Abstract
Long non-coding RNAs (lncRNAs) comprise the most representative transcriptional units of the mammalian genome. They are associated with organ development linked with the emergence of cardiovascular diseases. We used bioinformatic approaches, machine learning algorithms, systems biology analyses, and statistical techniques to define co-expression modules linked to heart development and cardiovascular diseases. We also uncovered differentially expressed transcripts in subpopulations of cardiomyocytes. Finally, from this work, we were able to identify eight cardiac cell-types; several new coding, lncRNA, and pcRNA markers; two cardiomyocyte subpopulations at four different time points (ventricle E9.5, left ventricle E11.5, right ventricle E14.5 and left atrium P0) that harbored co-expressed gene modules enriched in mitochondrial, heart development and cardiovascular diseases. Our results evidence the role of particular lncRNAs in heart development and highlight the usage of co-expression modular approaches in the cell-type functional definition.
Similar content being viewed by others
Introduction
Approximately 90% of the mammalian genome is transcribed in a cell-specific manner into RNA [1], but only 2–3% is transcribed to protein-coding messenger RNA (mRNA) [2, 3]. Most RNA transcripts are classified as non-coding RNAs (ncRNAs) and can be divided into small (sncRNAs) and long ncRNAs (lncRNAs) [2,3,4,5]. These ncRNAs play a crucial role in controlling the flow of genetic information through various gene regulation mechanisms [5, 6], LncRNAs exhibit cell type- and tissue-specific expression and have the ability to influence lineage fates and cell subtypes [7, 8]. Although the involvement of ncRNAs in cardiovascular diseases has been extensively studied [3, 6, 9, 10], our understanding of the expression and function of lncRNAs in heart development remains limited [11]. Investigating the single-cell expression patterns of lncRNAs could unveil novel biological functions associated with different cell populations. Therefore, accurately measuring the abundances of lncRNAs in individual cells is critical for identifying cell-type specificities and determining single-cell functions [12].
A new type of lncRNA, known as positionally conserved lncRNAs (pcRNAs), has been identified based on the evolutionary conservation of their promoters and their syntenic positions between humans and mice [6]. These pcRNAs are apparently involved in developing cardiovascular tissue, arterial morphogenesis, and smooth muscle tissue cell differentiation. A subset of them manifested potential importance in chromatin organization, as evidenced by their localization overlapping with the binding sites for the CTCF chromatin organizer, the anchor points of chromatin loops, and the borders of topologically associating domains [6].
The failing adult heart resembles the fetal heart, through the ‘re-expression of the fetal gene expression program’ [13, 14]. Our study aims to dissect patterns and cell-specific distributions of coding and non-coding RNAs during development using single-cell data. This information will enable us to unveil novel regulatory processes occurring during cardiac development and potentially lead to improved therapeutic targeting [2,3,4].
Results
Expression quantification and cell-type classification
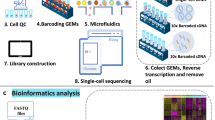
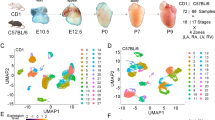
We investigated the gene expression patterns during cardiac development using single-cell RNA sequencing (scRNA-seq) data. The dataset encompassed four embryonic and four postnatal stages, covering five distinct myocardial anatomical areas (Fig. 1). Employing a bioinformatic pipeline (See Supplementary Methods), we retrieved the expression levels of all transcripts normalized using FPKM values, requiring that they must be expressed in at least 5% of cells to be considered in subsequent analyses. Table 1 shows the expression profiles of coding and long non-coding transcripts identified in specific compartments and time-points. The number of lncRNAs ranged from 738 (left ventricle P21) to 2,636 (left ventricle P7) transcripts, with P21 exhibiting fewer than 1000. These findings aligned with previously reported studies related to numbers of lncRNAs in breast cancer, cardiac fibrosis, and heart development [15,16,17,18].
Circular diagram reports the number of single cells for each data point. The bar graph shows the number of expressed transcripts color-coded by anatomical area (yellow: ventricle, red: atrium, purple: right ventricle, green: left ventricle, blue: left atrium). The left ventricle was the only compartment with data collected across all time points.
To characterize the cell types and assign them based on expression values, we performed cluster analysis. The Silhouette method [19] combined with hierarchical clustering determined the optimal number of clusters for each sample. Chi-squared and adherence tests assessed cluster significance and assigned cell types. Cluster visualization was achieved through t-SNE plots for identification of different cell populations, illustrating the spatial distribution of cell types based on transcript expression patterns (Fig. 2). t-SNE uses a Gaussian probability function to calculate how likely a cell will pick another cell as its neighbor and repeats this step for all cells. Heatmaps (Supplementary Fig. 1) visually represented expression patterns by cell type, confirming t-SNE cell assignments (Fig. 2). Postnatal stages revealed a maximum of five potential cell clusters, while embryonic ventricle stages showed four clusters. Supplementary File 1 contains a list of potential gene markers identified based on expression patterns.
t-SNE plots in which each square represents a compartment at a specific development stage. Each dot represents a cell, and each color describes a particular cell-type: Blue - Cardiomyocyte I; Light blue - Cardiomyocyte II; Salmon - Dendritic; Red - Endothelial; Green - Fibroblast and Myofibroblast; Purple - Macrophage and Monocyte II; Black - Mural, Vascular smooth muscle and Pericytes; Myoblast - Yellow; Brown - T-cell; and Gray - Unknown cell-types (i.e. can be a different cell-type in which we don’t have markers). In addition, each number inside the squares is related to the silhouette coefficient score for each compartment and time point.
Then, we classified clusters into specific cell types based on the expression patterns of known marker transcripts, integrating information from three [20,21,22] and the PanglaoDB database [23], creating a comprehensive heart cell marker database. M3Drop [24], fitting a Michaelis-Menten model, identified significantly variable genes that were expressed among the cell clusters, which were then cross-referenced with our heart cell markers database. Chi-squared and adherence tests determined cluster assignments significance. Eight distinct cell-types were identified in all compartments and time-points: cardiomyocytes, myoblasts, endothelial cells, vascular smooth muscle cells and pericytes, fibroblasts and myofibroblasts, T-cells, macrophages, monocytes and dendritic cells (Fig. 2). Four compartments and time-points exhibited two subpopulations of cardiomyocytes, and left ventricle P3 showed two macrophages and monocyte subpopulations. Supplementary File 1 provides markers identified for each cell-type at each stage. Supplementary Fig. 1 presents heatmaps and hierarchical clusters of the expression patterns for each heart chamber and time-point, corroborating each subtype assignment.
scRNA-seq reveals lncRNAs as potential cell markers for eight cell-types during heart development
The transcript cell markers determined above were divided into three classes, including coding mRNAs, lncRNAs, and pcRNAs (Fig. 3, Supplementary File 1), which were further explored to analyze their potential involvement in the heterogeneity of cell populations across various time points and chambers during heart development. PcRNAs served as markers for multiple cell-types. We found at least one lncRNA (cardiomyocytes, left atrium E14.5) as a cell marker in all time-points and chambers, reaching up to 197 in cardiomyocytes of the right ventricle E14.5. Our data confirmed the presence of cardiomyocytes in all compartments at all development stages [20,21,22,23, 25, 26], while also revealing region- and developmental stage-specific cell populations. For instance, a cell population with a dendritic cell-like transcriptional signature was observed exclusively in E9.5 atrium; meanwhile, we identified a vascular smooth muscle-like population in E14.5 right ventricle and a T cell-like population in E18.5 left ventricle. Interestingly, fibroblasts and myofibroblast transcriptional signatures were detected in E14.5, a stage at which fibroblasts from the epicardium are known to undergo epithelial-to-mesenchymal transition (EMT) [26]. Cell populations manifesting monocyte and macrophage profiles appeared only at late embryonic and postnatal stages, matching previously reported evidence [27]. Here, findings highlight the relevance of different lncRNAs and pcRNAs signatures in region- and stage-specific cellular populations during heart development.
The numbers inside each color-coded circle (green: coding genes, orange: lncRNAs, red: pcRNAs) represent the number of marker’s transcripts. Each compartment is represented by a color: Yellow - Ventricle; Red - Atrium; Purple - Right Ventricle; Green - Left Ventricle; and Blue - Left Atrium. In addition, we found marker’s through 8 different cell-types represented by each column.
We manually explored the cell types that can be identified in the same anatomical region (e.g. fibroblasts and myofibroblasts in the left atrium) that were previously reported [28]. At the E9.5, non-fibroblast-enriched cells were identified in atrium or ventricle compartments, consistent with a prior report [20]. We argue that our cell-type analysis based on coding and lncRNA expression patterns may reflect unique aspects of myocardial developmental events. In addition, it not only matches the analysis based on the coding transcriptome alone [20] but also reveals new cell populations not uncovered in that analysis (Fig. 3). These results were consistent with the previous report but also provided a novel insight by uncovering additional cell types that were not previously reported, offering a comprehensive understanding of the cellular composition under heart development.
scRNA-seq identifies specific signatures for coding genes and lncRNAs for the same cell-type at different heart compartments
To understand the dynamics of transcript markers during myocardial development, we compared the expression patterns of cell-type specific markers across cardiac chambers. To achieve this, we evaluated the presence/absence of each marker of the four most abundant cell populations (cardiomyocytes, endothelial cells, fibroblasts, macrophages, and monocytes) per time during the course of myocardial development (Fig. 4). Our analysis unveiled distinct coding and non-coding RNA (lncRNAs, pcRNAs) signatures for the same cell-type in different chambers, suggesting the same cell-type adopts novel compartment-specific gene expression patterns per anatomical milieu. The proportions of shared and unique transcripts were similar between coding and non-coding profiles across anatomical areas and time-points, suggesting no bias towards one transcript type being more specific to a particular anatomical origin.
Venn diagrams are color coded to represent a compartment of the heart: Yellow - Ventricle; Red - Atrium; Purple - Right Ventricle; Green - Left Ventricle; and Blue - Left Atrium. The diagram was created from E9.5 to P0, since P3, P7 and P21 only had left ventricle cells; thus, the compartment’s comparisons were not possible in these 3-time points. The number of transcripts is noted in the diagram through 4 different cell-types (Cardiomyocytes; Endothelial; Fibroblasts and Myofibroblasts; Macrophages and Monocytes) represented by each column. A This analysis shows the number of shared and unique coding transcripts of each compartment. B This analysis shows the number of shared and unique non-coding transcripts (lncRNA and pcRNA) of each compartment.
Differential expression and functional enrichment analysis explored cardiomyocyte subpopulations
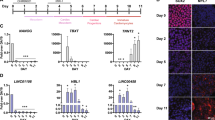
To pursue differences within the identified cardiomyocyte subpopulations, we analyzed differential expression to characterize biological variation between the two subpopulations (Cardiomyocyte 1 and 2) identified in each chamber and developmental stage, using DESeq2 [29] and Volcano plot for data visualization. Different cardiomyocyte cell-types were identified in distinct chambers and time-points (Fig. 5), with the ventricle population of cells presenting greater number of differentially expressed (DE) genes across both cardiomyocyte types, with 3,568 coding and 244 lncRNAs detected as DE at embryonic stage E9.5 (ventricle); 806 coding and 53 lncRNAs in the embryonic stage E11.5 (left ventricle); and 3173 coding and 222 lncRNAs at E14.5 (right ventricle). We also identified distinct cardiomyocyte subpopulations in the left atrium at the post-natal time-point P0, with 84 coding and 7 lncRNAs identified as DE (Supplementary File 2). In summary, the ventricle population presents a greater number of DE genes and a set of DE genes were identified in each chamber and developmental stage.
The left side (A, C, E, G) are related to coding transcripts in ventricle E9.5, left ventricle E11.5, right ventricle E14.5 and left atrium P0, respectively. The right side (B, D, F, H) are related to lncRNAs transcripts in ventricle E9.5, left ventricle E11.5, right ventricle E14.5 and left atrium P0, respectively. The blue and red colors are related to down- and up-regulated transcripts, respectively; the numbers are related to differentially expressed transcripts in each condition (down- and up-regulated); and the numbers in parenthesis are the number of differentially expressed genes.
To facilitate the identification of specific subpopulations, we named the cardiomyocytes by adding at the front of the cell type (e.g. cardiomyocyte (CM)) the name of the coding gene with the highest expression for each chamber and time-point. The ventricle E9.5 subpopulations were named “Mt-Atp8-CM” and “Myl3-CM”; left ventricle E11.5 were named “Pln-CM” and “Tpm1-CM”; right ventricle E14.5 as “Hba-a1-CM” and “Tnnt2-CM”; and the left atrium P0 were named “Mth6-CM” and “Sln-CM for P0”. All these genes have been described in the literature as involved in multiple mitochondrial and cardiovascular key processes and diseases. The blue and red colors are related to down- and up-regulated transcripts, respectively, relative to one another; the numbers are related to DE transcripts in each condition (down- and up-regulated); and the numbers in parenthesis are the number of DE lncRNAs, when presenting fold-change and p value cut-offs of 1.5 and 0.05, respectively. Notably, in all four time-points one cardiomyocyte subpopulation tended to dominate with a higher number of positively expressed gene transcripts compared to another for both the coding and lncRNA transcripts, except for lncRNAs P0 where the abundance was nearly equal. This suggests distinct gene expression profiles between the subpopulations.
To gain insights into the functional characteristics and potential disease associations of the DE transcripts, we performed enrichment analysis through EnrichR [30], using a corrected p-value cut-off of 0.05, and considering the databases Gene Ontology (GO) Biological Process [31], Jensen Diseases [32] and KEGG pathways [33] (Fig. 6 and Supplementary File 3). Figure 6 represents the GO Biological Processes and Jensen Diseases for each cardiomyocyte subpopulations top five terms. In the E9.5 ventricle, cell populations manifested associations with RNA processing and regulation (Fig. 6A). “Mt-Atp8-CM” cardiomyocytes manifested enrichment in transcripts related to cardiomyopathies and neurological disorders known to be associated with cardiomyopathies and heart defects. The “Myl3-CM” cardiomyocytes are enriched in transcripts associated with muscular diseases such as spinal muscular atrophy, myelofibrosis, and anterior ischemic optic neuropathy (Fig. 6B).
The gene ratio size was computed through the number of overlap (observed/expected) genes. We named the cardiomyocyte’ populations according to the coding gene with higher expression in each chamber and time point. A Biological processes from GO analysis of each cardiomyocyte subpopulation. B Diseases enrichment analysis of each cardiomyocyte subpopulation according to Jansen Disease Database. The blue and red colors are related to down- and up-regulated transcripts, respectively.
Regarding the left ventricle E11.5, the “Pln-CM” subpopulation was similarly associated with RNA processing, but also with GO terms linked to the insulin receptor signaling pathway (Fig. 6A). It was also enriched with genes related to hypertrophic and mitochondrial myopathies, pseudosarcomatous fibromatosis, Leigh disease, and Leber hereditary optic neuropathy (Fig. 6B). The “Tpm1-CM” subpopulation is enriched with transcripts associated with mitochondrial translational termination and elongation (Fig. 6A), and with different metabolic disorders such as tyrosinemia, Reye syndrome, hypermethioninemia, CREST syndrome, and carnitine palmitoyltransferase II deficiency (Fig. 6B).
In the right ventricle, the E14.5 “Hba-a1-CM” subpopulation was enriched with genes related to protein targeting the membrane and the endoplasmic reticulum (Fig. 6A). They were also associated with Leigh disease, as well as with blood-related diseases, mitochondrial complex I deficiency, and Bowen-Conradi syndrome. “Tnnt2-CM” cardiomyocytes manifested enrichment in genes associated with the regulation of protein localization to the Cajal body, regulation of protein ubiquitination, mitotic spindle organization, and nuclear division. They are associated with myelofibrosis, cardiomyopathies, and combined oxidative phosphorylation deficiency (Fig. 6B).
P0 left atrium cardiomyocytes “Myh6-CM” manifested enrichment in genes associated with mitochondrial function and respiratory electron transport chain (Fig. 6A), with a direct link with mitochondria-related diseases (Fig. 6B). The “Sln-CM” subpopulation was enriched with genes associated with the cell cycle, protein modification process, and cytokine-mediated signaling pathway (Fig. 6A). They were also linked with cardiomyopathies, besides cystic fibrosis, and cerebrovascular disease (Fig. 6B).
In summary, our differential expression and enrichment analyses uncovered distinct cardiomyocyte subpopulations with unique gene expression profiles and functional characteristics. These findings provide novel insights into the roles of different subpopulations in disease and contribute to a better understanding of cardiomyocyte heterogeneity in different chambers and developmental stages. Supplementary File 3 contains additional information on enriched GO terms, KEGG pathways and Jensen Diseases associated with each cardiomyocyte subpopulation.
Modular expression analysis reveals cell-specific functional insights for lncRNAs during myocardial development
We decided to investigate whether transcriptional modules based on co-expression patterns can shed light on novel functional connections between lncRNAs and mRNAs [34] during heart development in a single-cell perspective. We employed CEMiTool to identify expression modules at each time-point, derive GO terms over-represented in each module, retrieve protein-protein and co-expression interactions within module transcripts and perform cell-specific gene set enrichment analysis (GSEA) [35] to identify the functional significance of the co-expressed gene modules in a subpopulation in heart development. Each module represents a group of transcripts that exhibit similar expression across cell-types, and the interactions enable us to explore for co-regulation among the genes. CEMiTool was able to generate modules for 13 out of 15-time points and heart compartments, without being able to retrieve modular patterns based on guilt-by-association for atrium E9.5 and left ventricle E14.5 (Supplementary File 3).
Once the modules were identified, we examined their enrichment in distinct cell-types and functional processes (Fig. 7A–C). Figure 7A illustrates the associations between the enriched modules and specific cell types or functional categories, highlighting modules related to cardiac tissue development, mitochondrial process, cardiac functioning, and circulatory function. Notably, Module 1 (M1) from the left ventricle E11.5 is enriched in mitochondrial processes, and the GSEA analysis shows a clear difference in the module expression between cardiomyocyte subpopulations Pln-CM and Tpm1-CM (Fig. 7B). GSEAs were undertaken separately for each time-point and heart compartment, revealing cell-specific expression patterns for some of these modules (Fig. 7C). This latter analysis revealed modules harboring different expression patterns in cardiomyocyte and macrophage subpopulations (Supplementary File 4). In summary, we identified specific modules that were differentially enriched in distinct functional processes, cell-types and subpopulations.
A Biological processes from GO analysis of the modules that are related to the development of cardiac tissue, mitochondrial processes, cardiac functioning, and circulatory functioning. We observe the compartment and time point to which the adjusted p-value corresponds. B Module 1 (M1) from the Left ventricle E11.5 is enriched in mitochondrial processes and GSEA shows a difference between cardiomyocyte subpopulations 1 and 2. The co-expression network shows the gene´s interaction based on their expression patterns and protein-protein interaction, the gene’s names that appear are the hub genes that have more connectivity with others. C GSEAs were undertaken separately for each time point and heart compartment, revealing a cell-specific expression pattern for some of these modules. The blue and red colors are related to down- and up-regulated transcripts, respectively.
By the findings above, we selected enrichment analysis and modules directly related to the development of cardiac tissue. Figure 8A focuses on right ventricle samples from E14.5, and demonstrates a mitochondrial module with two cardiomyocyte subpopulations, with Hba-a1-CM being under-represented and Tnnt2-CM over-represented in the module. Figure 8B highlights some lncRNAs that are part of the module depicted in Fig. 8A. Some of these lncRNAs have been previously reported as directly involved in heart processes [2, 36,37,38,39,40,41,42,43,44]. Figure 8C presents the module’s enrichment analysis according to GO Biological Processes, providing valuable information about the functional categories associated with the genes within the specific module.
A Co-expression and protein-protein interaction network from Module 2, right ventricle samples from E14.5. The small heatmap shows the GSEA analysis. The numbers 469, 43 and 3 are related to the number of coding, lncRNAs and pcRNAs that compounds in this module. B The cell’s fraction and FPKM mean expression of 17 selected lncRNAs which compound the module. C Represents the top 10 GO terms based on the module’s enrichment analysis.
We observed Tln1 FPKM expression means of 42.7 and 47.55 in Hba-a1-CM and Tnnt2-CM, respectively, while endothelial cells exhibited an expression at 70.72 (Fig. 8). Tln1 showed low expression levels in normal CMs compared to other cell-types. Additionally, we found a Snhg5 FPKM expression mean of 265.8 in mural cells, greater than the other cell-types. This pcRNA contributes to angiogenesis in acute myelogenous leukemia (Supplementary File 4).
Notably, cardiomyocyte subpopulations were observed in specific stages and compartments of myocardial development, and these subpopulations manifested distinct co-expression patterns, as revealed by t-SNE plots and heatmaps (e.g. Fig. 2, Supplementary Fig. 1). Clusters classified as “unknown” (left ventricle P0 and left ventricle P3) manifested unique co-expression behaviors compared to other types in the same condition. These cells likely represent another cell type for which specific markers have not been identified. Based on the markers defined by the M3Drop, only one was determined for these unknown cells (Qk—left ventricle P0; and ENSMUST00000118613.6—left ventricle P3).
Previous studies have described cardiomyocyte subpopulations [45] and lncRNAs associated with these subpopulations [45, 46]. Notably, Gas5 and Sghrt (previously annotated as 1810058i24Rik) have been implicated in cardiac hypertrophy in a rat model [46], and certain regulatory lncRNAs appear to influence cell cycle arrest [45]. We also detected that these two lncRNAs are DE throughout heart development and cardiomyocyte subpopulations (Supplementary File 2).
Discussion
Gene expression is a fundamental mechanism for transmitting genetic information [1]. Gene expression encyclopedias catalog genes that are co-expressed in different cell-types, facilitating the understanding of their functions and regulatory processes [6]. LncRNAs comprise the most representative transcriptional units in mammalians and are associated with different organs development like corpus luteum formation, heart, skeletal muscle, brain and certain diseases [11, 47,48,49]. The study of heart development is of significance, given the toll taken by cardiovascular diseases.
We identified eight different cell types from different regions and stages of heart development, more than that described in the original scRNA-seq study, but the authors discuss that it would be possible to obtain additional cell lineages by improving the analysis of the dataset they obtain [20]. Interestingly, the fibroblasts and myofibroblasts transcriptional signatures were detected at E14.5, the stage at which fibroblasts from the epicardium are known to undergo EMT into the myocardium [26]. Cell populations with monocytes and macrophages profiles appeared only at late embryonic and postnatal stages, consistent with evidence in the literature of monocyte-derived macrophages increasing in the myocardium post-birth [27].
New marker transcripts distributed in coding genes and lncRNAs were identified, in addition, some found specifically in certain compartments, such as Myl3 that is known as a ventricular light chain myosin with increased expression in slow-twitch skeletal muscle fibers; and Sln, which is restricted to the atrial lineage during mouse heart development. Other transcripts are shared between compartments such as Tpm1 with established roles in cardiac looping, atrial septation, and ventricular trabeculae formation.
Different cardiomyocyte subpopulations were identified in distinct chambers and time-points (E9.5, E11.5, E14.5 and P0), with the ventricle and left atrium presenting greater differences within both cardiomyocyte types. We named the cardiomyocyte populations according to the coding gene with higher expression in each chamber and time-point. Of note, some of these genes are associated with heart diseases, Mt-Atp8, for example, is enriched in cardiomyocytes and is involved in many diseases in the ventricle and neuropathies [47]. Myl3 was classified as a cardiac protein that was abundantly expressed in the heart (>260 TPM) with mosaic patterns of expression in skeletal muscle [48]. Additionally, Myl3, known as a ventricular light chain myosin with increased expression in slow-twitch skeletal muscle fibers, is robustly and homogeneously present in ventricular cardiomyocytes [48]. Myl3 was described as DE in the ventricle and is associated with muscle atrophy and neuromuscular diseases [49]. A study in a mouse model of Huntington’s disease also reported elevated serum levels of Myl3 concomitant with skeletal muscle atrophy [50].
Pln, overexpressed in the left ventricle and called cardiac phospholamban, modulates calcium re-uptake during muscle relaxation and plays an essential role in calcium homeostasis in the heart [51]. Moreover, mutations in Pln have been associated with dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM) [52], and arrhythmogenic right ventricular cardiomyopathy (ARVC) [52]. Tpm1 plays multiple roles in heart development and the formation of congenital heart defects [53]. Mutations in Tpm1 have been associated with both HCM, DCM [53, 54], and with peripartum cardiomyopathy (PPCM) [54].
Hba − a1 is up-regulated in response to chronic stress along with genes associated with the vascular system. A reduced expression of Hba − a1 is related to a neuroprotective role [55]. Tnnt2 is a cardiac muscle troponin T that is expressed throughout heart development and in the post-natal heart, and in the left and right ventricle [56]. Tnnt2 is also associated with hypertrophic cardiomyopathy, dilated cardiomyopathy, and restrictive cardiomyopathies [56]. Myh6 is a cardiac muscle myosin that is preferentially expressed in the atrial chambers and its mutation causes atrial septal defect [57]. Myh6 was described in differentiation in adult cardiac precursor cells (CPCs), which were characterized by up-regulation of Myh6 and, therefore, massively differentiated into cardiomyocytes [58]. Studies have also reported additional mutations in Myh6 linked to both hypertrophic cardiomyopathy and dilated cardiomyopathy [59].
The expression of Sln is restricted to the atrial lineage during mouse heart development, and this pattern is conserved in other mammals, including humans [60]. Sln is involved with Ca2+ handling that plays a crucial role in the contraction and relaxation of cardiomyocytes [60]. Moreover, Sln is involved with dilated cardiomyopathy, cardiomyopathy [61], and heart failure [60].
Our examination revealed transcriptional gene modules related to the development of cardiac tissue, mitochondrial processes, cardiac functioning, and circulatory functioning. Our cardiomyocyte populations were enriched in mitochondrial functions, but we also identified differences between cardiomyocyte subpopulations according to GSEA. We identified some lncRNAs that are already described in the literature as directly related to heart processes: Mhrt is involved with cardiac hypertrophy and heart failure [36, 37], and also protects cardiomyocytes against H2O2-induced apoptosis [38]; Tln1 is expressed at low levels in normal CMs compared with other cell-types, and combined deletion of CMs Tln1 and Tln2 destabilized the myocardium, leading to heart failure [39]. Figure 8 demonstrates a Tln1 FPKM expression mean of 42.7 and 47.55 in “Hba-a1-CM” and “Tnnt2-CM”, respectively, while in endothelial cells, we observed expression at 70.72, evidencing a cell-specific higher expression pattern. Hand2os1 orchestrates heart development; its locus dampens Hand2 expression to restrain cardiomyocyte proliferation, thereby orchestrating balanced development of cardiac cell lineages [(40)]; Dancr is a lncRNA that has coding potential in the human heart, encoding putative micropeptides and is associated with cardiovascular diseases [41]; Mbnl1 regulates isoproterenol‐induced myocardial remodeling in vitro and in vivo [62]; Meg3 prevents cardiac fibrosis and diastolic dysfunction [2, 62]; AA465934 is associated with early diabetic cardiomyopathy [42]. Finally, Snhg5 upregulation induced by YY1 contributes to angiogenesis in acute myelogenous leukemia [63]. We observed a Snhg5 FPKM expression mean of 265.8 in mural cells, greater than the other cell-types (Fig. 8). Mural cells are integral components of brain blood vessels that play important roles in vascular formation, regulation of regional cerebral blood flow, regulation of vascular stability, and homeostasis [64]. Snhg5 has also been reported to participate in the occurrence and development of glioma [44].
However, even though we can describe different subpopulations of cardiomyocytes, there are some limitations. These subpopulations could only be identified in certain chambers of the heart and in some stages of development. Experimental techniques such as Fluorescence-activated Cell Sorting (FACS) of cell surface markers or in situ hybridization with specific transcripts will need to be used to individually isolate these two potential cardiomyocyte subpopulations. However, using single cell and hierarchical clustering strategies allows the identification of possible cell-types in other tissues during development.
Our results evidence the role of lncRNAs in heart development and highlights the usage of co-expression modular approaches in the cell-type functional definition. Perspectives of the present work are: knowledge generated here unveils potential processes guided by these lncRNAs during cardiac differentiation or development, which can lead to the development of improved therapeutics for cardiovascular diseases targeting genes potentially involved with the re-expression of the fetal gene expression program. In our analysis, several coding and non-coding transcripts were identified, and they can be used as early biomarkers for the prognosis for several cardiomyopathies, for example, Mhrt in cardiac hypertrophy and heart failure, Meg3 in cardiac fibrosis, Snhg5 in vascular cerebral homeostasis, among others. Then, future studies should investigate how these lncRNAs regulate other RNAs, like miRNAs, in cardiac development and diseases.
Methods
The online version contains supplementary detailed methods available at X.
Data availability
The authors declare that all single-cell sequencing data reported in this article have been deposited are available within the paper, Supplementary Information files and deposited in the GNomEx database (https://b2b.hci.utah.edu/gnomex/) under accession numbers 272 R, 274 R, 275–292 R, 439 R, and 440 R.
References
Li Z, Zhao W, Wang M, Zhou X. The role of long noncoding RNAs in gene expression regulation. Vlachakis D, editor. 2019.
Hobuß L, Bär C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol. 2019;10:1–9.
Hermans-Beijnsberger S, van Bilsen M, Schroen B. Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res. 2018;3:118–30.
Turton N, Swan R, Mahenthiralingam T, Pitts D, Dykes IM. The functions of long non-coding RNA during embryonic cardiovascular development and its potential for diagnosis and treatment of congenital heart disease. J Cardiovasc Dev Dis. 2019;6:1–22.
Bhatia G, Sharma S, Upadhyay SK, Singh K. Long non-coding RNAs coordinate developmental transitions and other key biological processes in grapevine. Sci Rep. 2019;9:1–14.
Amaral PP, Leonardi T, Han N, Viré E, Gascoigne DK, Arias-Carrasco R, et al. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018;19:32:1–21.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Gil N, Ulitsky I. Regulation of gene expression by cis -acting long non-coding RNAs. Nat Rev Genet. 2019;21:102–17.
Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–8.
Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–46.
Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12:415–25.
Chen G, Ning B, Shi T. Single-cell RNA-Seq technologies and related computational data analysis. Front Genet. 2019;10:317:1–13.
Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191–98.
Dirkx E, da Costa Martins PA, De Windt LJ. Regulation of fetal gene expression in heart failure. Biochim Biophys Acta. 2013;1832:2414–24.
Shaath H, Vishnubalaji R, Elango R, Khattak S, Alajez NM. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Discov. 2021;7:1–14.
Aghagolzadeh P, Bernasconi R, Nemir M, Khalil H, Pulido C, Chouvardas P, et al. Single-cell analysis of the long noncoding RNA transcriptome identifies novel therapeutic targets for cardiac fibrosis. Arch Cardiovascular Dis Suppl. 2020;12:243–45.
Touma M, Kang X, Zhao Y, Cass AA, Gao F, Biniwale R, et al. Decoding the Long Noncoding RNA During Cardiac Maturation: A Roadmap for Functional Discovery. Circ Cardiovasc Genet. 2016;9:395–407.
Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. 2015;36:353–68a.
Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65.
DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39:480–90.
Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018;138:166–80.
Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8:e43882.
Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database. 2019;2019:1–9.
Andrews TS, Hemberg M. M3Drop: dropout-based feature selection for scRNASeq. Bioinformatics. 2019;35:2865–7.
Derks W, Bergmann O. Cycling cardiomyocytes: scarce but important in recovery from heart infarction? Circ Res. 2021;128:169–71.
Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–49.
Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104.
Sánchez J, Gomez JF, Martinez-Mateu L, Romero L, Saiz J, Trenor B. Heterogeneous Effects of Fibroblast-Myocyte Coupling in Different Regions of the Human Atria Under Conditions of Atrial Fibrillation. Front Physiol. 2019;10:847:1–13.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550:1–21.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9.
Grissa D, Junge A, Oprea TI, Jensen LJ. Diseases 2.0: a weekly updated database of disease-gene associations from text mining and data integration. Database. 2022:1–8.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
de Lima DS, Cardozo LE, Maracaja-Coutinho V, Suhrbier A, Mane K, Jeffries D, et al. Long noncoding RNAs are involved in multiple immunological pathways in response to vaccination. Proc Natl Acad Sci USA. 2019;116:17121–6.
Russo PST, Ferreira GR, Cardozo LE, Bürger MC, Arias-Carrasco R, Maruyama SR, et al. CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinforma. 2018;19:56:1–13.
Wu C, Arora P. Long Noncoding Mhrt RNA. Circ Cardiovasc Genet. 2015:213–215.
Xu Y, Luo Y, Liang C, Zhang T. LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis. J Mol Cell Cardiol. 2020;139:47–61
Zhang J, Gao C, Meng M, Tang H. Long Noncoding RNA MHRT Protects Cardiomyocytes against H2O2-Induced Apoptosis. Biomol Ther. 2016;24:19–24.
Manso AM, Okada H, Sakamoto FM, Moreno E, Monkley SJ, Li R, et al. Loss of mouse cardiomyocyte talin-1 and talin-2 leads to β-1 integrin reduction, costameric instability, and dilated cardiomyopathy. Proc Natl Acad Sci USA. 2017;114:E6250–9.
Han X, Zhang J, Liu Y, Fan X, Ai S, Luo Y, et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development. 2019;146:1–16.
Das S, Shah R, Dimmeler S, Freedman JE, Holley C, Lee JM, et al. Noncoding RNAs in Cardiovascular Disease: Current Knowledge, Tools and Technologies for Investigation, and Future Directions: A Scientific Statement From the American Heart Association. Circulation: Genomic and Precision Medicine. 2020;13:350–72.
Pant T, Dhanasekaran A, Bai X, Zhao M, Thorp EB, Forbess JM, et al. Genome-wide differential expression profiling of lncRNAs and mRNAs associated with early diabetic cardiomyopathy. Sci Rep. 2019;9:1–16.
Kotrys AV, Cysewski D, Czarnomska SD, Pietras Z, Borowski LS, Dziembowski A, et al. Quantitative proteomics revealed C6orf203/MTRES1 as a factor preventing stress-induced transcription deficiency in human mitochondria. Nucleic Acids Res. 2019;47:7502–17.
Li X, Liu Q, Wang K, Luo W, Liang T, Yuan S, et al. Retracted Article: LncRNA SNHG5 regulates the cell viability and apoptosis of glioma cells by the miR-1297/KPNA2 axis. RSC Adv. 2020;10:1498–506.
Chaudhry F, Isherwood J, Bawa T, Patel D, Gurdziel K, Lanfear DE, et al. Single-cell RNA sequencing of the cardiovascular system: new looks for old diseases. Front Cardiovascular Med. 2019;6:173:1–14.
See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225:1–13.
Li S, Pan H, Tan C, Sun Y, Song Y, Zhang X, et al. Mitochondrial dysfunctions contribute to hypertrophic cardiomyopathy in patient iPSC-derived cardiomyocytes with MT-RNR2 mutation. Stem Cell Rep. 2018;10:808-21.
Wang TY, Lee D, Fox-Talbot K, Arking DE, Chakravarti A, Halushka MK. Cardiomyocytes have mosaic patterns of protein expression. Cardiovasc Pathol. 2018;34:50-57.
Mukund K, Mathewson M, Minamoto V, Ward SR, Subramaniam S, Lieber RL. Systems analysis of transcriptional data provides insights into muscle’s biological response to botulinum toxin. Muscle Nerve. 2014;50:744-58.
Will Y, Eric McDuffie J, Olaharski AJ, Jeffy BD. Drug discovery toxicology: from target assessment to translational biomarkers. John Wiley & Sons; 2016. 584 p.
Haghighi K, Bidwell P, Kranias EG. Phospholamban interactome in cardiac contractility and survival: a new vision of an OLD friend. J Mol Cell Cardiol. 2014;0:160-67.
Fish M, Shaboodien G, Kraus S, Sliwa K, Seidman CE, Burke MA, et al. Mutation analysis of the phospholamban gene in 315 South Africans with dilated, hypertrophic, peripartum and arrhythmogenic right ventricular cardiomyopathies. Sci Rep. 2016;6:1–8.
England J, Granados-Riveron J, Polo-Parada L, Kuriakose D, Moore C, David, et al. Tropomyosin 1: Multiple roles in the developing heart and in the formation of congenital heart defects. J Mol Cell Cardiol. 2017;106:1–13.
Ramaccini D, Montoya-Uribe V, Aan FJ, Modesti L, Potes Y, Wieckowski MR, et al. Mitochondrial function and dysfunction in dilated cardiomyopathy. Front Cell Dev Biol. 2021;8:1-21.
Sannino G, Pasqualini L, Ricciardelli E, Montilla P, Soverchia L, Ruggeri B, et al. Acute stress enhances the expression of neuroprotection- and neurogenesis-associated genes in the hippocampus of a mouse restraint model. Oncotarget. 2016;7:8455–65.
England J, Pang KL, Parnall M, Haig MI, Loughna S. Cardiac troponin T is necessary for normal development in the embryonic chick heart. J Anat. 2016;229:436–49.
Warkman AS, Whitman SA, Miller MK, Garriock RJ, Schwach CM, Gregorio CC, et al. Developmental expression and cardiac transcriptional regulation of Myh7b, a third myosin heavy chain in the vertebrate heart. Cytoskeleton. 2012;69:324-35.
Cardiomyocyte lineage specification in adult human cardiac precursor cells via modulation of enhancer-associated long noncoding RNA expression. JACC: Basic to Translational Science. 2016;1:472–93.
Farrell ET, Grimes AC, de Lange WJ, Armstrong AE, Ralphe JC. Increased postnatal cardiac hyperplasia precedes cardiomyocyte hypertrophy in a model of hypertrophic cardiomyopathy. Front Physiol. 2017;8:414:1–13.
Zhihao L, Jingyu N, Lan L, Michael S, Rui G, Xiyun B, et al. SERCA2a: a key protein in the Ca 2+ cycle of the heart failure. Heart Fail Rev. 2019;25:523–35.
Mareedu S, Pachon R, Thilagavathi J, Fefelova N, Balakrishnan R, Niranjan N, et al. Sarcolipin haploinsufficiency prevents dystrophic cardiomyopathy in mice. Am J Physiol Heart Circ Physiol. 2021;320:H200–10.
Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121:575–83.
Li ZJ, Cheng J, Song Y, Li HH, Zheng JF. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab Invest. 2020;101:341–52.
Tong L, Hill RA, Damisah EC, Murray KN, Yuan P, Bordey A, et al. Imaging and optogenetic modulation of vascular mural cells in the live brain. Nat Protoc. 2020;16:472–96.
Acknowledgements
TARR received a PhD degree fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil.
Funding
This work was supported by grants from the NIH: HL-128215 (JAH), HL-147933 (to JAH), HL-155765 (to JAH), Leducq grant number 11CVD04 (to JAH), Cancer Prevention and Research Institute of Texas grant RP110486P3 (to JAH) and by Agencia Nacional de Investigación y Desarrollo (ANID, Chile), FONDAP 15130011 and 1523A0008 (to SL and VMC), FONDECYT 1200490 (to SL) and FONDECYT 1211731 (to VMC).
Author information
Authors and Affiliations
Contributions
TARR conceived the project, designed, and performed the experiments, conducted the analyses and wrote the manuscript. SYK and SUZ validated the experiments and contributed to manuscript preparation. SL and TGG contributed to the experimental design and manuscript preparation. JAH conceived the project and contributed to manuscript preparation. VMC, SL and TGR conceived the project, designed the experiments, supervised the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Gerry Melino
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ramos, T.A.R., Urquiza-Zurich, S., Kim, S.Y. et al. Single-cell transcriptional landscape of long non-coding RNAs orchestrating mouse heart development. Cell Death Dis 14, 841 (2023). https://doi.org/10.1038/s41419-023-06296-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-06296-9
This article is cited by
-
RNA structure: implications in viral infections and neurodegenerative diseases
Advanced Biotechnology (2024)