 mutant (GMR > Aβ42+ miR-277 mutant; Fig. 3H, J). Hence, TUNEL data suggests that the miR-277 might downregulate cell death caused due to Aβ42-mediated neurodegeneration.
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay is employed to mark the cells undergoing cell death. A Wild-type (Canton-S) and B GMR-Gal4 eye imaginal discs show a few TUNEL-positive nuclei. C Misexpression of Aβ42 using GMR-Gal4 driver (GMR > Aβ42) shows increased levels of TUNEL positive nuclei as compared to (A) wild-type eye imaginal disc. D GMR>miR-277, E GMR>miR-277 mutant, and F GMR > Df(3 R)miR-277KO serve as controls. G Gain-of-function of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277) significantly rescue the cell death as compared to GMR > Aβ42. Loss-of-function of miR-277 in GMR > Aβ42 flies using H miR-277 mutant (GMR > Aβ42+ miR-277 mutant), and (I) Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO) show elevated levels of TUNEL positive nuclei. A–I TUNEL-positive nuclei were counted within yellow dotted line, the region of interest, for the statistical analysis. A, C, G–J TUNEL-positive nuclei in photoreceptor cells of five eye imaginal discs per genotype were counted (1. Canton-S, 2. GMR-Gal4, 3. GMR > Aβ42, 4. GMR>miR-277, 5. GMR > Aβ42+ miR-277, 6.GMR> miR-277 mutant, 7. GMR > Aβ42+ miR-277 mutant, 8. GMR > Df(3 R)miR-277KO, 9. GMR > Aβ42 + Df(3 R)miR-277KO). Statistical analysis was performed using student’s t test for independent samples. GMR > Aβ42+miR-277 exhibits significant reduction in TUNEL-positive nuclei as compared to GMR > Aβ42 (n = 5; p = 0.000000014) whereas GMR > Aβ42+ miR-277 mutant (n = 5; p = 0.083) and GMR > Aβ42 + Df(3 R)miR-277KO (n = 5; p = 0.056) show slight increase in TUNEL positive nuclei as compared to GMR > Aβ42. The data plotted shows mean ± SEM (Standard Error of Mean), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05 respectively. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. Scale bar = 100 μm.
To investigate further if miR-277 can inhibit apoptosis during later stages of development, we performed pupal retina staining at 48 h after pupa formation (APF). During late larval development, the photoreceptor and cone cells are determined, following which the interommatidial cells (IOC’s) are specified to become pigment cells. Interommatidial (or pigment cells) surround the centrally located photoreceptor and cone cells, generating a precise, repeating hexagonal structure (ommatidium), which can be visualized during the pupal stage. This hexagonal lattice is formed due to programmed cell death (PCD), which occurs during 24–40 h APF to eliminate extra interommatidial cells [18, 65]. We dissected the pupal retina at 48 h APF, when PCD was over. The number of photoreceptor nuclei and pigment cells from five individual areas was selected from five different pupal retinae for each genotype for analysis. The control GMR-Gal4 shows hexagonal arrangement of ommatidia and a monolayer of secondary and tertiary pigment cells (Fig. 4A, E, F). In contrast, the basic ommatidial organization is disrupted in GMR > Aβ42 pupal retina due to cell death, which results in the fusion of photoreceptors among neighboring ommatidia as evident from ELAV staining that marks the photoreceptor nuclei (Fig. 4B, E). In addition, the membrane of primary and secondary pigment cells is lost, as marked by Dlg (Fig. 4B, F). We observed multiple layers of secondary and tertiary pigment cells resulting in extra pigment cells when miR-277 was misexpressed in the GMR domain (GMR>miR-277; Fig. 4C, F). Since miR-277 is an anti-apoptotic miRNA, in GMR>miR-277 background, extra IOC’s accumulate between the ommatidia of the pupal retina as programmed cell death is inhibited. Interestingly the disruption of cell membrane, number of pigment cells, and fused ommatidia were significantly restored to near wild-type when miR-277 was misexpressed in the GMR > Aβ42 background (GMR > Aβ42+miR-277; Fig. 4D–F). Therefore, these results further validated our hypothesis that higher levels of miR-277 suppress the neurodegenerative phenotype of Aβ42 aggregate accumulation. Hence, miR-277 could be an anti-apoptotic miRNA. It has been previously reported that Aβ42 aggregate triggers the production of reactive oxygen species (ROS) [58, 66, 67].
Pupal retinae were stained with Discs large (Dlg) which marks the membrane (green), and a pan neural marker ELAV (red) which marks photoreceptors. A Pupal retina of wild-type shows hexagonal ommatidia and single layer of interommatidial cells. B GMR > Aβ42 shows disrupted pupal retina as compared to A wild-type pupal retina. C pupal retina of GMR>miR-277 shows excess interommatidial cells as compared to the control (A) wild-type pupal retina. D GMR > Aβ42+ miR-277 restores the number and shape of ommatidial and interommatidial cells. E Bar graph represents the number of photoreceptors within the area of 500px × 500px. We examined five pupal retinae per genotype (n = 5) (1. GMR-Gal4, 2. GMR > Aβ42, 3. GMR>miR-277, 4. GMR > Aβ42+ miR-277). Statistical analysis was performed using student’s t test for independent samples. The number of photoreceptors is significantly restored in GMR > Aβ42+ miR-277 (n = 5; p = 0.0000018) as compared to GMR > Aβ42. F Bar graph represents the number of pigment cells (secondary, tertiary, and bristle cells) within the area of 500px × 500px. We examined five pupal retinae per genotype (n = 5) (1. GMR-Gal4, 2. GMR > Aβ42, 3. GMR>miR-277, 4. GMR > Aβ42+ miR-277). Statistical analysis was performed using student’s t test for independent samples. The number of pigment cells is significantly restored in GMR > Aβ42+ miR-277 (n = 5; p = 0.000064) as compared to GMR > Aβ42. Error bars show standard error of mean (mean ± SEM), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, * p value < 0.05, and not significant (ns), p value > 0.05 respectively. Scale bar = 50 μm.
Gain-of-function of miR-277 downregulates ROS production
The accumulation of amyloid plaques triggers oxidative stress in the neurons resulting in an imbalance in the generation of reactive oxygen species (ROS) and antioxidant defense mechanism [17, 58, 68, 69]. Excessive generation of ROS levels leads to oxidative modification of biomolecules in postmitotic neurons that are associated with AD pathology [58, 70, 71]. Hence, we measured ROS levels using dihydroethidium (DHE) staining in eye-antennal imaginal discs when miR-277 levels were modulated in the background of GMR > Aβ42 flies. DHE is oxidized by superoxide radical to form 2-hydroxyethidium, which intercalates with DNA and provides signal at 550 nm in cells where ROS is produced [58, 72, 73]. The ROS puncta were calculated (within the yellow dotted line that marks ROI) from five imaginal discs per genotype and were used for statistical analysis (Fig. 5A–F). Misexpression of Aβ42 (GMR > Aβ42; Fig. 5B, F) results in prominent ROS production as compared to minimal ROS levels in wild-type control Canton-S (Fig. 5A, F). Interestingly, gain-of-function of miR-277 in the GMR > Aβ42 background (GMR > Aβ42 + miR-277; Fig. 5C, F) shows reduced or similar levels of ROS signal as compared to the wild-type (Fig. 5A, F). Loss-of-function of miR-277 in the background of GMR > Aβ42 either by miR-277 mutant (GMR > Aβ42 + miR-277 mutant; Fig. 5D, F) and Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO; Fig. 5E, F) show increased ROS levels as compared to the GMR > Aβ42 (Fig. 5B, F). Hence, high levels of miR-277 can downregulate ROS levels in GMR > Aβ42 flies.
Dihydroethidium (DHE) is employed to detect ROS production in cells. A Wild-type (Canton-S) eye imaginal discs show a few ROS puncta. B GMR > Aβ42 shows elevated ROS puncta as compared to A wild-type eye imaginal disc. C Gain-of-function of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277) significantly reduced the ROS production as compared to GMR > Aβ42. Loss-of-function of miR-277 in GMR > Aβ42 background using D miR-277 mutant (GMR > Aβ42+ miR-277 mutant), and E Df(3 R) miR-277KO [GMR > Aβ42 + Df(3 R)miR-277KO] result in increased ROS production. A–F ROS puncta were counted within yellow dotted line, the region of interest, for the statistical analysis. ROS puncta were quantified in photoreceptor cells of five eye imaginal discs per genotype (1. Canton-S, 2. GMR > Aβ42, 3. GMR > Aβ42+ miR-277, 4. GMR > Aβ42+ miR-277 mutant, 5. GMR > Aβ42 + Df(3 R)miR-277KO). Statistical analysis was performed using student’s t test for independent samples. GMR > Aβ42+miR-277 exhibits significant reduction in ROS puncta as compared to GMR > Aβ42 (n = 5; p = 0.000024), whereas GMR > Aβ42+ miR-277 mutant (n = 5; p = 0.1) shows slight increase and GMR > Aβ42 + Df(3 R)miR-277KO (n = 5; p = 0.02) shows significant increase in ROS puncta as compared to GMR > Aβ42. The data plotted shows mean ± SEM (Standard Error of Mean), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05, respectively. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. Scale bar = 100 μm.
Gain-of-function of miR-277 enhances the eclosion rate of Aβ42 expressing flies
Since overexpression of miR-277 rescues the neurodegeneration phenotype of flies expressing Aβ42, we wanted to check if miR-277 can rescue the defects in eclosion rate observed in AD flies. To address this, we overexpressed miR-277 in the central nervous system by using Elav>Gal4 driver [42]. All wild-type flies that serve as control did not show any lethality and had 100% eclosion rate (Fig. 6A, n = 270, 100%). Conversely, a high mortality rate was observed in Elav > Aβ42 as only 37% (n = 270) of the flies could hatch out and survive whereas the remaining 63% population failed to eclose as adults. However, overexpression of miR-277 resulted in significant improvement in the eclosion rate of Elav > Aβ42 flies (Elav > Aβ42+ miR-277; Fig. 6A, n = 270) as 86.6% of flies hatched. Whereas when miR-277 was downregulated using miR-277 mutant (Elav > Aβ42+ miR-277 mutant) or Df(3 R)miR-277KO deficiency (Elav > Aβ42 + Df(3 R)miR-277KO), the eclosion rates reduced to 35.3% and 30% respectively (Fig. 6A, n = 270). Thus, there is a significant improvement in eclosion rates when miR-277 is overexpressed in Elav > Aβ42 background.
A The bar graph represents the number of flies eclosed. We compared the number of flies eclosed in 1. Elav Gal4, 2. Elav > Aβ42, 3. Elav > Aβ42+ miR-277 and 4. Elav > Aβ42+ miR-277 mutant 5. Elav > Aβ42 + Df(3 R)miR-277KO background and validated that overexpression of miR-277 in the Elav > Aβ42 background rescues the Elav > Aβ42 mortality rate. We counted 270 flies in three independent biological sets from each background and plotted it on a graph in GraphPad Prism. The data plotted shows mean ± SEM (Standard Error of Mean), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05, respectively. B Improved climbing activity when miR-277 is overexpressed in AD flies. We compared the number of flies which crossed 10 cm from the bottom of the vial. Graph represents the climbing activity of the flies from day 1–day 30.
miR-277 rescues locomotor defects in AD flies
We assessed the locomotor function of AD flies in the miR-277 gain-of-function background using the climbing assay. Since we observed lethality with Elav > Aβ42 flies, we used OK107 Gal4 to drive human Aβ42 misexpression in the mushroom body neurons and their axons [51, 74,75,76]. Previous studies have shown that OK107 Gal4 can be used to assess the locomotion [74,75,76]. First, we overexpressed Aβ42 in mushroom body of larvae and adult using OK-Gal4 driver line and assayed the locomotor dysfunction in AD flies [40, 41]. We then compared the locomotor dysfunction of OK > Aβ42 flies with the ones where the levels of miR-277 expression were modulated in OK > Aβ42 background. We checked their climbing ability from day 1 to day 30 of their eclosion as AD is an age-dependent disorder. The flies were cultured from the embryonic stage to adult stage on regular food. We performed climbing assay of one-day-old to one-month-old flies. We found climbing defects in OK > Aβ42 flies on day 1 and further worsened through day 30 as compared to OK-Gal4 (Fig. 6B). Overexpression of miR-277 in OK > Aβ42 (OK > Aβ42+ miR-277) flies exhibit significant rescue in climbing ability as compared to the control OK > Aβ42 flies (Fig. 6B). The climbing activity of OK > Aβ42+ miR-277 slightly decreased from day 20 to day 30 due to aging (Fig. 6B). Conversely, downregulation of miR-277 in OK > Aβ42 using miR-277 mutant (OK > Aβ42+ miR-277 mutant) or Df(3 R)miR-277KO (OK > Aβ42 + Df(3 R)miR-277KO) flies showed severe climbing defects as compared to OK > Aβ42 control flies which progressively worsened with the age (Fig. 6B). This data further demonstrates that miR-277 overexpression can restore the climbing defects of the OK > Aβ42 flies.
hid is one of the targets of miR-277
To discern the molecular mechanism underlying miR-277-mediated rescue of GMR > Aβ42 neurodegenerative phenotype, we used bioinformatics tools like TargetScanFly and BLAST to predict targets of miR-277. It is known that miRNAs carry out their function by targeting the 3′UTR of their target mRNAs and thereby regulate gene expression. TargetScanFly predicts the targets of fly miRNA by matching the seed sequence (consensus sequence) of 3’untranslated region of the miRNA and their target mRNA [77] (http://www.targetscan.org/fly_72/). TargetScanFly predicted the proapoptotic factor- head involution defective (hid) as one of the mRNA targets with 0.24 PCT (Probability of conserved targeting) score and eight complementary consensus nucleotides (Supplementary Figure 3). Other software tools like DIANA and PicTar also predicted hid as one of the targets of miR-277 with the scores of 0.998 and 18, respectively. Additionally, BLAST tool predicted three different unique consensus sequences that were complementary between miR-277 and hid. Hence, we employed a genetic approach in order to validate whether hid and/or other proapoptotic factors is/are the target(s) of miR-277.
Gain-of-function of hid, rpr, and grim in the developing eye by using GMR-Gal4 driver results in a strong phenotype of the highly reduced eye (Fig. 7A–C, G–I). We investigated if gain-of-function of miR-277 can rescue the reduced eye phenotype of GMR>hid, GMR>rpr, and GMR>grim. Surprisingly, gain-of-function of miR-277 in the GMR>hid+miR-277 background exhibit significant phenotypic rescue with frequency (n = 600, 246/600 = 41%) respectively (Fig. 7D, G, H, I). However, gain-of-function of miR-277 in GMR>rpr+miR-277 (Fig. 7E, G, H, I) and GMR>grim+miR-277 (Fig. 7F–I) did not show any rescue. This result ruled out the possibility that the other proapoptotic factors: rpr and grim being the targets of miR-277. Hence, gain-of-function of miR-277 in the GMR>hid flies significantly suppressed the eye degenerative phenotype (Fig. 7H) and increased the surface area of the eye (Fig. 7I), further validating our finding that hid is the target of miR-277.
A GMR>hid, B GMR>rpr, and C GMR>grim show reduced eye phenotype due to caspase-mediated cell death. D GMR>hid+ miR-277 results in significant rescue as compared to GMR>hid flies. E GMR>rpr+ miR-277 and F GMR>grim+ miR-277 do not show a significant change in eye phenotype as compared to their respective controls B GMR>rpr and C GMR>grim. G Bar graph shows frequency as average between three repetitions. Two hundred flies were counted per repetition (200 × 3 = 600) to calculate the frequency for each genotype (1. Canton-S, 2. GMR-Gal4, 3. GMR>hid, 4. GMR>hid+ miR-277, 5. GMR>rpr 6. GMR>rpr+ miR-277, 7. GMR>grim, 8. GMR>grim+ miR-277). Statistical analysis was performed using the student’s t test for independent samples. H Quantitative analyses of severity score of neurodegenerative phenotypes(s) in eye. Flies from each genotype were randomly selected for scoring according to criteria described in the methods section. Comparisons were made using non-parametric: Mann–Whitney t test. I Quantitative analyses of area of the eye. The surface area of the eye (within white dotted line) was calculated using Image J. Statistical analysis was performed using the student’s t-test for independent samples. The surface area of GMR>hid+ miR-277 is significantly rescued (n = 5; p = 0.0000000073) as compared to GMR>hid. Error bars show standard error of mean (mean ± SEM). The orientation of all imaginal discs is identical with posterior to the left and dorsal up. Scale bar = 100 μm.
Additionally, we also tested ROS by DHE staining in GMR>hid and observed high levels of ROS puncta (Supplementary Figure 4A). Interestingly, number of ROS puncta significantly reduced when miR-277 was misexpressed (GMR>hid+miR-277; Supplementary Figure 4B) as compared to the GMR>hid alone (Supplementary Figure 4A). The loss-of-function of miR-277 in GMR>hid background either by using miR-277 mutant (GMR>hid +miR-277 mutant; Supplementary Figure 4C) or Df(3 R)miR-277KO (GMR>hid + Df(3 R)miR-277KO; Supplementary Figure 4D) show significantly increased number of ROS puncta. This suggests that miR-277 downregulate ROS production in GMR>hid background.
drice and Dark are not the targets of miR-277
We also explored if other caspases like drice or Dark get regulated by miR-277. We examined the relative luciferase activity of drice, Dark, and hid by luciferase assay in S2 cells. The positive miRNA control used was miR-1, which was tagged to different pTub-ffLuc-3′UTR constructs. The value of relative luciferase activities of different pTub-ffLuc-3′UTR constructs with miR-277 was calculated from the normalization of the luciferase activities of control: miR-1. If any of the caspases are the target of miR-277, it will bind to the 3′UTR of its target mRNA, degrades the target mRNA, and hence will exhibit less luciferase activity. The relative luciferase activity of hid with miR-277 was significantly reduced by 50% as compared to the positive control (Fig. 8A). The relative luciferase activity of dark with miR-277 was significantly reduced by 25% as compared to the positive control (Fig. 8A). Whereas the relative luciferase activity of drice with miR-277 was slightly increased as compared to the positive control (Fig. 8A). Hence, luciferase activity suggested that hid is the target of miR-277 and not the other caspases.
A In vitro dual-luciferase reporter assays of miR-mRNA interactions in S2 cell. The mean ± SEM of the relative luciferase expression ratio (firefly luciferase/Renilla luciferase, Luc/R-luc) was calculated for four biological replicates and compared with the control miRNA, miR-1. Statistical analysis was performed using student’s t test for independent samples. The results of dual-luciferase reporter assays of target 3′UTR of drice, hid, and Dark have showed that miR-277 can efficiently target hid and Dark except for drice. B Relative expression of hid at the transcriptional level using quantitative PCR (qPCR) in genotypes (1. GMR-Gal4 2. GMR > Aβ42, 3. GMR > Aβ42+ miR-277, 4. GMR > Aβ42+ miR-277 mutant, 5. GMR > Aβ42 + Df(3 R)miR-277KO). Triplicate was used for the calculation. Statistical analysis was performed using student’s t test for independent samples. hid transcript levels were significantly downregulated in GMR > Aβ42+ miR-277 (n = 3; p = 0.001) whereas slightly increased in GMR > Aβ42+ miR-277 mutant (n = 3; p = 0.14) and GMR > Aβ42 + Df(3 R)miR-277KO) (n = 3; p = 0.1) as compared to GMR > Aβ42. C Bar graph represents average intensity of hid GFP levels within yellow dotted line, region of interest, of five eye imaginal discs per genotype (n = 5) (1. hid5’-GFP, 2. GMR > Aβ42+ hid5’-GFP, 3. GMR > Aβ42+ miR-277+ hid5’-GFP, 4. GMR > Aβ42+ miR-277 mutant+ hid5’-GFP, 5. GMR > Aβ42 + Df(3 R) miR-277KO+ hid5’-GFP). Statistical analysis was performed using student’s t test for independent samples. hid GFP reporter is significantly downregulated in GMR > Aβ42+ miR-277+ hid5’-GFP (n = 5; p = 0.002) whereas slightly upregulated in GMR > Aβ42+ miR-277 mutant+ hid5’-GFP (n = 5; p = 0.75) and GMR > Aβ42 + Df(3 R)miR-277KO+ hid5’-GFP (n = 5; p = 0.29) as compared to GMR > Aβ42+ hid5’-GFP. Error bars show standard error of mean (mean ± SEM), and symbols above the error bar signify as ***p value < 0.001, **p value < 0.01, *p value < 0.05, and not significant (ns), p value > 0.05 respectively. E GMR > Aβ42+ hid5’-GFP results in elevated levels of hid-GFP levels as compared to the wild-type (D) hid5’-GFP. F GMR > Aβ42+ miR-277+ hid5’-GFP results in the significant downregulation of hid-GFP levels, whereas G GMR > Aβ42+ miR-277 mutant+ hid5’-GFP (H) GMR > Aβ42 + Df(3 R)miR-277KO+ hid5’-GFP result in the upregulation of hid-GFP levels. The orientation of all imaginal discs is identical with posterior to the left and dorsal up. Scale bar = 100 μm.
hid transcript levels are downregulated by miR-277
We investigated hid transcript levels by qPCR approach. The hid transcript levels were significantly increased by ̴1.75-fold in GMR > Aβ42 eye imaginal discs as compared to the control GMR-Gal4 eye imaginal discs (Fig. 8B). hid mRNA levels were significantly downregulated by ̴2.3-fold in GMR > Aβ42+ miR-277 eye imaginal discs as compared to the GMR > Aβ42 alone (Fig. 8B). However, loss-of-function of miR-277 in GMR > Aβ42 background by miR-277 mutant (GMR > Aβ42 +miR-277 mutant) and Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO) resulted in ̴1.8 and ̴1.9-fold increase in hid mRNA levels respectively as compared to the control GMR-Gal4 (Fig. 8B). Hence, our results strongly suggest that miR-277 ameliorates Aβ42-mediated neurodegeneration by post-transcriptionally regulating hid expression in GMR > Aβ42 flies.
To validate if hid is the target of miR-277, we employed hid5’F-WT-GFP reporter, which was generated to characterize the role of E2F binding site (present in 5′ ends of hid) in hid transcription [43]. A 2.2Kb fragment containing the hid 5′ E2F binding site was cloned into the pH-stinger GFP vector to generate hid5’F-WT GFP transgenic line [43, 78]. The hid GFP intensity within the yellow dotted line was calculated from five imaginal discs of each genotype and was used for statistical analysis (Fig. 8C–H). Minimal levels of GFP reporter expression were observed in wild-type hid5’F-WT eye-antennal imaginal discs (Fig. 8C, D) [43]. In comparison to the hid5’F-WT GFP control (Fig. 8C, D), ~2.5-fold high levels of hid-GFP were observed in GMR > Aβ42 eye discs (Fig. 8C, E). Gain-of-function of miR-277 in the GMR > Aβ42 (GMR > Aβ42+miR-277) background (Fig. 8C, F) exhibits significant downregulation of hid-GFP intensity as compared to hid5’F-WT GFP control and GMR > Aβ42 alone (Fig. 8C–E). Loss-of-function of miR-277 in GMR > Aβ42 flies using miR-277 mutant (GMR > Aβ42+miR-277 mutant; Fig. 8C, G) and Df(3 R)miR-277KO (GMR > Aβ42 + Df(3 R)miR-277KO; Fig. 8C, H), showed upregulation of hid GFP reporter intensity as compared to the hid5’F-WT GFP (Fig. 8C, D). Hence, overexpression of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277), targets 3′UTR of hid mRNA, silences its expression, and hence rescues the reduced eye phenotype seen in GMR > Aβ42 flies (Fig. 9).
Misexpression of Aβ42 (GMR > Aβ42 flies) results in the accumulation of amyloid plaques and causes aberrant activation of caspase Hid resulting in neurodegenerative adult eye phenotype. Misexpression of miR-277 in the background of GMR > Aβ42 (GMR > Aβ42+ miR-277), targets 3′UTR of hid mRNA, silences its expression, and hence rescues the reduced eye phenotype observed in GMR > Aβ42 flies.
Discussion
AD, a progressive neurodegenerative disorder, is caused by the accumulation of amyloid plaques and intracellular neurofibrillary tau tangles (NFTs). As per the Amyloid cascade hypothesis, the formation and accumulation of amyloid plaques and NFTs initiate other biochemical changes like oxidative stress, synaptic dysfunction due to aberrant signaling that eventually leads to neuronal cell death [79,80,81]. However, the mechanism(s) underlying AD-mediated neurodegeneration is not been fully understood [26, 82]. To understand the pathophysiology of AD, many animal models have been generated, including Drosophila melanogaster, which has proven very valuable due to the genetic conservation, ease of handling, and the large genetic repository and experimental tools [12, 13, 25]. Human Aβ42 is ectopically misexpressed in differentiating retinal neurons of fly and mimics AD-like neuropathology [17]. Drosophila AD models allow us to explore and test the signaling pathways involved in AD by genetic approaches. Numerous signaling pathways like the JNK pathway, Hippo pathway, caspases, GSK pathway etc. are aberrantly activated or dysregulated in AD [17, 83, 84]. Besides, members of these pathways involved in AD also cross-talk with each other making the disease even more complicated.
In eukaryotic systems, microRNAs (miRNAs) are highly conserved and are considered as essential components of gene regulatory networks. miRNAs play crucial roles in the regulation of gene expression and are implicated in various biological processes, including development, metabolism, and disease. miRNAs significantly affect signaling pathways and cause alterations of cellular signaling that can impact human diseases [85, 86]. We identified a highly conserved miR-277, as one of the genetic modifiers, which can rescue the Aβ42-mediated neurodegeneration using Drosophila eye model. miR-277 expression is enriched in larval brain, gut, fat body, and adult head, brain, eyes, gut, fat body (https://flybase.org/reports/FBgn0262419.html). The mechanism(s) underlying the effects of miR-277 targets involved in AD is still unknown.
In AD, synaptic dysfunction occurs due to neuronal cell death and impaired axonal targeting [87, 88]. Using the Drosophila eye model, we found that gain-of-function of miR-277 restores the axonal targeting defects observed in AD [16, 17]. Additionally, miR-277-mediated rescue is not restricted to morphological rescue only but it also promotes functional rescue as evidenced from behavioral assays like the eclosion rate and rescue of climbing defects. The Drosophila AD model shows robust cell death that is rescued by the gain-of-function of miR-277. Moreover, AD is a progressive neurodegenerative disease where the neurodegenerative phenotype progressively worsens with time, such as in pupal retina, photoreceptors are fused due to excessive cell death. However overexpression of miR-277 restores the hexagonal structure of ommatidia and a number of pigment cells by inhibiting the apoptosis present in GMR > Aβ42 pupal retina. Recently, the anti-apoptotic function of miR-277 has been reported where gain-of-function of miR-277 rescues the tumor suppressor gene, lethal giant larvae (lgl), mutant clones in third instar larvae, which would otherwise be eliminated due to cell death [89], and miR-277 is also expressed in hub cells and may contribute to their protection [90, 91].
Furthermore, to discern the molecular genetic mechanism(s) for miR-277 neuroprotective function, we screened for its target mRNA and identified hid, a proapoptotic gene, as one of its targets. Earlier, we have shown that aberrant activation of evolutionarily conserved JNK signaling pathway induces hid expression, which triggers cell death response seen in Aβ42-mediated neurodegeneration [17]. In GMR > Aβ42 background there is a significant decrease in hid transcript levels, as evident from hid-GFP reporter studies and qPCR assays. However, gain-of-function of miR-277 in GMR > Aβ42 significantly reduces the hid transcript levels. Based on our results from hid transcript level studies as well as luciferase reporter assay, we propose a model where miR-277 binds to 3′UTR of hid and degrades the expression of hid transcript (Fig. 9). Hence, we show here for the first time that miR-277 achieves its neuroprotective function by downregulating hid transcript levels thereby ameliorating Aβ42-mediated neurodegeneration. miR-277 has also been implicated in neurodegenerative disease like Fragile X-associated tremor/ataxia syndrome (FXTAS) [92]. However, its role in other neurodegenerative disorders like ALS, Parkinson’s, Huntington, etc is not yet determined. Further studies on the regulatory network and downstream targets of miR-277 will deepen our understanding of its biological significance and potential implications in human health and other neurodegenerative diseases. Additionally, microRNAs (miRNAs), which serve as invaluable tool for manipulating target gene expression [93, 94], have gained attention as potential biomarkers in various diseases, including neurodegenerative disorders like AD [95]. MiRNAs, carried by exosomes and microparticles in the blood and exhibiting greater stability than mRNAs, are valuable diagnostic biomarkers to enhance the accuracy of AD diagnoses [35, 96]. Insights into the roles of miRNAs in AD, particularly through affecting the signaling pathways, can make miRNAs attractive tools for novel therapeutic approaches.
Mutations affecting three proapoptotic genes: hid, rpr and grim, nearly completely eliminate the process of apoptosis during development [97]. However, research suggests that the overexpression of hid leads to cell death in various tissues of transgenic animals, as well as in cultured insect and mammalian cells [98]. Previous study demonstrates that role of hid as an inducer of cell death in Drosophila is conserved in mammalian cells, suggesting the possible existence of a mammalian homolog of this crucial apoptosis regulator [97]. When co-transfected with certain inhibitors, apoptosis is significantly reduced, indicating that Bcl2-type anti-apoptotic genes can inhibit Hid-induced apoptosis in mammalian cells. The inhibition of Hid’s proapoptotic activity by DIAP1 in mammalian cells mirrors a similar regulatory process as in insect cells, and the involvement of a mammalian IAP, XIAP, further indicates the evolutionary conservation of IAP-mediated inhibition of Hid activity [97]. Drosophila RHG proteins are functional counterparts to the mammalian Smac/DIABLO proteins, and they work by binding to and reducing the levels of IAP [99, 100]. Hence, the presence of an apoptosis pathway activated by hid in mammalian cells, regulated by conserved molecular components, strongly suggests the existence of a vertebrate hid counterpart.
Based on the conserved seed sequence, the human ortholog of miR-277 is hsa-miR-3660 (https://www.mirbase.org/). hsa-miR-3660 is expressed in the nervous system (brain and cortex) and reproductive system (testis and uterus) as shown in RNA seq data (https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR3660). It has been shown by GWAS studies that hsa-miR-3660 is implicated in human diseases such as attention deficit hyperactivity disorder [101], rheumatic heart disease [102], and lung cancer [103,104,105]. Moreover, it has been shown that hsa-miR-3660 is dysregulated in pseudoexfoliation glaucoma (PEXG) [106]. However, its role in Alzheimer’s Disease has not been shown to date. A multiple sequence alignment analysis has identified a putative binding site for hsa-miR-3660 within the 3′ untranslated region (3′UTR) of the gene hid. It is possible that hsa-miR-3660 could bind to the mammalian component of hid and degrade it. One of the potential targets predicted of hsa-miR-3660 is associated with the CARD domain (https://www.targetscan.org/vert_80/). The CARD domain, which stands for Caspase Activation and Recruitment Domain, is a protein module involved in initiating and regulating various signaling pathways, particularly those related to apoptosis (programmed cell death) and inflammation [107, 108]. The CARD domain in caspase 2 and caspase 9, initiator caspases, facilitate the formation of protein complexes involved in the apoptotic signaling cascade [107, 108]. In AD, the dysregulation of caspase 2 and caspase 9 activation may contribute to the loss-of neurons and the progression of the disease [107]. Hence, the identification of specific targets regulated by hsa-miR-3660 holds significant promise for advancing therapeutic strategies for AD. Further exploration of hsa-miR-3660 in AD mammalian model systems and AD patient’s brain samples and biofluids can help shed light on the etiology of AD or potential role of hsa-miR-3660 (an ortholog of fly miR-277) as a biomarker and as a druggable target of AD. In the future, hsa-miR-3660 inhibitor, can be employed to specifically bind to and inhibit the activity of endogenous hsa-miR-3660.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Crews L, Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum Mol Genet. 2010;19:R12–20.
Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66.
Yankner BA, Duffy LK, Kirschner DA. Neurotrophic and neurotoxic effects of amyloid beta protein: reversal by tachykinin neuropeptides. Science. 1990;250:279–82.
Calhoun ME, Wiederhold KH, Abramowski D, Phinney AL, Probst A, Sturchler-Pierrat C, et al. Neuron loss in APP transgenic mice. Nature. 1998;395:755–6.
Wei W, Wang X, Kusiak JW. Signaling events in amyloid beta-peptide-induced neuronal death and insulin-like growth factor I protection. J Biol Chem. 2002;277:17649–56.
van Dyck CH, Swanson CJ, Aisen P, Bateman RJ, Chen C, Gee M, et al. Lecanemab in early Alzheimer’s disease. N. Engl J Med. 2023;388:9–21.
Alexander AG, Marfil V, Li C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front Genet. 2014;5:279.
Newman M, Ebrahimie E, Lardelli M. Using the zebrafish model for Alzheimer’s disease research. Front Genet. 2014;5:189.
Leeanne M. Drosophila as an in vivo model for human neurodegenerative disease. Genetics. 2015;201:377–02.
Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol Neurodegener. 2017;12:89.
Yeates CJ, Sarkar A, Kango-Singh M, Singh A. Unravelling Alzhiemer’s disease using drosophila. In: Mutsuddi M, and Mukherjee A, eds. Insights into human neurodegeneration: lessons learnt from drosophila. Singapore: Springer. 2019;251–77.
Tsuda L, Lim YM. Alzheimer’s disease model system using drosophila. Adv Exp Med Biol. 2018;1076:25–40.
Singh A, Irvine KD. Drosophila as a model for understanding development and disease. Dev Dyn. 2012;241:1–2.
Singh A. Neurodegeneration- a means to an end. J Cell Sci Ther. 2012;3:10000e107.
Moran MT, Tare M, Kango-Singh M, Singh A. Homeotic Gene teashirt (tsh) has a neuroprotective function in amyloid-beta 42 mediated neurodegeneration. PLoS One. 2013;8:e80829.
Steffensmeier AM, Tare M, Puli OR, Modi R, Nainaparampil J, Kango-Singh M, et al. Novel neuroprotective function of apical-basal polarity gene crumbs in amyloid beta 42 (abeta42) mediated neurodegeneration. PLoS One. 2013;8:e78717.
Tare M, Modi RM, Nainaparampil JJ, Puli OR, Bedi S, Fernandez-Funez P, et al. Activation of JNK signaling mediates amyloid-ß-dependent cell death. PLoS One. 2011;6:e24361.
Rusconi JC, Hays R, Cagan RL. Programmed cell death and patterning in Drosophila. Cell Death Differ. 2000;7:1063–70.
Brachmann CB, Cagan RL. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–6.
Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–40.
Singh A, Lim, J, and Choi, K-W Dorso-ventral boundary is required for organizing growth and planar polarity in the Drosophila eye. In: Mlodzik M, editor. “Planar cell polarization during development: advances in developmental biology and biochemistry”: Elsevier Science & Technology Books. 2005;59–91.
Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–20.
Singh A, Shi X, Choi K-W. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development. 2006;133:4771.
Tare M, Puli OR, and, Singh A. Molecular genetic mechanisms of axial patterning: mechanistic insights into generation of axes in the developing eye. In: Singh A, and, Kango-Singh M, editors. Molecular genetics of axial patterning, growth and disease in the drosophila eye. I. Springer NewYork Heidelberg Dordrecht London: Springer. 2013; 37–75.
Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23.
Sarkar A, Irwin M, Singh A, Riccetti M, Singh A. Alzheimer’s disease: the silver tsunami of the 21(st) century. Neural Regen Res. 2016;11:693–7.
Cutler T, Sarkar A, Moran M, Steffensmeier A, Puli OR, Mancini G, et al. Drosophila eye model to study neuroprotective role of CREB binding protein (CBP) in Alzheimer’s disease. Plos One. 2015;10:e0137691.
Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15.
Cline EN. The amyloid-β oligomer hypothesis: beginning of the third decade. J. Alzheimers Dis. 2018:64:S567–S610.
Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–82.
Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–708.
White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–83.
Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–9.
Ben-Hamo R, Efroni S. MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget. 2015;6:1594–604.
Vaghf A, Khansarinejad B, Ghaznavi-Rad E, Mondanizadeh M. The role of microRNAs in diseases and related signaling pathways. Mol Biol Rep. 2022;49:6789–801.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–39.
Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–93.
Yao KM, White K. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63:41–51.
Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, et al. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–7.
Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76.
Sarkar A, Gogia N, Glenn N, Singh A, Jones G, Powers N, et al. A soy protein Lunasin can ameliorate amyloid-beta 42 mediated neurodegeneration in Drosophila eye. Sci Rep. 2018;8:13545.
Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2009;326:347–56.
Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–6.
Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P. The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum Mol Genet. 2011;20:2144–60.
Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29:135–6.
Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–22.
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82.
Li HH, Li JC, Su MP, Liu KL, Chen CH. Generating mutant. STAR Protoc. 2021;2:100432.
Wittkorn E, Sarkar A, Garcia K, Kango-Singh M, Singh A. The Hippo pathway effector Yki downregulates Wg signaling to promote retinal differentiation in the Drosophila eye. Development. 2015;142:2002–13.
Deshpande P, Chimata AV, Snider E, Singh A, Kango-Singh M, Singh A. N-Acetyltransferase 9 ameliorates Abeta42-mediated neurodegeneration in the Drosophila eye. Cell Death Dis. 2023;14:478.
Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129:4271–80.
Zipursky SL, Venkatesh TR, Teplow DB, Benzer S. Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell. 1984;36:15–26.
Singh A, Gopinathan KP. Confocal microscopy: a powerful technique for biological research. Curr Sci. 1998;74:841–51.
McCall K, Peterson JS. Detection of apoptosis in Drosophila. Methods Mol Biol. 2004;282:191–205.
Chimata AV, Deshpande P, Mehta AS, Singh A. Protocol to study cell death using TUNEL assay in Drosophila imaginal discs. STAR Protoc. 2022;3:101140.
Fogarty CE, Diwanji N, Lindblad JL, Tare M, Amcheslavsky A, Makhijani K, et al. Extracellular reactive oxygen species drive apoptosis-induced proliferation via drosophila macrophages. Curr Biol. 2016;26:575–84.
Deshpande P, Gogia N, Chimata AV, Singh A. Unbiased automated quantitation of ROS signals in live retinal neurons of. Biotechniques. 2021;71:416–24.
Jin Y, Chen Z, Liu X, Zhou X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol. 2013;936:117–27.
Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60.
Mehta A, Singh A. Real-time quantitative PCR to demonstrate gene expression in an undergraduate lab. Drosoph Inf Serv. 2017;100:225–30.
Mehta AS, Luz-Madrigal A, Li JL, Tsonis PA, Singh A. Comparative transcriptomic analysis and structure prediction of novel Newt proteins. PLoS One. 2019;14:e0220416.
Tayler TD, Garrity PA. Axon targeting in the Drosophila visual system. Curr Opin Neurobiol. 2003;13:90–5.
Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401.
Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–35.
Guo T, Zhang D, Zeng Y, Huang TY, Xu H, Zhao Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol Neurodegener. 2020;15:40.
Bouter Y, Dietrich K, Wittnam JL, Rezaei-Ghaleh N, Pillot T, Papot-Couturier S, et al. N-truncated amyloid beta (Abeta) 4-42 forms stable aggregates and induces acute and long-lasting behavioral deficits. Acta Neuropathol. 2013;126:189–205.
Butterfield DA, Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci. 2019;20:148–60.
Forman HJ, Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat Rev Drug Discov. 2021;20:689–09.
Cioffi F, Adam RHI, Broersen K. Molecular mechanisms and genetics of oxidative stress in Alzheimer’s disease. J Alzheimers Dis. 2019;72:981–1017.
Markesbery WR. The role of oxidative stress in Alzheimer disease. Arch Neurol. 1999;56:1449–52.
Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, et al. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci USA. 2005;102:5727–32.
Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, et al. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci USA. 2006;103:15038–43.
Mabuchi I, Shimada N, Sato S, Ienaga K, Inami S, Sakai T. Mushroom body signaling is required for locomotor activity rhythms in Drosophila. Neurosci Res. 2016;111:25–33.
Melicharek DJ, Ramirez LC, Singh S, Thompson R, Marenda DR. Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum Mol Genet. 2010;19:4253–64.
Silva B, Goles NI, Varas R, Campusano JM. Serotonin receptors expressed in Drosophila mushroom bodies differentially modulate larval locomotion. PLoS One. 2014;9:e89641.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques 2004;36:436–40.
Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608.
Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–5.
Ricciarelli R, Fedele E. The amyloid cascade hypothesis in Alzheimer’s disease: it’s time to change our mind. Curr Neuropharmacol. 2017;15:926–35.
Deshpande P, Gogia N, Singh A. Exploring the efficacy of natural products in alleviating Alzheimer’s disease. Neural Regen Res. 2019;14:1321–9.
Irwin M, Tare M, Singh A, Puli OR, Gogia N, Riccetti M, et al. A positive feedback loop of hippo- and c-jun-amino-terminal kinase signaling pathways regulates amyloid-beta-mediated neurodegeneration. Front Cell Dev Biol. 2020;8:117.
Sofola O, Kerr F, Rogers I, Killick R, Augustin H, Gandy C, et al. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 2010;6:e1001087.
Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9:276.
Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–32.
Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29:12776–86.
Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–85.
Bejarano F, Chang CH, Sun K, Hagen JW, Deng WM, Lai EC. A comprehensive in vivo screen for anti-apoptotic miRNAs indicates broad capacities for oncogenic synergy. Dev Biol. 2021;475:10–20.
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36.
Volin M, Zohar-Fux M, Gonen O, Porat-Kuperstein L, Toledano H. microRNAs selectively protect hub cells of the germline stem cell niche from apoptosis. J Cell Biol. 2018;217:3829–38.
Tan H, Poidevin M, Li H, Chen D, Jin P. MicroRNA-277 modulates the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2012;8:e1002681.
Fan J, Feng Y, Zhang R, Zhang W, Shu Y, Zeng Z, et al. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–37.
Giza DE, Vasilescu C, Calin GA. Key principles of miRNA involvement in human diseases. Discoveries. 2014;2:e34.
Zhao Y, Jaber V, Alexandrov PN, Vergallo A, Lista S, Hampel H, et al. microRNA-based biomarkers in Alzheimer’s Disease (AD). Front Neurosci. 2020;14:585432.
Feng Y, Yu X. Cardinal roles of miRNA in cardiac development and disease. Sci China Life Sci. 2011;54:1113–20.
Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci USA. 1999;96:4936–41.
Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–97.
Chew SK, Chen P, Link N, Galindo KA, Pogue K, Abrams JM. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature. 2009;460:123–7.
Creagh EM, Murphy BM, Duriez PJ, Duckett CS, Martin SJ. Smac/Diablo antagonizes ubiquitin ligase activity of inhibitor of apoptosis proteins. J Biol Chem. 2004;279:26906–14.
Rao S, Baranova A, Yao Y, Wang J, Zhang F. Genetic relationships between attention-deficit/hyperactivity disorder, autism spectrum disorder, and intelligence. Neuropsychobiology. 2022;81:484–96.
Li N, Lian J, Zhao S, Zheng D, Yang X, Huang X, et al. Detection of differentially expressed MicroRNAs in rheumatic heart disease: miR-1183 and miR-1299 as potential diagnostic biomarkers. Biomed Res Int. 2015;2015:524519.
Kussainova A, Bulgakova O, Aripova A, Khalid Z, Bersimbaev R, Izzotti A. The role of mitochondrial miRNAs in the development of radon-induced lung cancer. Biomedicines. 2022;10:428.
Zhu R, Guo W, Xu XJ, Zhu L. An integrating immune-related signature to improve prognosis of hepatocellular carcinoma. Comput Math Methods Med. 2020;2020:8872329.
Liao X, Zhu G, Huang R, Yang C, Wang X, Huang K, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803.
Czop M, Gasinska K, Kosior-Jarecka E, Wrobel-Dudzinska D, Kocki J, Zarnowski T. Twenty novel MicroRNAs in the aqueous humor of pseudoexfoliation glaucoma patients. Cells. 2023;12:737.
Bredesen DE. Neurodegeneration in Alzheimer’s disease: caspases and synaptic element interdependence. Mol Neurodegener. 2009;4:27.
Park HH. Caspase recruitment domains for protein interactions in cellular signaling (Review). Int J Mol Med. 2019;43:1119–27.
Acknowledgements
We thank Bloomington Drosophila Stock Center (BDSC) for Drosophila strains and the Developmental Studies Hybridoma Bank (DSHB) for antibodies. Confocal microscopy was supported by the core facility at the University of Dayton. We would like to thank Aditi Singh for comments on the manuscript. AS is supported by 1RO1EY032959-01 from NIH, the Schuellein Chair Endowment Fund, and the STEM Catalyst Grant from the University of Dayton. MKS is supported by 1RO1EY032959-01 from NIH.
Author information
Authors and Affiliations
Contributions
PD, C-YC, AVC, J-CL, and CY performed experiments. C-HC contributed resources. MKS and AS were involved in designing the study. PD, MKS, and AS analyzed the data. PD and AS wrote the manuscript with input from all authors. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Massimiliano Agostini
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deshpande, P., Chen, CY., Chimata, A.V. et al. miR-277 targets the proapoptotic gene-hid to ameliorate Aβ42-mediated neurodegeneration in Alzheimer’s model. Cell Death Dis 15, 71 (2024). https://doi.org/10.1038/s41419-023-06361-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-06361-3