Abstract
Mycobacterium tuberculosis (Mtb) is known to evade host immune responses and persist in macrophages for long periods. A mechanism that the host uses to combat Mtb is xenophagy, a selective form of autophagy that targets intracellular pathogens for degradation. Ubiquitination of Mtb or Mtb-containing compartments is a key event to recruit the autophagy machinery and mediate the bacterial delivery to the lysosome. This event relies on the coordinated and complementary activity of different ubiquitin ligases, including PARKIN, SMURF1, and TRIM16. Because each of these factors is responsible for the ubiquitination of a subset of the Mtb population, it is likely that additional ubiquitin ligases are employed by macrophages to trigger a full xenophagic response during Mtb infection. In this study, we investigated the role TRIM proteins whose expression is modulated in response to Mtb or BCG infection of primary macrophages. These TRIMs were ectopically expressed in THP1 macrophage cell line to assess their impact on Mtb replication. This screening identified TRIM32 as a novel player involved in the intracellular response to Mtb infection, which promotes autophagy-mediated Mtb degradation. The role of TRIM32 in xenophagy was further confirmed by silencing TRIM32 expression in THP1 cells, which causes increased intracellular growth of Mtb associated to impaired Mtb ubiquitination, reduced recruitment of the autophagy proteins NDP52/CALCOCO2 and BECLIN 1/BECN1 to Mtb and autophagosome formation. Overall, these findings suggest that TRIM32 plays an important role in the host response to Mtb infection through the induction of autophagy, representing a promising target for host-directed tuberculosis therapies.
Similar content being viewed by others
Introduction
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), has evolved a wide variety of strategies to survive within host macrophages [1]. In most cases, the host immune system can contain Mtb infection in a non-sterilizing manner thanks to the contribution of the innate and adaptive immune responses, which results in an infection that can persist whole life time [2, 3]. TB disease occurs in 3–10% of patients, in which bacterial replication overcomes the immune defenses resulting in an increase in bacterial burden and tissue damage [4]. Recent evidence indicates that the spectrum from TB infection to TB disease is more complex, including a "continuum" of subclinical situations, which also contribute to Mtb transmission [5].
Molecular mechanisms that Mtb has developed to evade host innate immunity include cytosolic escape, restricted production of antimicrobial peptides, blockade of phagosome maturation and antigen presentation, inflammasome activation, and modulation of cell death and autophagy [6, 7]. Autophagy is a main catabolic process that guarantees cellular fitness by mediating turnover of intracellular components in both basal and stress conditions [8]. Materials to be degraded are sequestered in double-membrane vesicles called autophagosomes, which eventually fuse with lysosomes for degradation [8]. In addition to mediate self-digestion, autophagy contributes to immune defense during pathogen infection by delivering intracellular pathogens to the lysosomal compartment, a selective type of autophagy referred as xenophagy [9,10,11,12]. This event leads to both bacterial killing and the accessibility of pathogen components to innate and adaptive immune receptors [11]. In particular, an important role of autophagy in restricting Mtb replication in macrophages has been recently confirmed using both human and mouse models [13,14,15]. On the other hand, Mtb has evolved different strategies to modulate the autophagic activity through the ESX-1 secretion system and PE_PGRS and PE/PPE-containing proteins [7, 12, 16,17,18]. Interestingly, genetic variability of Mtb may account for a different ability to modulate the autophagy response in primary macrophages [19].
As for other types of selective autophagy, recognition of cargos in xenophagy is achieved through different types of autophagy adapters, which physically link cargos and autophagosomal proteins of the Atg8/LC3 family [20]. The best characterized type of autophagy adapters are those belonging to the sequestosome 1/p62-like receptors (SLR) family, which deliver ubiquitinated structures to autophagosomes, containing both ubiquitin-associated and LC3 interacting domains [21, 22]. In the case of Mtb infection, two ubiquitin ligases responsible for ubiquitination of Mtb-containing structures have been reported. The ubiquitin ligase Smurf1 transfers preferentially K48-linked Ub chains on Mtb, targeting it to the autophagosomes via the autophagy adapter NBR1 [23], while Parkin mediates K63-linked Ub chains, which are recognized by the p62/SQSTM1 and NDP52/ CALCOCO2 adapter proteins [24]. Intriguingly, host xenophagy is also triggered by ubiquitin recruited to Mtb in an ubiquitin ligase-independent manner by direct binding to the surface protein PE_PGRS29 [25].
Along with ubiquitin, autophagy adapters are recruited to Mtb by galectins, which bind to cytosolic glycan residues exposed by phagosomes damaged by the Mtb secretion systems, as shown for TAX1BP1 and Galectin-8 [26], and by ubiquilin-1, an ubiquitin-like, and ubiquitin-binding domain protein, which promotes ubiquitin, p62, and LC3 accumulation around Mtb in IFN-γ activated macrophages [27]. Interestingly, recent evidence showed that ubiquitination is also required for the membrane repair of vacuoles damaged by the M. marinum, a close relative of Mtb, to prevent its cytosolic escape through the recruitment of the ESCRT machinery and the nascent autophagosome vesicles [28].
How the autophagy core machinery responsible for autophagosome formation is activated and recruited to Mtb during infection remains less characterized. NEDD4, a HECT type of ubiquitin ligase, has been shown to increase BECLIN 1 stability via ubiquitination and to promote the autophagic response to Mtb infection [29].
The tripartite motif (TRIM) proteins are a large family of ubiquitin ligases with several members playing a role in the regulation of immune responses [30] by interacting with pattern recognition receptors, immune adapter molecules and kinases involved in innate immune signaling pathways [31, 32]. For example, regarding Mtb, TRIM27 functions as a potential restriction factor suppressing the intracellular bacterial survival by enhancing host immune-inflammatory responses and apoptosis mediated by JNK/p38 pathways [33]. Conversely, TRIM25 expression promotes Mtb survival in macrophages possibly by activating the p38 MAPK pathway and suppressing the p65 NF-kB pathway [34].
In addition, several members of TRIM family have been shown to play their roles by modulating the autophagic response by acting at different steps of this process, ranging from upstream signaling pathways and autophagosome formation to cargo recognition and transcriptional regulation of autophagy genes [35]. TRIM5α, 6, 16, 20, 21, 32, 49, and 50 associate with ULK1 and/or BECLIN 1 to promote their activation [31, 36,37,38,39]. Moreover, TRIM28, TRIM37, and TRIM19 control autophagy by targeting upstream signaling pathways, such as those regulated by AMPK and mTOR [40,41,42]. Instead, TRIM22 promotes autophagy by regulating LC3 and BECLIN 1 gene expression through NF‐κB [43]. There is also a subset of TRIM proteins, like TRIM17 or TRIM59, that negatively regulates the autophagy initiation by preferably acting on the BECLIN 1 complex [44, 45].
An important autophagic contribution of TRIM proteins to the innate immune response is their ability to bind pathogens structures and deliver them to the autophagosomes for degradation [35]. Regarding Mtb infection, TRIM-mediated recognition occurs indirectly by binding to galectins that are recruited to bacteria-containing damaged phagosomes, as in the case of TRIM16, which binds to galectin 3 and promotes Mtb ubiquitination and autophagosome engulfment [38].
In this work, we aimed at broadening our knowledge of the role of TRIM proteins in the intracellular response to Mtb infection by combining a transcriptomic (RNA-seq) analysis of TRIM expression in primary macrophages and the effect of ectopic expression of TRIMs on Mtb infection using THP1 macrophage cells. This screening identified TRIM32 as a novel player that is involved in the intracellular immune response to Mtb infection by promoting xenophagy.
Results
Modulation of TRIM expression levels in macrophage-derived monocytes infected with Mtb or BCG
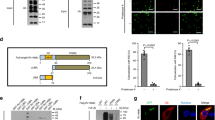
To evaluate the role of TRIM ubiquitin ligases in regulating the innate immune response to Mtb, we analyzed the modulation of their expression levels by RNA-sequencing analysis. Human monocyte-derived macrophages (hMDM) purified from peripheral blood mononuclear cells (PBMC) from healthy donors were in vitro infected with Mtb strain H37Rv or BCG and transcriptomic changes evaluated after 24 h by RNA-seq (Fig. 1A). Experiments were performed with hMDM isolated from three different donors. Differential expression analysis showed that, among 55,766 transcripts identified, 1043 genes were significantly deregulated (fold change≥1.3; p ≤ 0.05) by BCG and 2219 genes by Mtb (Table S1). Among them, we identified 17 distinct TRIM genes, whose levels were modulated upon infection (Table S2). In particular, Mtb infection was associated to a 2.8-fold increased expression of TRIM9 and >1.3 fold reduction of TRIM2, 3, 5, 6, 14, 22, 32, 34, 36, 44, 68, 69 levels when compared to uninfected cells, while BCG increased the expression of TRIM 5, 22, 55, 56 and decreases that of TRIM2. Comparison of TRIM levels in Mtb- versus BCG-infected cells showed an increased TRIM9 expression in Mtb vs BCG infected cells, while the levels of TRIM2, 5, 14, 21, 22, 34, 44, 55, 65, 69 were decreased.
A Experimental scheme: human monocyte-derived macrophages purified from peripheral blood mononuclear cells from healthy donors were infected with the Mtb strain H37Rv and transcriptomic changes evaluated after 24 h by RNA-seq. We have identified 17 TRIM genes, whose levels are modulated upon infection. cDNAs of identified TRIMs were cloned in a lentiviral vector in frame with the coding sequence for the Green Fluorescence Protein (GFP). Lentiviral particles were produced and used to stably transduce the monocyte cell line THP-1 cells. GFP TRIM expressing cells were differentiated to macrophages using PMA and infected with Mtb H37Rv DsRed. Twenty-four hours post infection, cells were fixed and the percentage of Mtb-infected cells among those positive for GFP were evaluated by confocal microscopy. B GFP TRIM expressing THP-1 cells were infected with Mtb H37Rv DsRed. Twenty-four hours post infection, cells were fixed and the percentage of Mtb-infected cells among those positive for GFP were evaluated by confocal microscopy. Ectopic expression of TRIM22 and TRIM32 reduces the percentage of Mtb-infected THP1, while TRIM36 and TRIM56 have the opposite effect. *P < 0.05, **P < 0.01.
Identification of TRIM genes that control Mtb replication in THP-1 cells
To elucidate if the TRIM genes that are modulated during Mtb/BCG infection play a role in the macrophage antibacterial response, we analyzed the effect of lentivirus-mediated TRIM ectopic expression on Mtb infection of THP-1 monocyte cell line. This approach was chosen because most of the modulated TRIMs were downregulated during Mtb infection. cDNAs of selected TRIMs were cloned in a lentiviral vector in frame with the coding sequence for the Green Fluorescence Protein (GFP). Lentiviral particles were produced and used to stably transduce THP-1 cells. GFP TRIM-expressing cells were in vitro differentiated to macrophages using PMA and infected with Mtb H37Rv DsRed. Twenty-four hours post in vitro infection, cells were fixed and the percentage of Mtb-infected cells among those positive for GFP were evaluated by confocal microscopy. We found that the ectopic expression of TRIM22 and TRIM32 reduces the percentage of Mtb-infected THP1 compared to GFP control cells, while TRIM36 and TRIM56 have the opposite effect (Fig. 1B). Interestingly, one of the two TRIMs that we found to restrict Mtb replication, TRIM22, was previously reported to participate to the host response to viral and bacterial infection [46], including Mtb [43], thus corroborating the reliability of the screening results. We therefore focused on TRIM32, since evidences on its contribution on Mtb infection have not been reported so far.
TRIM32 potentiates the autophagy response in Mtb-infected THP-1 macrophages
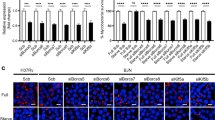
First, we validate the ability of TRIM32 to restrict Mtb infection by a colony forming units (CFU) counts, the gold-standard for quantifying viable Mtb. As shown in Fig. 2A, a significant reduction in the number of CFU were observed in TRIM32 expressing THP-1 when compared to GFP expressing THP-1 upon infection with Mtb for 4 days.
A TRIM32-GFP and control GFP expressing THP-1 were differentiated for 24 h with PMA and infected with the Mtb strain H37Rv. Ectopic expression of TRIM32 was monitored by western blot using an anti-GFP antibody (Left Panel). Four days after infection cells were lysed to measure the number of viable bacteria by plating for determining CFU (Right Panel). Values are expressed as a mean of three independent experiments; *P < 0.05. B TRIM32-GFP and control GFP expressing THP-1 were differentiated for 24 h with PMA and infected with Mtb strain H37Rv for 24 h or left uninfected. Two hours before lysis, cells were incubated with the lysosome inhibitors E64d and Pepstatin A (E64d/PepA), as indicated. LC3 levels were analyzed by western blot. HSP90 was included as a loading control. The graph reporting means ± SD of LC3-II/HSP90 values from three independent experiments is shown in C *P = < 0.05. A.U.: Arbitrary Units. D, E TRIM32-GFP and control GFP expressing THP-1 were transduced with control shRNA, shAMBRA1 (D) or shBECLIN1 (E). AMBRA1, BECLIN 1, and TRIM32-GFP levels were analyzed by western blot; HSP90 was included as a loading control. F TRIM32-GFP and GFP control cells silenced for BECLIN 1 or AMBRA1 were infected with Mtb H37Rv and, 4 days after infection, cells were lysed to measure the number of viable bacteria by CFU counting (shCTR: cells infected with unrelated shRNA lentivirus). Values are expressed as a mean of three independent experiments. *P = < 0.05, **P = < 0.01. Full western blots are reported in Supplemental Fig. 2A.
To elucidate how TRIM32 affects Mtb infection, we started by analyzing whether its ectopic expression results in increased autophagy levels in THP-1 cells, since we and others have previously demonstrated that TRIM32 is involved in the regulation of autophagy [47,48,49]. GFP- and TRIM32-transduced THP-1 cells were infected with Mtb and autophagy flux was analyzed upon 24 h by comparing the levels of the autophagosome protein LC3-II in the presence or absence of lysosome inhibitors. Immunoblot analysis showed a significant increase in the rate of LC3-II degradation in TRIM32-expressing cells following infection with Mtb (Fig. 2B, C).
Autophagy inhibition rescues Mtb clearance in TRIM32 expressing macrophages
These results prompted us to assess if autophagy was required for the ability of TRIM32 to restrict Mtb infection. To this aim, GFP and TRIM32 expressing THP-1 were silenced for the expression of two upstream autophagy genes, BECLIN 1 and AMBRA1, infected with Mtb and bacterial replication analyzed by CFU counts 4 days post infection. As shows in Fig. 2D–F, restriction in intracellular Mtb replication in TRIM32 THP-1 was abolished when the expression of BECLIN 1 and AMBRA1 was inhibited, indicating that TRIM32 potentiates the autophagic response in Mtb-infected cells. We also verified that the observed rescue was not due to a reduction of TRIM32 expression in autophagy gene silenced cells (Fig. 2D, E).
Increased ubiquitination and autophagosomal engulfment of Mtb in TRIM32 expressing THP1
To confirm the hypothesis that the ubiquitin ligase TRIM32 is involved in xenophagy induction in Mtb-infected cells, we analyzed three sequential events required for the autophagosomal engulfment of Mtb: (i) ubiquitination of Mtb (or Mtb-containing structures); (ii) recruitment of ubiquitin-binding autophagy adapters to Mtb; (iii) recruitment of the autophagosomal markers to Mtb.
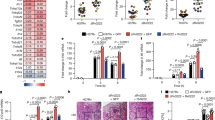
To this aim, we carried out a colocalization analysis of Mtb DsRed with: (i) ubiquitin (Fig. 3A, D); (ii) the ubiquitin-binding autophagy adapter NDP52 (Fig. 3B, E), which is also recruited to Mtb upon K63-linked polyubiquitination mediated by Parkin [24]; and (iii) the autophagosome marker LC3 (Fig. 3C, F) in GFP and TRIM32 expressing THP-1 24 h post infection by confocal microscopy. Remarkably, all analyzed autophagy markers showed increased colocalization with Mtb in TRIM32 expressing cells compared to controls, confirming that TRIM32 promotes the Mtb clearance mediated by autophagy. Consistent with the involvement of TRIM32 in bacterial ubiquitination, we observed a mean of 10% colocalization of GFP-TRIM32 signal, mainly characterized by dotted structures, with Mtb (Fig. S1), a percentage in line with what reported for Parkin in Mtb-infected macrophages [24].
TRIM32-GFP (right) and control GFP (left) expressing THP-1 were infected with Mtb H37Rv DsRed. After 24 h cells were fixed and analyzed for Ubiquitin (A) or NDP52 (B) or LC3 (C) localization with Mtb DsRed, by confocal microscopy using specific antibodies. Merging of the three fluorescence signals is shown in the large panels. Scale bar, 10 μm. Colocalization rate was calculated using the ImageJ software. Graphs reporting the quantification of the experiments in A–C, are shown in D–F, respectively. The results represent the mean ± SEM of three independent experiments (***P < 0.001).
TRIM32 is required for triggering an efficient autophagy response to Mtb infection in THP-1 macrophages
Based on the results obtained with THP-1 cells ectopically expressing TRIM32, we investigated the contribution of endogenous TRIM32 in the control of Mtb replication. TRIM32 expression was downregulated by using two different lentiviral short hairpin RNAs (shTRIM32a and shTRIM32b) in THP1 (Fig. 4A), infected with Mtb and bacterial growth analyzed by CFU counts 4 days after infection. Notably, Mtb replication is significantly increased in TRIM32 silenced THP-1 (Fig. 4B), indicating that TRIM32 contributes to the control of Mtb replication in macrophages.
A THP-1 cells were transduced with control short hairpin RNAs (shRNA) and two independent lentiviral different (shTrim32a and shTrim32b) and silencing verified by western blot using a TRIM32 antibody. B Cells described in A were infected with the Mtb H37Rv and, 4 days after infection, cells were lysed to measure the number of viable bacteria by CFU counting. Values are expressed as a mean of three independent experiments. *P < 0.05, **P < 0.01. C shCTR, shTRIM32a and shTRIM32b THP-1 cells were infected with Mtb H37Rv for 24 h. Two hours before lysis, cells were incubated with the lysosome inhibitors E64d and pepstatin A (E64d/PepA), as indicated, to evaluate autophagic flux. LC3 levels were analyzed by western blot. β-Actin was included as a loading control. The graph reports means ± SD of LC3-II/β-Actin values from three independent experiments is shown in D; *P < 0.05. **P < 0.01. A.U. arbitrary units. Full western blots are reported in Supplemental Fig. 2B.
Then, we assessed if TRIM32 is required for promoting the autophagic response during Mtb infection by analyzing LC3-II levels by immunoblotting in Mtb-infected cells, in the presence or absence of lysosome inhibitors. As shown in Fig. 4C, D, autophagy levels are critically reduced when TRIM32 expression is inhibited by two lentiviral shRNAs both in basal conditions and upon Mtb infection, indicating that TRIM32 is required to sustain the autophagic activity in THP-1 cells. Consistently, colocalization of Mtb with LC3-positive autophagosomes is significantly reduced (Fig. 5A, D). Furthermore, we analyzed if TRIM32 deficiency affects early steps of xenophagy i.e.: Mtb ubiquitination and the recruitment of autophagy adapters by confocal microscopy. Confocal analysis showed that colocalization of Mtb with ubiquitin (Fig. 5B, E) and NDP52 (Fig. 5C, F) is drastically reduced in cells silenced for TRIM32.
shTRIM32 (left) and shCTR (right) THP-1 were infected with Mtb H37Rv DsRed. Cells were fixed and analyzed for LC3 (A), NDP52 (B), or Ubiquitin (C) localization with Mtb H37Rv DsRed, by confocal microscopy, using specific antibodies. Merging of the two fluorescence signals is shown in the large panels. Scale bar, 10 μm. Colocalization rate was calculated using the ImageJ software. Graphs reporting a quantification of the experiments in A–C, are shown in D–F, respectively. The results represent the mean ± SEM of three independent experiments (*P ≤ 0.05, **P ≤ 0.01, ***P < 0.001).
Finally, based on the ability of TRIM32 to stimulate the activity of ULK1 and BECLIN 1, we assessed if the impaired autophagy response observed in TRIM32-silenced macrophages is associated to a reduced recruitment of the autophagy core machinery to intracellular Mtb [47, 50]. By confocal analysis, we observed, in control cells (shCTR), an extensive translocation of BECLIN 1 on Mtb positive structures (Fig. 6), while ULK1 colocalization was very limited (data not shown), suggesting that ULK1 does not translocate to Mtb or this interaction is highly dynamic. Notably, in TRIM32-silenced cells (shTRIM32) the colocalization of Mtb with BECLIN 1 (Fig. 6) was significantly reduced.
A shTRIM32 (lower panel) and shCTR (upper panel) THP-1 were infected with Mtb H37Rv DsRed. Cells were fixed and analyzed for BECLIN 1 localization with Mtb H37Rv DsRed by confocal microscopy using specific antibodies. Merging of the two fluorescence signals is shown on the large panels. Scale bar, 10 μm. Colocalization rate was calculated by ImageJ software. A graph reporting a quantification of the experiments is shown in B. The results represent the mean ± SEM of three independent experiments (**P < 0.01).
Altogether, these results indicate that TRIM32 mediates the recruitment to intracellular Mtb of the autophagy machinery required for autophagosome formation and cargo engulfment.
Discussion
This study shows that the ubiquitin ligase TRIM32 plays an important role in the regulation of xenophagic response to Mtb infection. We have initially identified TRIM32 in an RNA-seq analysis aimed at identifying TRIM genes whose expression is modulated during ex vivo infection of human primary macrophages with Mtb or BCG. This screening identified 17 TRIMs, of which TRIM32, together with TRIM22, showed the largest extent of downregulation in Mtb-infected cells. We have then characterized the role of the identified TRIMs in the host response to Mtb infection by ectopic expression approaches in THP-1 cells and identified TRIM32, together with TRIM22, as TRIM proteins with anti-Mtb replication activity. Further characterization of the molecular mechanisms by which TRIM32 controls Mtb replication showed that TRIM32 induces xenophagy in Mtb-infected cells and autophagy core machinery proteins are required for the antibacterial activity of TRIM32. In addition, using both ectopic expression and RNA interference approaches, we showed that TRIM32 contributes to xenophagy by promoting both the recruitment of BECLIN 1 to Mtb for autophagosome formation and the autophagosome engulfment of Mtb-mediated by ubiquitination.
TRIM32 is a gene mutated in limb-girdle muscular dystrophy 2H, a late-onset autosomal recessive myopathy [51]. TRIM32 is ubiquitously expressed and several evidences point at the role of TRIM32 in regulating cellular processes beyond muscle cells. TRIM32 was originally identified as an interactor of HIV-1 TAT protein. A role of TRIM32 in the innate immunity has been reported in response to viral infection, as in the case of influenza and Herpes Simplex Viruses [52, 53]. In this regard, TRIM32 has been identified as an important regulator of the cGAS–STING pathway of cytosolic DNA sensing [54].
Multiple roles of TRIM32 in the regulation of autophagy have recently emerged. We have shown that the ubiquitin ligase activity of TRIM32 regulates upstream autophagy events in muscle cells upon atrophy induction to protect them from excessive ROS production and consequent induction of the atrophic genes [47]. TRIM32 stimulates autophagy by interacting with ULK1 and AMBRA1 and stimulating ULK1 kinase activity through the formation of unanchored K63-linked polyubiquitin chains. TRIM32 has also been reported to act further downstream in the autophagy process. It positively regulates p62/SQSTM1 activity upon nutrient deprivation, favoring p62 dot formation and autophagosome sequestration via monoubiquitination [49]. Moreover, TRIM32 interacts with TRIF, a critical adapter protein of Toll-like receptors 3/4, and mediates its autophagic degradation in an E3 activity-independent manner by binding to the autophagy cargo receptor TAX1BP1 [48].
Our results suggest that TRIM32 may also act at different steps in xenophagy. TRIM32 silenced cells show a significant decrease in the ubiquitination of Mtb, a prerequisite for the binding of autophagy cargo receptors and autophagosome engulfment. Previous studies have identified the ubiquitin ligases SMURF1 and PARKIN responsible for Mtb ubiquitination and autophagy engulfment [23, 24], highlighting how Mtb ubiquitination could be mediated by the cooperative activity of various ubiquitin ligases. It remains to be elucidated whether TRIM32 is required for the xenophagic activity of SMURF1 and PARKIN or acts independently of them. In addition to a direct role in Mtb ubiquitination, TRIM32 may also facilitate the autophagosome engulfment of Mtb by acting directly as a cargo receptor, as in the case of TRIF [48], or by stimulating the activity of SQSTM1 family proteins through monoubiquitination [49]. Moreover, we found that the inhibition of TRIM32 expression impairs the translocation of BECLIN 1 to Mtb-positive structures, suggesting that TRIM32 is involved in the activation of the core machinery for autophagosome formation in pathogen proximity. In this regard, based on the previously reported role of TRIM32 in cGAS/STING-activated signaling pathways [54], TRIM32 could be involved in transducing pathogen recognition signaling to the autophagy core machinery upon Mtb infection.
In conclusion, our experimental approaches led to the identification of TRIM32 as a novel protein playing a role in the intracellular response to Mtb infection by potentiating autophagy-mediated clearance, whose expression is downregulated in Mtb-infected cells. These results candidate TRIM32 as a novel immune response regulator, whose impairment of expression or activity may be evaluated as a biomarker for TB stages stratification or as a target for host directed therapies. Noteworthy, TRIM32 expression was recently reported to be decreased in leukocytes from TB disease patients in comparison with healthy controls and subjects with TB infection [55].
Materials and methods
Cell culture
Macrophage-derived monocytes (MDM) were obtained as described previously [19]. THP-1 cell line (ATCC TIB-202) were cultured in RPMI 1640 (Sigma-Aldrich, R8758) supplemented with 10% of inactivated fetal bovine serum (Gibco, 10270), 2 mM l-glutamine, and 1% penicillin–streptomycin solution (Sigma-Aldrich, G7513/P0781) and maintained in a humidified atmosphere containing 5% CO2 at 37 °C. Cells were screened for mycoplasma contamination by PCR (ABMgood, G238). THP-1 cells were induced to differentiation in RPMI 1640 containing 20 ng/ml of phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) for 2 days to obtain THP-1 macrophages. Depending on the experimental requirements, different amounts of cells were used. In details, for the western blot experiments, Thp-1 cells were seeded in 6-well flat bottom tissue culture plates (1 × 106 cells/well, 2 mL/well; Corning). For the confocal experiments, THP-1 cells were seeded in glass slide chambers (Nunc, Lab-Tek) (5 × 105 cells/well, 1 mL/well). For the colony forming unit (CFU) experiment, THP-1 cells were seeded in 24-well flat bottom tissue culture plates (5 × 105 cells/well, 1 mL/well; Corning). For RNA-seq experiments macrophage-derived monocytes were seeded in 6-well flat bottom tissue culture plates (3 × 106 cells/well, 2 mL/well; Corning). Autophagy was evaluated 24 h after infection. To assess the autophagy flux, the lysosomal inhibitors E64d/Pepstatin A (5 µg/mL Santa Cruz Biotechnology, sc-201280A and sc-45036), were added 2 h before lysis.
Bacteria
The M. tuberculosis strain H37Rv, M. tuberculosis strain H37RvDsRED, and M. bovis Bacillus Calmette et Guerin (BCG) were cultured at the Fondazione Policlinico Gemelli IRCCS, Università Cattolica del Sacro Cuore, as previously described [56]. In brief, the strains were grown in Middlebrook 7H9 (BD NY) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; BD 212240), with 0.2% glycerol (Sigma-Aldrich G7757) and 0.05% Tween 80 (Sigma-Aldrich P8074) at 37 °C. Mycobacterial cultures were harvested at late log phase, glycerol was added at 20% final concentration, and 1-ml aliquots stored at −80 °C. All experiments with Mtb strains were carried out in biosafety laboratory level 3 (BSL-3), following standard safety procedures. Macrophages were infected with H37Rv or H37Rv Ds-red or BCG at a multiplicity of infection (MOI) of 5:1 for RNA-Seq analysis, for western blot analysis and CFU and at a MOI of 2:1 for confocal experiments. Two hours after the in vitro infection, macrophages were washed once with phosphate-buffered saline (PBS) and then fresh medium was added.
Colony-forming unit assay
To determine the intracellular bacterial load, CFUs of infected macrophages were measured in triplicate and determined at 0 h and 4 days post-infection, as described [57]. Briefly, infected cell cultures were lysed in PBS 0.1% Triton X-100 (Sigma-Aldrich, T9284) and the serial dilution was prepared in PBS 0.05% Tween 80 (Sigma-Aldrich P8074) Fifty-microliter aliquots of each dilution were plated on 7H11/ OADC (BD) agar plates. Plates were incubated for 3 weeks.
Total RNA isolation and RNA-seq analysis
RNA was extracted from BCG and H37Rv infected MDM cells, at 24 h p.i. and from non-infected cells as control, using TRIzol reagent (Invitrogen, 15596-018) according to manufacturer’s guidelines. RNA integrity was evaluated by using the Agilent 2100 Bioanalyzer (Agilent Technologies). Next-generation sequencing experiments were performed by Genomix4Life S.r.l. Indexed libraries were prepared from purified RNA with the TruSeq Total Stranded RNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies). Index-tagged samples were equimolar and the overall concentration was 2 nM. The pooled samples were subject to cluster generation and sequenced using an Illumina HiSeq 2500 System (Illumina) in a 2 × 100 paired-end format at a final concentration of 8 pmol. The resulting short reads were aligned against the GRCh38.p14 genome assembly, using STAR (ver. 2.6.1a). Piled-up reads were counted with htseq-count. Read-count normalization and comparisons were performed using the edgeR R package. Genes were considered differentially expressed between groups if their expression values significantly differed by ≥ 1.3 folds. Correction for multiple tests was achieved by the Benjamini–Hochberg procedure. The significance threshold was set to 0.05.
Plasmids and cloning
TRIM ORFs were PCR amplified from Genscript vector (TRIM2 OHu30750D, TRIM6 OHu03491D, TRIM34 OHu13090D, and TRIM65 OHu11804), from Sino Biological vector (TRIM14 HG24800-UT, TRIM22 HG19229-UT), from Proteo Genix vector (TRIM21 ID 6737, TRIM36 ID 55521). TRIM5 and TRIM32 were amplified from pcDNA3.1TRIM5 vector and pcDNA3.1TRIM32 vector, respectively, and TRIM 3, 9, 44, 55, 56, 68, and 69 were PCR amplified from a pool of cDNA from Hela, HEK293T, and SHSY5Y cells. All the TRIMs were cloned into a lentiviral pBOB-GFP (Addgene #12337) vector using appropriate oligonucleotides followed by in-frame insertion into the vector.
Lentiviral transduction
Lentiviral production was performed using 293T as previously described [58] and THP1 cells infected using 50 μL of viral suspension in a medium supplemented with 4 μg/ml polybrene (Sigma-Aldrich, TR-1003) for 8 h. For stable human TRIM32 RNA interference, two lentiviral TRIM32 shRNA–targeting pLKO.1 plasmids were used (TRCN0000273103 and TRCN0000273176; Sigma-Aldrich). For stable human AMBRA1 mRNA interference, a lentiviral AMBRA1 mRNA–targeting pLKO.1 plasmid was used (TRCN0000168652; Sigma-Aldrich). For stable human BECN-1 mRNA interference, a lentiviral BECN-1 mRNA–targeting pLKO.1 plasmid was used (TRCN0000299864; Sigma-Aldrich). A pLKO.1 containing a non-mammalian shRNA was used as a negative control (Sigma-Aldrich).
Western blot analysis
Cells were collected 24 h post infection and lysed in CelLytic buffer (Sigma-Aldrich, C3228) plus the following protease and phosphatase inhibitors: Protease Inhibitor Cocktail plus (Sigma- Aldrich, P8340), 5 mM sodium fluoride (Sigma-Aldrich, S-7920), 0.5 mM sodium orthovanadate (Sigma-Aldrich, S6508), 1 mM sodium molybdate (Sigma-Aldrich, S-6646), 50 mM 2-chloroacetamide (Sigma-Aldrich, C0267), 2 mM 1,10-phenanthroline monohydrate (Sigma-Aldrich, 320056), and 0.5 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, P7626). Proteins were separated on SDS PAGE gels and electroblotted onto nitrocellulose (Whatman Amersham, 10600041) or PVDF (Millipore, IPVH20200) membranes. Blots were incubated with primary antibodies in 5% nonfat dry milk (Biosigma, 711160) in PBS (Thermo Fisher Scientific, 18912– 0149) plus 0.1% Tween-20 (Sigma-Aldrich, P1379) overnight at 4 °C. Detection was achieved using horseradish peroxidase–conjugated secondary antibodies (anti-goat 705–036-147, anti-rabbit 711–036-152, and anti-mouse 715–036-150, Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence (ECL, Immobilon Classico WBLUC0500 and Immobilon Crescendo Western HRP substrate WBLUR0500, Millipore). Signals were acquired using a ChemiDoc imaging system. The primary antibodies used in this study were rabbit anti-LC3 (Cell Signaling, Danvers, MA), rabbit anti–BECLIN 1 (Santa Cruz Biotechnology, sc-11427 (H-300)), rabbit anti-AMBRA1 (Millipore, ABC131), rabbit anti-TRIM32 (Thermo Fisher Scientific, PA5-22316), mouse anti-HSP90 (Santa Cruz Biotechnology, sc-13119; F-8), mouse anti β-Actin (Santa Cruz Biotechnology, sc-47778 C4) and mouse anti GFP (Santa Cruz Biotechnology, sc-9996 B-2).
Confocal microscopy
Confocal microscopy analyses were carried out as previously described [59]. In brief, twenty-four hours after infection, cells were fixed with 4% paraformaldehyde (CARLO ERBA Reagents, 387507) in PBS followed by permeabilization with 0.2% triton X-100 (Sigma-Aldrich, T9284) in PBS. Cells were labeled with the primary antibody anti-LC3 (Cosmo Bio Ltd, CTB-LC3-2-IC), anti-NDP52 (Ab-184688, Abcam), anti-Ubiquitin (FK2) (ST1200, Millipore) for 1 h at room temperature and visualized by means of Cy5-conjugated secondary antibodies (Jackson Immunoresearch) or Alexa Fluor-488 secondary antibodies (Thermo Fisher Scientific). Coverslips were mounted in Prolong Gold antifade (P36935, Life Technologies) and examined under a confocal microscope (Leica TCS SP2). Digital images were acquired with the Leica software and studies of colocalization were performed by using appropriated tools of ImageJ software [60]. A minimum of 200 cells per sample experimental condition were counted for triplicate samples per condition in each experiment.
Statistical analysis
Statistical analysis of confocal microscopy data were carried out using the Mann-Whitney test (independent samples, two-sided, Graph-Pad, San Diego, CA), while CFU and immunoblotting data were analyzed using unpaired, two-tailed Student’s t test (Excel software). Values are shown as mean ± standard deviation of at least 3 independent experiments. Densitometric analysis of immunoblots was performed using the Adobe Photoshop software. The control ratio was arbitrarily defined as 1.00. P-values < 0.05, <0.01, <0.001 were marked by *, **, ***, respectively. Normal distribution was assumed on the basis of the appearance of the data, since n < 5. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. No exclusion criteria were applied to exclude samples from analysis.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Chai Q, Wang L, Liu CH, Ge B. New insights into the evasion of host innate immunity by Mycobacterium tuberculosis. Cell Mol Immunol 2020;17:901–13.
Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell Mol Immunol 2017;14:963–75.
Goletti D, Delogu G, Matteelli A, Migliori GB. The role of IGRA in the diagnosis of tuberculosis infection, differentiating from active tuberculosis, and decision making for initiating treatment or preventive therapy of tuberculosis infection. Int J Infect Dis. 2022;124:S12–19.
Dutta NK, Karakousis PC. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev 2014;78:343–71.
Migliori GB, Ong CWM, Petrone L, D’Ambrosio L, Centis R, Goletti D. The definition of tuberculosis infection based on the spectrum of tuberculosis disease. Breathe (Sheff). 2021;17:210079–2021.
Chandra P, Grigsby SJ, Philips JA. Immune evasion and provocation by Mycobacterium tuberculosis. Nat Rev Microbiol 2022;20:750–66.
Goletti D, Petruccioli E, Romagnoli A, Piacentini M, Fimia GM. Autophagy in Mycobacterium tuberculosis infection: a passepartout to flush the intruder out? Cytokine Growth Factor Rev. 2013;24:335–43.
Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet 2023;24:382–400. https://doi.org/10.1038/s41576-022-00562-w.
Vargas JNS, Hamasaki M, Kawabata T, Youle RJ, Yoshimori T. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol 2023;24:167–85.
Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–11.
Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35.
Romagnoli A, Etna MP, Giacomini E, Pardini M, Remoli ME, Corazzari M, et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy. 2012;8:1357–70.
Aylan B, Bernard EM, Pellegrino E, Botella L, Fearns A, Athanasiadi N, et al. ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages. Nat Microbiol 2023;8:803–18.
Deretic V, Wang F. Autophagy is part of the answer to tuberculosis. Nat Microbiol 2023;8:762–3.
Golovkine GR, Roberts AW, Morrison HM, Rivera-Lugo R, McCall RM, Nilsson H, et al. Autophagy restricts Mycobacterium tuberculosis during acute infection in mice. Nat Microbiol 2023;8:819–32.
De Maio F, Berisio R, Manganelli R, Delogu G. PE_PGRS proteins of Mycobacterium tuberculosis: a specialized molecular task force at the forefront of host-pathogen interaction. Virulence. 2020;11:898–915.
Sharma T, Alam A, Ehtram A, Rani A, Grover S, Ehtesham NZ, et al. The Mycobacterium tuberculosis PE_PGRS protein family acts as an immunological decoy to subvert host immune response. Int J Mol Sci. 2022;23:525. https://doi.org/10.3390/ijms23010525.
Petruccioli E, Romagnoli A, Corazzari M, Coccia EM, Butera O, Delogu G, et al. Specific T cells restore the autophagic flux inhibited by Mycobacterium tuberculosis in human primary macrophages. J Infect Dis. 2012;205:1425–35.
Romagnoli A, Petruccioli E, Palucci I, Camassa S, Carata E, Petrone L, et al. Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy. Cell Death Dis 2018;9:624–8.
Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013;13:722–37.
Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16.
Lamark T, Johansen T. Mechanisms of selective autophagy. Annu Rev Cell Dev Biol 2021;37:143–69.
Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017;21:59–72.
Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6.
Chai Q, Wang X, Qiang L, Zhang Y, Ge P, Lu Z, et al. A Mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat Commun 2019;10:1973–8.
Bell SL, Lopez KL, Cox JS, Patrick KL, Watson RO. Galectin-8 senses phagosomal damage and recruits selective autophagy adapter TAX1BP1 to control mycobacterium tuberculosis infection in macrophages. mBio. 2021;12:e0187120–20. Epub 2021 Jul 6
Sakowski ET, Koster S, Portal Celhay C, Park HS, Shrestha E, Hetzenecker SE, et al. Ubiquilin 1 promotes IFN-γ-induced xenophagy of Mycobacterium tuberculosis. PLoS Pathog. 2015;11:e1005076.
Raykov L, Mottet M, Nitschke J, Soldati T. A TRAF-like E3 ubiquitin ligase TrafE coordinates ESCRT and autophagy in endolysosomal damage response and cell-autonomous immunity to Mycobacterium marinum. Elife. 2023; 12. https://doi.org/10.7554/eLife.85727.
Pei G, Buijze H, Liu H, Moura-Alves P, Goosmann C, Brinkmann V, et al. The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy. 2017;13:2041–55.
Versteeg GA, Benke S, García-Sastre A, Rajsbaum R. InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014;25:563–76.
Kimura T, Jain A, Choi SW, Mandell MA, Johansen T, Deretic V. TRIM-directed selective autophagy regulates immune activation. Autophagy. 2017;13:989–90.
van Gent M, Sparrer KMJ, Gack MU. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol 2018;5:385–405.
Wang J, Teng JL, Zhao D, Ge P, Li B, Woo PC, et al. The ubiquitin ligase TRIM27 functions as a host restriction factor antagonized by Mycobacterium tuberculosis PtpA during mycobacterial infection. Sci Rep. 2016;6:34827.
Liu H, Zhu T, Li Q, Xiong X, Wang J, Zhu X, et al. TRIM25 upregulation by Mycobacterium tuberculosis infection promotes intracellular survival of M.tb in RAW264.7 cells. Microb Pathog 2020;148:104456.
Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, Fimia GM. TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020;27:887–902.
Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell 2014;30:394–409.
Fusco C, Mandriani B, Di Rienzo M, Micale L, Malerba N, Cocciadiferro D, et al. TRIM50 regulates Beclin 1 proautophagic activity. Biochim Biophys Acta Mol Cell Res 2018;1865:908–19.
Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell 2016;39:13–27.
Di Rienzo M, Antonioli M, Fusco C, Liu Y, Mari M, Orhon I, et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci Adv 2019;5:eaau8857.
Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–28.
Wang W, Xia Z, Farré JC, Subramani S. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy. 2018;14:1574–85.
Missiroli S, Bonora M, Patergnani S, Poletti F, Perrone M, Gafà R, et al. PML at mitochondria-associated membranes is critical for the repression of autophagy and cancer development. Cell Rep. 2016;16:2415–27.
Lou J, Wang Y, Zheng X, Qiu W. TRIM22 regulates macrophage autophagy and enhances Mycobacterium tuberculosis clearance by targeting the nuclear factor-multiplicity κB/beclin 1 pathway. J Cell Biochem. 2018;119:8971–80.
Mandell MA, Jain A, Kumar S, Castleman MJ, Anwar T, Eskelinen EL, et al. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J Cell Sci. 2016;129:3562–73.
Han T, Guo M, Gan M, Yu B, Tian X, Wang JB. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy. 2018;14:2035–48.
Pagani I, Poli G, Vicenzi E. TRIM22. A multitasking antiviral factor. Cells. 2021;10:1864. https://doi.org/10.3390/cells10081864.
Di Rienzo M, Piacentini M, Fimia GM. A TRIM32-AMBRA1-ULK1 complex initiates the autophagy response in atrophic muscle cells. Autophagy. 2019;15:1674–6.
Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13:e1006600.
Overå KS, Garcia-Garcia J, Bhujabal Z, Jain A, Øvervatn A, Larsen KB, et al. TRIM32, but not its muscular dystrophy-associated mutant, positively regulates and is targeted to autophagic degradation by p62/SQSTM1. J Cell Sci. 2019;132:jcs236596. https://doi.org/10.1242/jcs.236596.
Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66.
Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2H and the role of TRIM32. Handb Clin Neurol. 2011;101:125–33.
Wu S, Zhang J, Xue Q, Liu J, Huang B, He Z, et al. Duck TRIM32 functions in IFN-β signaling against the infection of H5N6 highly pathogenic avian influenza virus. Front Immunol 2020;11:377.
Cui H, Liu Y, Huang Y. Roles of TRIM32 in corneal epithelial cells after infection with herpes simplex virus. Cell Physiol Biochem 2017;43:801–11.
Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–55.
Chen Y, Cao S, Sun Y, Li C. Gene expression profiling of the TRIM protein family reveals potential biomarkers for indicating tuberculosis status. Microb Pathog 2018;114:385–92.
Palucci I, Matic I, Falasca L, Minerva M, Maulucci G, De Spirito M, et al. Transglutaminase type 2 plays a key role in the pathogenesis of Mycobacterium tuberculosis infection. J Intern Med 2018;283:303–13.
Palucci I, Salustri A, De Maio F, Pereyra B, Maria Del C, Paglione F, Sali M, et al. Cysteamine/cystamine exert anti-mycobacterium abscessus activity alone or in combination with amikacin. Int J Mol Sci. 2023;24:1203. https://doi.org/10.3390/ijms24021203.
Di Rienzo M, Romagnoli A, Ciccosanti F, Refolo G, Consalvi V, Arena G, et al. AMBRA1 regulates mitophagy by interacting with ATAD3A and promoting PINK1 stability. Autophagy. 2022;18:1752–62.
Reali V, Mehdawy B, Nardacci R, Filomeni G, Risuglia A, Rossin F, et al. Reticulon protein-1C is a key component of MAMs. Biochim Biophys Acta. 2015;1853:733–45.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Acknowledgements
This study was supported by grants from Ministry of Health: Ricerca Corrente Linea 4 and Ricerca Finalizzata RF-2021-12373231 to GMF and GD, GR-2011-02350886 to AR. We acknowledge co-funding from Next Generation EU through the Italian Ministry of University and Research, in the context of the National Recovery and Resilience Plan, Investment PE8 – Project Age-It: “Ageing Well in an Ageing Society” to Gian Maria Fimia, co-financed by the Next Generation EU (DM 1557 11.10.2022), and PE6 “Heal Italia" to Mauro Piacentini CUP E83C22004670001. The views and opinions expressed are only those of the authors and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them. We also acknowledge funds from 5 × 1000 tax donation to MDR, TM, and GMF.
Author information
Authors and Affiliations
Contributions
GMF, MP, DG, GM, and GD contributed to the study conception and design. Material preparation, data collection, and analysis were performed by AR, MDR, EP, CF, IP, LM, and TM. The first draft of the manuscript was written by GMF, with the help of AR, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
COMPETING INTERESTS
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Gerry Melino
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Romagnoli, A., Di Rienzo, M., Petruccioli, E. et al. The ubiquitin ligase TRIM32 promotes the autophagic response to Mycobacterium tuberculosis infection in macrophages. Cell Death Dis 14, 505 (2023). https://doi.org/10.1038/s41419-023-06026-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-06026-1
This article is cited by
-
Multipronged regulation of autophagy and apoptosis: emerging role of TRIM proteins
Cellular & Molecular Biology Letters (2024)