Abstract
The cytidine deaminase, Apolipoprotein B mRNA editing enzyme catalytic subunit 3B (APOBEC3B, herein termed A3B), is a critical mutation driver that induces genomic instability in cancer by catalyzing cytosine-to-thymine (C-to-T) conversion and promoting replication stress (RS). However, the detailed function of A3B in RS is not fully determined and it is not known whether the mechanism of A3B action can be exploited for cancer therapy. Here, we conducted an immunoprecipitation-mass spectrometry (IP-MS) study and identified A3B to be a novel binding component of R-loops, which are RNA:DNA hybrid structures. Mechanistically, overexpression of A3B exacerbated RS by promoting R-loop formation and altering the distribution of R-loops in the genome. This was rescued by the R-loop gatekeeper, Ribonuclease H1 (RNASEH1, herein termed RNH1). In addition, a high level of A3B conferred sensitivity to ATR/Chk1 inhibitors (ATRi/Chk1i) in melanoma cells, which was dependent on R-loop status. Together, our results provide novel insights into the mechanistic link between A3B and R-loops in the promotion of RS in cancer. This will inform the development of markers to predict the response of patients to ATRi/Chk1i.

Similar content being viewed by others
Introduction
Apolipoprotein B mRNA editing enzyme catalytic subunit 3B (APOBEC3B, herein termed A3B) belongs to the cytidine deaminase APOBEC3 family, which can cause the accumulation of cytosine-to-thymine (C-to-T) mutations in DNA and induce genome instability [1]. Thus, A3B activation usually leads to a mutator phenotype that is commonly observed in cancers [2, 3]. Genomic uracil lesions catalyzed by A3B are responsible for a large proportion of both dispersed and clustered mutations in breast, lung, cervix, head, neck and bladder cancers [4,5,6]. With defined roles in mutagenesis, A3B is reported to promote both initiation and progression of cancer, and overexpression of A3B is usually associated with poor clinical outcomes of cancer patients [7,8,9], including primary head/neck mucosal melanomas (MMs) [10]. Recently, emerging evidence has demonstrated that A3B activation was positively correlated with elevated levels of DNA RS, pointing to the additional function of A3B in promoting cancer [3, 11, 12]. RS often occurs in response to various intrinsic or extrinsic stimuli, such as chemotherapy drugs, hypoxia, ultraviolet light and other forms of radiation [13, 14]. The replication stress response (RSR) at stalled replication forks is governed by Ataxia telangiectasia and Rad3-related protein (ATR), a key molecule in modulating DNA damage repair [15]. ATR phosphorylates and activates Checkpoint kinase 1 (Chk1), resulting in the amplification of RSR signaling [16]. The RSR signaling cascades subsequently stabilize the stalled replication forks and promote their restart [17]. In contrast to the well-established mutagenic functions, however, the underlying mechanisms through which A3B exerts the modulation of RS have not yet been characterized. It was noticed that A3B-exacerbated RS might be through replication fork slowing and incomplete replication of DNA as suggested by accumulated FANCD2-flanked ultrafine bridges and 53BP1 foci [3]. But it remains elusive whether these phenomena mediated by A3B were entangled with transcriptional apparatus.
R-loops are intermediates of transcription consisting of a complementary RNA:DNA hybrid plus a displaced DNA strand (ssDNA) [18]. They are involved in various biological processes, such as transcription initiation and termination, telomere homeostasis and class-switching recombination [19, 20]. The co-transcriptionally formed R-loop structures could halt DNA replication if they occurred in the opposite direction to replication fork elongation [21,22,23], highlighting the necessity to tightly control the formation and distribution of R-loops. Otherwise, unscheduled R-loops interfere with DNA replication and transcription, resulting in transcription elongation defects and genome instability [19, 20, 24]. Among the regulators of R-loop, RNASEH1 is a gatekeeper which cleaves the RNA moiety of the RNA:DNA hybrid to prevent R-loop formation [25]. A recent study has demonstrated the involvement of fork cleavage by endonuclease MUS81, transcriptional reactivation and fork relegation by LIG4/XRCC4 complex to resolve the stalled replication forks by co-transcriptional R-loops [23]. Unexpectedly, we identified A3B to be a novel interacting component of R-loops through an IP-MS, suggesting the functional connectivity between A3B and R-loop modulation. In this study, we aimed to investigate the mechanisms of A3B in modulating RS through its interaction with R-loops, and to explore the therapeutic opportunities by exploiting these mechanisms.
Results
A3B is overexpressed in melanoma
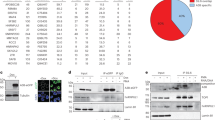
We here used melanoma as a model to elucidate the biological function of A3B. In order to evaluate the expression of A3B, we performed immunohistochemical (IHC) staining in ocular melanoma (n = 67), adjacent normal (n = 5) and benign nevi tissues (n = 14). Concordant with previous reports [10, 26], A3B staining was mainly found in the nuclei. Strong staining was detected in melanoma tissues, while adjacent normal and nevi tissues had only weak or mild immunopositivity (Fig. 1A). Statistical analysis demonstrated that A3B levels were significantly higher in melanoma tissue than in adjacent normal (P = 0.0029) and nevi tissues (P < 0.0001) (Fig. 1B). Similarly, the percentage of A3B-positive cells was also higher in melanoma tissue (Fig. 1C). We next examined the expression of A3B in a variety of melanoma cell lines. Both qPCR and western blotting revealed that the expression of A3B was dramatically elevated in melanoma cell lines with lines OMM2.3 and 92.1 exhibiting the highest levels (Fig. S1A, B). Also, APOBEC-driven C to T mutation is the main single base substitution in Skin cutaneous Melanoma (SKCM) and Uveal Melanoma (UVM) (Fig. S1C).
A Representative IHC staining of A3B in nevus, ocular melanoma intratumor and peritumor tissues. Scale bar, 50 μm. Images on the right are 5-fold magnification of the selected area. Quantification of A3B H-scores B and percentage of A3B positive cells C of the indicated tissues. Mean values are indicated by black lines (unpaired t-test). D Co-IP of R-loop using anti-S9.6 antibody. Endogenous A3B, p-RPA32 and RPA70 were detected by western blot. Cell lysates (1%) were loaded as input. E Schematic representation of full-length A3B (1-382 aa) with two domains named CDD1 (29-138 aa) and CDD2 (210-326 aa) and plasmid CDD1_Flag and CDD2_Flag. CDD, cytidine deaminase domain. F Co-IP of R-loop using anti-S9.6 antibody. 92.1 cells were transfected with Flag-tagged CDD1 or CDD2. Flag was detected by western blot. Cell lysates (1%) were loaded as input. G Volcano plot of differentially expressed R-loop binding proteins before (blue) or after A3B depletion (red) in OMM2.3 cell as determined by IP-MS. Protein targets (p < 0.05) were shown. H Bar graph of gene ontology analysis of the proteins in (G). blue: R-loop binding proteins before A3B depletion; red: R-loop binding proteins after A3B depletion.
A3B interacts with R-loops and regulates the interactome
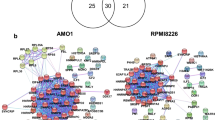
Among various cancer types, including UVM and SKCM, we observed a positive correlation between the expression of A3B and RNASEH1, a well-established nuclease that resolves R-loops [27] (Fig. S2). By analyzing the R-loop interactors from previous reports [28, 29], we noticed the involvement of APOBEC family members such as APOBEC3A (A3A), APOBEC3C (A3C) and APOBEC3D (A3D) in a context-dependent manner. This result prompted us to hypothesize that A3B might also be linked to R-loops in our scenario. We tested this by IP-MS analysis with a monoclonal antibody (S9.6) to investigate R-loop interacting components. R-loops interact with Replication protein A (RPA), a key member of the ssDNA-binding complex responsible for DNA replication and damage repair [30]. As expected, we observed this interaction; the phosphorylated form of RPA32 (p-RPA32 S4/S8) and RPA70 were detected in our IP-MS and IP-western blotting assays in multiple cancer cell lines (Fig. 1D and Fig. S3A, Table S1). Surprisingly, A3B was identified as a novel R-loop binding protein (Fig. 1D and Fig. S3A, B). To test if this interaction was restricted to melanoma, we examined the binding of A3B with R-loop in renal cancer cell lines 786 O and 769 P (Fig. S3A). A3B was a two-domain APOBEC3 protein with two cytidine deaminase domains (CDD) [31]. We further noticed that the N- terminal CDD1, which mediates viral RNA binding, was the R-loop binding domain (Fig. 1E, F). We then sought to explore the impact of A3B upon the R-loop interactome using control (shCtrl) and A3B-depleted (shA3B) OMM2.3 and 92.1 cells (Fig. S3C, D). IP-MS analysis of these cells revealed that 270 binding proteins completely lost their interactions with R-loops in the absence of A3B, including cell cycle-related regulatory proteins Cell division cycle 27 (CDC27) and Aurora kinase B (AURKB) (Fig. 1G and Fig. S3E, F). In addition, 301 proteins exhibited impaired interactions with R-loops after A3B-depletion because their protein amounts were markedly reduced (>2-fold change compared with control), including DNA replication-associated protein minichromosomal maintenance complex component 4 (MCM4). In contrast, 367 proteins gained the ability to interact with R-loops when A3B was depleted (Fig. 1G and Fig. S3E, F). Gene ontology analysis of the A3B-dependent R-loop interactome demonstrated the enrichment of proteins involved in regulating the cell cycle and cellular response to stress (Fig. 1H), consistent with previously established functions of A3B [32]. We further compared the R-loop interactome in OMM2.3 with reported R-loop interactors in MCF10A [29] and HeLa [28] cells and found 254 overlapped R-loop interactors (Fig. S3G). The overlapped interactors are mainly involved in RNA metabolism-related functions (Fig. S3H). In addition to RNA metabolism, OMM2.3 specific R-loop interactors also participate in cell cycle and DNA repair (Fig. S3I). These data indicated that A3B might maintain the R-loop interactome to modulate cell cycle, stress response or gene transcription.
A3B regulates the formation and genomic distribution of R-loops
To explore the regulatory function of A3B on R-loops, we calculated the number of R-loops by immunofluorescence staining using the S9.6 antibody in control and A3B knock-down cells. S9.6 nuclear signals were significantly reduced in A3B-depleted OMM2.3 (P < 0.001) and 92.1 (P < 0.001) cells (Fig. 2A–C). In contrast, A3B overexpression in MEL270 (P < 0.001) and MEL290 (P < 0.001) cells resulted in significant increases in S9.6 staining (Fig. 2D–F). Overexpression of RNASEH1 almost abrogated the formation of R-loops (Fig. 2B, C, E–F and Fig. S4A–F). The catalytically inactive form of RNASEH1, which harbored a D210N mutation in the catalytic domain but retained the binding affinity to R-loops, served as a negative control.
A Representative images of S9.6 staining in OMM2.3 and 92.1 cells in each condition. Scale bar, 4 μm. Quantification of nuclear S9.6 signal intensity of B OMM2.3 and C 92.1 cells in (A). D Representative images of S9.6 staining in MEL290 and MEL270 cells in each condition. Scale bar, 4 μm. Quantification of nuclear S9.6 signal intensity of MEL290 E and MEL270 F cells in (D). Data in (B, C, E, F) are presented as a scatter plot (n ≥ 100). Median values are indicated by black lines (two-tailed Mann–Whitney U-test). AU, arbitrary units.
We then applied the CUT&Tag approach to characterize the genomic distribution of R-loops in control and A3B-depleted OMM2.3 cells with RNASEH1_D210N_V5 expression (Fig. 3A). We used a V5 antibody to pull-down R-loops, and to harvest the DNA fragments from R-loop regions for sequencing. The R-loop peaks were mainly distributed near transcription start sites (TSS) (Fig. 3B, C). Knockdown of A3B markedly reduced the number of R-loop peaks, confirming our findings in Fig. 2A (Fig. 3B, C). Importantly, A3B depletion redistributed the genome-wide positioning of R-loops. The enrichment of R-loops at TSS was diminished and redirected to more diverse regions across the genome (Fig. 3C). Further annotation revealed that R-loops were preferentially localized at the promoter, intronic and distal intercellular regions of the genome (Fig. 3D), correlating with the highest R-loop signal intensity at promoter regions (Fig. S5A, B). This localization preference of R-loops was impaired by A3B knockdown (Fig. 3D).
A Workflow for R-loop mapping with CUT&Tag assay. Catalytically inactive RNASEH1 plasmids with V5 tags were transformed into A3B knockdown and control OMM2.3 cell lines. Schematic diagram of the structure of the tool plasmid ppyCAG_RNASEH1_D210N_V5 is shown on the right. B The heatmap presentation of R-loop signals in regions ± 3 Kb from TSS region. C The peak distribution of CUT&Tag signals for R-loop relative to TSS. The distribution ranges are marked according to the colors shown in the labels on the right. D Annotation of R-loop peaks captured by CUT&Tag assay. Genomic regions are coded on the right. UTR, untranslated region. E GO analysis of differential genes between A3B knock down and control cell line captured by CUT&Tag.
We then characterized the function of genes that showing different R-loop peaks after A3B knockdown. The R-loop peaks of a total of 459 genes were depleted by A3B knockdown, most of which function in stress response and cell cycle processes (Fig. 3E). From this observation, we reasoned that A3B may regulate RSR through stress-related, R-loop containing genes.
A3B coordinates R-loops to regulate the RSR
To further explore the relationship between A3B and R-loop, we performed ChIP-seq analysis of A3B. The profiles of A3B distribution of our study were in agreement with those of previous reports using the breast cancer cell line MCF7 (GSE56979) [33] (Fig. S5C–E). We performed an integrated analysis of R-loop CUT&Tag-seq and A3B ChIP-seq data. A Venn diagram showed that 8,167 (54.7%) out of 14,930 genes co-occurred in A3B and R-loop peak regions (Fig. S5F). It is worth noting that the overall C-to-T mutation rate in R-loop-unique but not R-loop-A3B regions was significantly decreased by A3B perturbation (P < 0.001), reflecting the potential involvement of other factors in the promotion of C-to-T conversion in these co-occurring regions and compensation for the effects on DNA sequence in the absence of A3B (Fig. S5G).
In fact, we found that both A3B and R-loop peaks occurred at the Cell division cycle protein 45 (CDC45) gene, which is important for initiation of DNA replication [34] (Fig. 4A). In addition, an aggregated distribution of A3B was observed at R-loop center locations (Fig. 4B). Among the 465 genes that R-loops peaks were altered by A3B knockdown, 276 of them were also detected with A3B ChIP-seq binding signals (Fig. 4C). Subsequent gene ontology analysis of the overlapped genes indicated involvement in stress response regulation (Fig. 4D), further validating our results in Fig. 3E.
A Genome browser view of R-loop and A3B signals at example loci. B The heatmap presentation of ChIP-seq signals for A3B in regions ±3 Kb from R-loop centers. C Venn diagram shown genome-wide co-occurrence across A3B target genes (blue) and R-loop-prone differential genes on A3B knock down (orange). D GO analysis of overlapping genes in (C). E Heatmap of gene expression of MEL290 RNA-seq. F GSEA pathway enrichment analysis of differential genes obtained by transcriptomic sequencing of stable transfection cell lines of MEL290. NES: normalized enrichment score. G Bar charts of RNASEH1 expression based on RNA-seq of A3B overexpression. H Bar charts of RSR signature score based on RNA-seq of A3B overexpression.
We also conducted transcriptomic sequencing in control and A3B-depleted melanoma cell lines. In 92.1 cells, 183 differentially expressed genes were detected in response to A3B depletion, among which 27 and 156 were up- and down-regulated, respectively. In contrast, A3B overexpression resulted in elevated expression of 379 genes in MEL290 cells. DNA replication and stress pathways were significantly altered by A3B perturbation (Figs 4E, F and Fig. S5H), possibly through a mechanism involving the regulation of RNASEH1 (Fig. 4G). To further assess the influence of RS by A3B, we employed a set of RSR signature genes as reported previously [35]. The RSR signature score was markedly increased by A3B overexpression, confirming exacerbated RS (Fig. 4H).
A3B overexpression confers R-loop dependent sensitivity to ATRi/Chk1i in melanoma cells
We sought to evaluate the functional consequences of A3B on R-loops in melanoma cells. Compared with control cells, the colony area was increased in A3B-depleted cells about 3-fold (Fig. 5A, B and Fig. S6A, B), but significantly reduced in A3B-overexpressing MEL270 (P = 0.0071) and MEL290 (P = 0.0032) cells (Fig. 5C, D). A3B suppression of colony number was substantially attenuated by the overexpression of RNASEH1 (Fig. 5A–D). These results demonstrated the critical role of R-loop status in A3B-mediated melanoma cell survival. Given the function of A3B and R-loops in RSR activation, we next tested their impact on sensitivity to ATRi/Chk1i, which lead to mitotic catastrophe and tumor suppression [36]. The survival of melanoma cells under of ATRi (VE-821/VE-822) or Chk1i (Rabusertib) treatment was significantly improved by A3B depletion, indicating an essential role of A3B in conferring sensitivity to these drugs (Fig. 5E–H). In contrast, overexpression of A3B resulted in significant reduction of melanoma cell survival under the treatment of VE-822 (Fig. S6C). We also tested the effects of BAY-1895344, a potent and selective ATRi that was currently under clinical evaluation [37]. Intriguingly, the A3B-mediated sensitivity to BAY-1895344 appeared to be R-loop-dependent because the addition of RNASEH1 almost abolished these phenotypes (Fig. 5I). Furthermore, the measurement of p-RPA32 (S4/S8) foci in control and A3B-depleted cells showed that the percentage of p-RPA32 positivity was significantly reduced by A3B ablation under ATRi (VE-822) treatment (OMM2.3: P = 0.0107; 92.1: P = 0.0008) (Fig. 5J, K). We also noticed that A3B depletion inhibited the baseline level of p-RPA32 foci (OMM2.3: P = 0.0224; 92.1: P = 0.0091) (Fig. 5J, K). Consistent with the observations in Fig. 5J, K, RNASEH1 substantially inhibited the formation of p-RPA32 foci that was augmented by A3B overexpression (Fig. 5L, M). However, it was unclear whether the A3B-mediated drug sensitivity was specific to ATRi/Chk1i [38, 39]. To address this, we tested a variety of conventional chemotherapy drugs for melanoma treatment, including cisplatin, carboplatin and dacarbazine. We found that the sensitivities to these selected drugs remained unchanged in control and A3B-depleted melanoma cells (Fig. S6D–S6K). These results demonstrated that, at least in our scenario, the A3B-mediated drug sensitivity was exclusive to ATRi/Chk1i.
A Images of representative colony formation assays of OMM2.3 cells in each condition. B Bar chart displayed the quantification of colony formation assay of (A) (n = 3 biological replicates, mean + SEM, unpaired t-test). FC, fold change. C Images of representative colony formation assays of MEL270 and MEL290 with or without RNASEH1 and/or A3B expression. D Bar chart displayed the quantification of colony formation assay of MEL270 and MEL290 with or without RNASEH1 and A3B expression (n = 3 biological replicates, mean + SEM, unpaired t-test). E Images of representative colony formation assays of OMM2.3 cell lines treated with ATRi VE-821. F Survival rate of OMM2.3 cell lines treated with VE-821 (n = 4 biological replicates, mean + SEM, unpaired t-test). G Images of representative colony formation assays of OMM2.3 cell lines treated with Chik1i rabusertib. H Survival rate of OMM2.3 cell lines treated with rabusertib (n = 4 biological replicates, mean + SEM, unpaired t-test). I Survival rate of MEL290 cell lines represented by cell viability treated with 2.5 nM BAY-1895344 for 72 h (n = 3 biological replicates, mean + SEM, unpaired t-test). J A3B knockdown and control cells were treated with 2 μM ATRi VE-822 or DMSO for 24 h. Representative images of each condition were shown. Scale bar, 20 μm. K Percentage of cells with more than 10 p-RPA32 foci in (J) was analyzed (n = 3 biological replicates, mean + SEM, unpaired t-test). L MEL290 cells with A3B and/or RNASEH1 overexpression were treated with 2 μM VE-822 for 24 h. Representative images of each cell lines were shown. Scale bar, 10 μm. M Percentage of cells with more than 10 p-RPA32 foci in (L) was analyzed (n = 3 biological replicates, mean + SEM, unpaired t-test).
The association between A3B and R-loops predicts levels of RSR and patient survival
To explore the clinical relevance of A3B for RSR, we measured RSR signature scores using The Cancer Genome Atlas (TCGA) datasets. Pearson correlation analysis revealed that A3B levels were positively correlated with RSR signature scores in both UVM and prostate adenocarcinoma (PRAD) datasets (Fig. 6A, B). Similar correlations were found in various other cancer types (Fig. 6C). Studies in tumour-adjacent (TCGA_Normal) and normal tissues (GTEx) also exhibited a positive correlation between A3B and RSR, but to a lesser extent (Fig. 6C, D, Table S2), indicating that the association of A3B with RS was not limited to melanoma. In addition, the expression of A3B appeared to be a negative prognostic marker for the prediction of patient survival in a variety of cancer types (Fig. 6E, F, Table S3). The contribution of R-loops to A3B-mediated survival correlation was further assessed by stratifying patients into four groups based on A3B and RNASEH1 levels. The group of patients with low A3B but high RNASEH1 levels exhibited the longest survival durations compared with the other three groups (Fig. 6G, H, Table S4). This indicates that higher R-loop levels are associated with poorer prognosis in UVM and PRAD patients when A3B expression is low. Accordingly, the calculation of RSR signature scores for the four patient clusters demonstrated that the A3B-high but RNASEH1-low group produced the highest RSR score, in contrast to the lowest score produced by the A3B-low but RNASEH1-high group (Fig. 6I, J, Table S5). To further investigate the roles of A3B and R-loops in ATRi/Chk1i sensitivity, we analyzed two sets of data with ChK1i responsiveness (GSE149724 and GSE143152). To our expect, we observed an expression pattern of high A3B but low RNASEH1 in prexasertib (Chk1i) sensitive ovarian cancer cell lines, in contrast to that of prexasertib-resistant cell lines (Fig. 6K). These findings further supported the notion that A3B and R-loop levels were associated with the strength of RSR and could be used to predict patients’ responses to ATRi/Chk1i.
Pearson correlation of A3B expression and RSR signature score in A UVM and B PRAD patients. Pearson correlation analysis between A3B and RS in C TCGA Cohort and D GTEx Cohort. E Overall survival and F disease-free survival of A3B were examined in TCGA Cohort. Hazard Ratio were calculated based on Cox proportional hazards model. Those with HR > 1 are marked in red, those with HR < 1 are marked in blue. Kaplan-Meier curve of overall survival in G UVM and H PRAD patients with A3B high/low and RNASEH1 high/low expression (log rank test). RSR signature score in I UVM and J PRAD patients with A3B high/low and RNASEH1 high/low expression. Mean values are indicated by black lines (unpaired t-test). K A3B and RNASEH1 expression of wildtype (WT) and Chk1i resistant cell lines in GSE149724 and GSE143152 datasets.
Discussion
A number of R-loop regulators are related to DNA damage modulation, including RPA [30], BRG1 [40] and BRCA2 [41]. Here, we report the discovery of A3B as a novel binding partner and regulator of R-loops, linking DNA editing family members to the modulation of R-loop. C-to-T mutagenesis is a critical feature of A3B function in cancer, and the kataegic mutational signature attributed to A3B is commonly observed [4, 6]. A3B mutagenesis occurs on the lagging-strand template during DNA replication or at stalled replication forks [42, 43]. However, it is unclear whether and how this mutagenic activity is involved in A3B-mediated RS. We found that A3B interacted with R-loops and was enriched in the proximity of R-loop regions. We, therefore, speculated that such recruitment might affect the C-to-T mutation rate at R-loop-A3B loci. Indeed, A3B depletion led to marked reduction of C-to-T ratio across R-loop loci, in agreement with the active role of A3B to catalyze C-to-T conversion at R-loop regions. Intriguingly, the C-to-T ratio at R-loop-A3B co-occurrent loci was largely retained when A3B was depleted compared with other regions, suggesting the involvement of other C-to-T mutagenic factors to complement the role of A3B at the co-occurrent regions. It seemed that A3B was more likely to function as a “road blocker” instead of “mutator” at R-loop loci to orchestrate the expression of neighboring genes. Though the mechanistic details were unknown, the recruitment of other binding components to R-loop may be involved as indicated by our IP-MS analysis.
Mounting evidence demonstrates that A3B is clinically significant in cancer. A3B overexpression was shown to confer resistance to drugs such as doxorubicin and tamoxifen [44, 45]. But on the other hand, A3B may enhance the sensitivity to immunotherapies. Studies have shown that patients with high levels of A3B are more likely to respond to immune checkpoint blockade [9, 46]. These apparently paradoxical observations highlight the fact that the function of A3B is fine-tuned in different contexts, and A3B alone is not sufficient to predict the overall benefits to patients. Perturbation of the ATR-Chk1 pathway induces deleterious consequences, thus can be exploited to treat cancer in which RS levels are always augmented because of oncogene hyperactivation [47, 48]. Accordingly, ATRi or Chk1i exhibit promising antitumor functions in pre-clinical models and are currently under evaluation in clinical trials [37, 49,50,51]. Our data revealed that both RS and sensitivity to ATRi elicited by A3B was dependent on R-loop formation because these effects could be largely alleviated by overexpression of RNASEH1. In this regard, A3B together with R-loop status could better define the level of RS and ATRi/ChK1i sensitivity. Recently, CRISPR-based screening has revealed several potential biomarkers, including ATM, ARID1A, POLE3/4, CDK8 and SMG1 [49, 52,53,54,55]. Most of them are related to DNA damage and cell cycle functions that ATR is primarily involved in. From the point of RS regulation, our results demonstrated that the combined expression pattern of A3B and RNASEH1 (A3B-high RNASEH1-low) instead of A3B alone may represent a robust metric to predict the therapeutic benefits to ATRi. And the vulnerability to ATRi may not be limited to melanoma but may also be applicable to other cancer types.
Taken together, we have shown mechanistic insights into A3B-coordinated R-loop, which exerts RS-promoting activity in cancer. ATRi sensitivity elicited by A3B expression and R-loop status may be exploited to guide ATRi treatment of cancer patients.
Materials and methods
Patient samples
A total of 67 human ocular melanoma tissues, 14 human nevi tissues and 5 adjacent normal tissues were collected for IHC from Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine from 2007 to 2017. Patients’ clinical and demographic features are detailed in Table S6.
Immunohistochemistry (IHC)
IHC staining of tissue slides was performed by Servicebio (Wuhan, China). Tissues were deparaffinized and rehydrated through an alcohol series, followed by antigen retrieval with sodium citrate buffer. Then tissue sections were blocked with 3% bovine serum albumin (BSA) 30 min at room temperature and then incubated with anti-A3B (abcam, Cambridge, England, ab184990, 1:100) antibody at 4 °C overnight. Finally, the tissues were covered with horseradish peroxidase (HRP) labeled secondary antibody and incubated at room temperature for 50 minutes. All immunostained slides were scanned on 3D Histech Quant Center (3D Histech, Hungary), and computerized image analysis was performed by Halo (Indica labs, USA). Immunostaining for A3B was analyzed in adjacent normal tissues, nevi tissues and melanoma tissues using percentage of positive cells and histochemistry score (H-score). H-score was determined based on the proportion of positive cells and the intensity of nuclear staining as previously described [56]. H-score = ∑(pi×i)= (percentage of cells of weak intensity ×1) + (percentage of cells of moderate intensity ×2)+ (percentage of cells of strong intensity ×3). In the formula, pi represents the percentage of positive cells in the slide; i represents the intensity of A3B staining.
Cell culture
The adult retinal pigment epithelium cell line ARPE-19 was obtained from the Cell Bank/Stem Cell Bank (Chinese Academy of Sciences). Human cutaneous melanocyte cell line PIG1 was obtained from the Department of Ophthalmology, Peking University Third Hospital. The human UVM cell lines OMM1, OMM2.3, MEL285 and MEL290 and conjunctiva melanoma cell lines CRMM1, CRMM2, and CM2005.1 were kind gifts from Prof. Martine J. Jager (Leiden University Medical Center, Leiden, The Netherlands). The human UVM cell line MUM2B and 92.1 were kindly supplied by Prof. John F. Marshall (Tumor Biology Laboratory, Cancer Research UK Clinical Center, John Vane Science Centre, London, UK). HEK293T human embryonic kidney cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). The cell lines used in this study were authenticated by short tandem repeat (STR) profiling. Human HEK293T, A375 and A2058 cells was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Massachusetts, USA). Other cells were cultured in RPMI1640 medium (Gibco). All mediums are supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin (P/S; Thermo Fisher Scientific). All cells were cultured in a 37 °C humidified incubator containing 5% CO2.
RNA extraction and quantification
RNA purification was performed using EZ-press RNA purification kit (EZBioscience, CA, USA, B0004DP) according to the manufacturer’s guidelines. Then, cDNA synthesis was achieved using the PrimeScript RT Master Mix (Takara, Kyoto, Japan). Finally, real-time quantitative PCR (RT-qPCR) was carried out using the SYBR Green qPCR Master Mix (Applied Biosystems). mRNA expression values were calculated using the ΔΔCt method and GAPDH gene as a control. A detailed list with primers used in the present study is provided in Table S7. CT values were shown in Supplementary Material 1.
Western blotting
Total cell lysates were prepared in RIPA lysis buffer (Beyotime, Shanghai, China, P0013B). Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). After blocking with 5% non-fat milk for 1 h at room temperature, the membranes were incubated with anti-p-RPA32 (BETHYL, TX, USA, A300-245A, 1:2000), anti-RPA70 (CST, MA, USA, 2267 S, 1:1000), anti-Flag (ABclonal, Wuhan, China, AE063, 1:1000), anti-A3B (Abcam, ab184990, 1:1,000), anti-GFP (Invitrogen, CA, USA, A-11122, 1:1000), anti-Myc (santa cruz, TX, USA, sc-40, 1:1000) or anti-V5 (Abcam, ab15828, 1:2000) antibodies overnight at 4 °C and then with the appropriate secondary antibodies conjugated to a fluorescent tag (Invitrogen) for 1 h at room temperature. Anti-β-Actin (Proteintech, IL, USA, 66009-1-Ig, 1:20000) antibody served as the loading control. The immunoblots were recorded with the Odyssey infrared imaging system (LI-COR Biosciences, NE, USA). Full and uncropped western blots were shown in Supplementary Material 2.
Co-immunoprecipitation (co-IP)
Cells were pelleted and washed with PBS, and then lysates were prepared in 500 μL lysis buffer containing 120 mM NaCl, 20 mM Tris-Cl, 2 mM EDTA, 1% NP40 and 5% Glycerol supplemented with 1× protease inhibitor cocktail (Roche, NJ, USA). Anti-S9.6 antibody (Kerafast, MA, USA, ENH001, 1:100) or normal mouse IgG (Santa cruz, sc-2025, 1:10000) was incubated with the cell lysates overnight at 4 °C, after which 30 μL protein A magnetic beads (CST, #73778) were added and incubated for 2 additional hours. Then, the magnetic beads were washed three times with lysis buffer. For IP-MS analysis, 100 μL glycine solution was used for each tube to elute the protein complexes from the beads, while 1 × SDS loading buffer (NCM, Suzhou, China) was used per sample for SDS-PAGE analysis.
Transfection and virus packaging
Three shRNA sequences targeting A3B were cloned into the pGIPZ-TurboGFP-puro vector. The A3B targeting sequences were: 5’-TAAAGTTGAAAGTGAATGTGTT-3’(shA3B_1); 5’-TTAAAGTTGAAAGTGAATGTGG-3’ (shA3B_2). The full length of human A3B (isoform NM_004900.5), Myc_RNASEH1_EGFP (isoform NM_002936.3) and CDD1/2_Flag were cloned into the pHAGE-puro vector, respectively for overexpression of A3B and RNASEH1. Primers used for cloning are shown in Table S8. PolyJet DNA In Vitro Transfection Reagent (SignaGen, MD, USA) was used for plasmid transfection following the manufacturer’s instructions. After lentiviral packaging with HEK293T cells, cells were infected with lentiviruses and selected by incubation with 2 μg/mL puromycin for 3 days. For transfection of RNASEH1 D210N mutant construct, cells were transfected with ppyCAG_RNASEH1_D210N_V5 plasmid (Addgene, MA, USA, 111904) and selected with 300 μg/mL Hygromycin B (Selleck, TX, USA, S2908) for 3 days.
Colony formation
A volume of 2 mL of complete medium containing 1000 cells was seeded in each well of a 12-well plate and incubated for about 2 weeks. For drug sensitivity colony formation assay, cells were treated with the indicated drug concentrations every 48 h. Once colonies formed in DMSO control conditions, plates were fixed with 4% paraformaldehyde, stained with crystal violet and the number of colonies was counted. Colony formation efficiency indicated by area of colonies treated relative to area of control colonies was used to measure survival between wells. RSR inhibitors and chemotherapy drugs for drug sensitivity assay were purchased from Selleck: VE-821 (S8007) targeting ATR, VE-822 (S7102) highly selective and potent derivatives of VE-821, Rabusertib (S2626) targeting Chk1, Cisplatin (S1166), Carboplatin (S1215), Dacarbazine (S1221). The quantifications presented were obtained from at least three independent biological replicates.
Cell viability
To determine cell viability, MEL290 cells were seeded in 96-well plates at a density of 1000 cells per well and treated with 2.5 nM BAY-1895344 (Selleck, S8666) or 2 μM VE-822. After incubation with 10 μL CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) per well, the absorbance was measured at a wavelength of 450 nm at the indicated time points. The data were recorded and analyzed. The quantifications presented were obtained from three independent biological replicates.
Immunofluorescence (IF)
S9.6 IF was performed essentially as described [57]. Briefly, cells were fixed with 100% ice-cold methanol, blocked with 2% BSA for 1 h at room temperature and incubated with S9.6 (Kerafast, ENH001; 1:200) overnight at 4 °C. Then coverslips were washed three times in PBS and incubated with Alexa Fluor 594 secondary antibody (Invitrogen, #A21203; 1:1000) for 1 h at room temperature. Finally, cells were stained with DAPI and mounted in ProLong Gold AntiFade reagent (Invitrogen). For DNA RS assessment by p-RPA32 (S4/S8) immunostaining, cells adhering to a glass slide were fixed with 4% paraformaldehyde (Biosharp, Beijing, China, 70071800) for 15 min, permeabilized with 0.1% Triton X-100 for 15 min and then blocked with 2% BSA solution for 1 h at room temperature. After incubation with primary antibody against p-RPA32 (S4/S8) (BETHYL, A300-245A, 1:2000) overnight, the cells were washed three times with PBS and subsequently incubated with Alexa Fluor 594 secondary antibody (Invitrogen, A11012, 1:1000) for 1 h. The coverslips were then mounted with ProLong Gold mounting medium with DAPI (Invitrogen, P36931) and observed under an inverted fluorescence microscope (Nikon, Tokyo, Japan). Images were acquired with Leica TCS SP8 confocal microscope (Leica, Mannheim, Germany). Nuclear intensity was quantified using FIJI (ImageJ) image processing package. The nuclear mean gray value for S9.6 (>100 cells) was measured for each condition. Percentage of cells with more than 10 p-RPA32 foci was quantified. The results presented were obtained from three independent biological replicates; at least 100 cells were measured per replicate.
RNA-seq
RNA sequencing was performed by Novogene (Beijing, China). Total RNA was harvested from MEL290 cells overexpressing A3B and/or RNASEH1 and 92.1 cells transfected with A3B knockdown or empty vector using Trizol. The integrity of the RNA was assessed by Agilent 2100 bioanalyzer (Thermo Fisher Scientific). Approximately 1 μg mRNA from each sample was used for RNA sequencing (Illumina HiSeq PE150 platform). Short reads were aligned to the reference human genome GRCh38 using STAR [58]. Gene expression in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM) of genes overlapping with gene annotations in Ensembl release 75 was calculated by Cufflinks [59]. Differential gene expression was determined using the Cufflinks Cuffdiff package [59]. Gene Set Enrichment Analysis (GSEA) was performed to interpret the function of regulated genes using the cancer hallmark gene sets, canonical pathways gene sets and GO biological process gene sets.
ChIP-seq
ChIP assay was performed as previously described, with minor modifications [60]. Briefly, OMM2.3 and 92.1 (1 × 107 cells) were cross-linked for 10 min at room temperature with 1% formaldehyde-containing medium. The reaction was then halted using a 1× glycine solution. Then cells were pelleted before resuspension in sonication buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA and 1 × protease inhibitor cocktail). The cellular chromatin was then sonicated to lengths between 100 and 600 bp. 10% chromatin for each ChIP reaction was kept as input DNA. The rest chromatin was then incubated with anti-A3B antibody (abcam, ab184990, 1:50) or normal rabbit IgG (CST, 2729, 1:500) at 4 °C overnight. 30 μL magnetic protein A/G beads (CST) were then added for another 1 h of incubation at 4 °C before going through a series of salt washes. Chromatin was then eluted from the magnetic beads in the elution buffer at 65 °C for 15 minutes while vortexing. The supernatant was removed and treated with RNase A followed by Proteinase K. ChIP DNA was then purified using MinElute PCR Purification Kit (QIAGEN, Dusseldorf, Germany, 28006). The ChIP-seq libraries were constructed by Novogene. Pair-end sequencing of the sample was performed on the Illumina platform (Illumina, CA, USA). Library quality was assessed on the Agilent Bioanalyzer 2100 system. Raw data of fastq format was processed using FastQC 0.11.9 software [61]. Clean reads were aligned to the reference genome GRCh38 using Bowtie2 2.2.5 [62] to generate bam files. After removing PCR duplicates with Sambamba 0.8.2 software [63], we used MACS2 2.2.7.1 to identify regions of IP enrichment over the background [64]. A p-value threshold of 0.05 was used for all data sets. Deeptools 3.5.1 was used to transfer deduplicated bam file to BigWig format and to generate heatmaps and density plot figures [65]. ChIPseeker package and ChIPpeakAnno package were used for peak annotation and visualization [66, 67].
CUT&Tag
CUT&Tag assay was performed using Hyperactive In-Situ ChIP Library Prep Kit for Illumina (Vazyme, Nanjing, China, TD901-TD902) following the manufacturer’s protocol. Briefly, 5 × 105 OMM2.3 cells transfected with V5 tagged RNASEH1_D210N were collected. After binding to concanavalin A–coated magnetic beads (ConA beads), bead-bound cells were incubated with anti-V5 antibody (Abcam, ab15828, 5ug/1:10) or normal rabbit IgG (CST, 2729, 0.5ug/1:100) for 2 h at room temperature. After brief wash with dig-wash buffer, cells were then incubated with goat antirabbit secondary antibody (Abcam, ab6702, 1:100) for 1 h at room temperature. When antibody binding procedures were finished, the bead-bound cells were then mixed with hyperactive pA-Tn5 transposon and tagmentated with tagmentation buffer. Tagmentated DNA was then extracted and amplified to form the sequencing-ready libraries. After the PCR reaction, libraries were purified with the DNA clean beads (Vazyme, N411) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina). The library preparations were sequenced on the Illumina Novaseq platform at Novogene and 150 bp paired-end reads were generated. Raw data was first processed using FastQC 0.11.9. Clean data was obtained by removing adapters using Cutadapt 4.1 with default parameters [68]. Paired-end reads were aligned to genome GRCh38 using Bowtie2 2.2.5 as follows: bowtie2 -local -very-sensitive -no-mixed -no-discordant -phred33 -I 10 -X 700. After removing the duplicated reads with Picard 2.27.3 (https://broadinstitute.github.io/picard/), we calculated the read coverage and depth with Samtools 1.15.1 [69]. To ensure that our data were of high quality and reproducibility, we filtered data with minQualityScoreå 2 using Samtools 1.15.1. Then we filtered and kept the mapped read pairs. Bedtools was used to convert files into bed file format and calculate reads coverage at the genome level. Peak calling was performed with SEACR 1.3 [70]. EdgeR package was used to get differential peaks [71]. BigWig file and reads distributions across genes were acquired using Deeptools 3.5.1. Peak annotation was performed with ChIPseeker and ChIPpeakAnno package as previously described. Gene ontology enrichment analysis was implemented by Metascape (www.metascape.org). In Venn diagrams, numbers represent genes co-occurring between conditions.
TCGA data analysis
Two TCGA data sets (PRAD and UVM) were used to test the correlation between the selected genes and patient survival. Based on the expression levels of A3B and RNASEH1 and their median expression values, respectively, patients were further discretized as four groups: “A3B High and RNASEH1 High”, “A3B High and RNASEH1 Low”, “A3B Low and RNASEH1 High” and “A3B Low and RNASEH1 Low”. The statistical analysis was performed by the R package “survival” and survival curves were fitted by the survfit function. RSR signature score was calculated based on the median expression levels of the RSR signature gene sets (Table S9) [35]. The somatic mutation signatures of 80 UVM and 472 SKCM patients from TCGA were deciphered using the R package SomaticSignatures [72].
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, CA, USA). Descriptive values are presented as mean + standard error of the mean (SEM) unless stated elsewhere. Differences between two groups were analyzed by unpaired Student’s t-test while differences among multiple groups were analyzed by one-way analysis of variance (ANOVA) with post-hoc intergroup comparisons with Turkey’s test. When data did not meet the normal distribution, the Mann–Whitney U-test was performed. Test details were indicated in the figure legends. Results were considered statistically significant when p < 0.05. Pearson correlation analysis was performed on GEPIA (http://gepia.cancer-pku.cn/).
Data availability
The data that support the findings of this study are available in Gene Expression Omnibus (GEO) with accession number GSE218751, GSE218753 and GSE218754.
References
Petljak M, Dananberg A, Chu K, Bergstrom EN, Striepen J, von Morgen P, et al. Mechanisms of APOBEC3 mutagenesis in human cancer cells. Nature. 2022;607:799–807.
Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci USA. 2015;112:2841–6.
Venkatesan S, Angelova M, Puttick C, Zhai H, Caswell DR, Lu WT, et al. Induction of APOBEC3 Exacerbates DNA Replication Stress and Chromosomal Instability in Early Breast and Lung Cancer Evolution. Cancer Disc. 2021;11:2456–73.
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93.
Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Tsuboi M, Yamane A, Horiguchi J, Yokobori T, Kawabata-Iwakawa R, Yoshiyama S, et al. APOBEC3B high expression status is associated with aggressive phenotype in Japanese breast cancers. Breast Cancer. 2016;23:780–8.
Serebrenik AA, Argyris PP, Jarvis MC, Brown WL, Bazzaro M, Vogel RI, et al. The DNA Cytosine Deaminase APOBEC3B is a Molecular Determinant of Platinum Responsiveness in Clear Cell Ovarian Cancer. Clin Cancer Res. 2020;26:3397–407.
Xia S, Gu Y, Zhang H, Fei Y, Cao Y, Fang H, et al. Immune inactivation by APOBEC3B enrichment predicts response to chemotherapy and survival in gastric cancer. Oncoimmunology. 2021;10:1975386.
Argyris PP, Naumann J, Jarvis MC, Wilkinson PE, Ho DP, Islam MN, et al. Primary mucosal melanomas of the head and neck are characterised by overexpression of the DNA mutating enzyme APOBEC3B. Histopathology. 2023;82:608–21.
Nikkila J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, et al. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br J Cancer. 2017;117:113–23.
Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016;17:185.
Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat Rev Cancer. 2018;18:586–95.
Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27.
Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66:801–17.
Simoneau A, Zou L. An extending ATR-CHK1 circuitry: the replication stress response and beyond. Curr Opin Genet Dev. 2021;71:92–8.
Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9.
Petermann E, Lan L, Zou L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat Rev Mol Cell Biol. 2022;23:521–40.
Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97.
García-Muse T, Aguilera A. R Loops: From Physiological to Pathological Roles. Cell. 2019;179:604–18.
Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:774–86.e19.
Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, et al. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell. 2017;170:787–99.e18.
Chappidi N, Nascakova Z, Boleslavska B, Zellweger R, Isik E, Andrs M, et al. Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-transcriptional R-Loops. Mol cell. 2020;77:528–41.e8.
Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24.
Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–88.
Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–72.
Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol. 2020;21:167–78.
Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018;23:1891–905.
McCann JL, Cristini A, Law EK, Lee SY, Tellier M, Carpenter MA, et al. R-loop homeostasis and cancer mutagenesis promoted by the DNA cytosine deaminase APOBEC3B. bioRxiv. 2021:2021.08.30.458235. https://doi.org/10.1101/2021.08.30.458235.
Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell. 2017;65:832–47.e4.
Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–31.
Hirabayashi S, Shirakawa K, Horisawa Y, Matsumoto T, Matsui H, Yamazaki H, et al. APOBEC3B is preferentially expressed at the G2/M phase of cell cycle. Biochem Biophys Res Commun. 2021;546:178–84.
Periyasamy M, Patel H, Lai CF, Nguyen VTM, Nevedomskaya E, Harrod A, et al. APOBEC3B-Mediated Cytidine Deamination Is Required for Estrogen Receptor Action in Breast Cancer. Cell Rep. 2015;13:108–21.
Szambowska A, Tessmer I, Prus P, Schlott B, Pospiech H, Grosse F. Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res. 2017;45:3217–30.
Dreyer SB, Upstill-Goddard R, Paulus-Hock V, Paris C, Lampraki EM, Dray E, et al. Targeting DNA Damage Response and Replication Stress in Pancreatic Cancer. Gastroenterology. 2021;160:362–77.e13.
Forment JV, O’Connor MJ. Targeting the replication stress response in cancer. Pharmacol Therap. 2018;188:155–67.
Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-Human Trial of the Oral Ataxia Telangiectasia and RAD3-Related (ATR) Inhibitor BAY 1895344 in Patients with Advanced Solid Tumors. Cancer Discovery. 2021;11:80–91.
Bader SB, Ma TS, Simpson CJ, Liang J, Maezono SEB, Olcina MM, et al. Replication catastrophe induced by cyclic hypoxia leads to increased APOBEC3B activity. Nucleic Acids Res. 2021;49:7492–506.
Buisson R, Lawrence MS, Benes CH, Zou L. APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition. Cancer Res. 2017;77:4567–78.
Bayona-Feliu A, Barroso S, Muñoz S, Aguilera A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat Genet. 2021;53:1050–63.
D’Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat Commun. 2018;9:5376.
Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–49.
Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–35.
Zhang X, Wu Z, Hao Y, Yu T, Li X, Liang Y, et al. Aberrantly Activated APOBEC3B Is Associated With Mutant p53-Driven Refractory/Relapsed Diffuse Large B-Cell Lymphoma. Front Immunol. 2022;13:888250.
Law E, Sieuwerts A, LaPara K, Leonard B, Starrett G, Molan A, et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci Adv. 2016;2:e1601737.
DiMarco AV, Qin X, McKinney BJ, Garcia NMG, Van Alsten SC, Mendes EA, et al. APOBEC Mutagenesis Inhibits Breast Cancer Growth through Induction of T cell-Mediated Antitumor Immune Responses. Cancer Immunol Res. 2022;10:70–86.
Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40:109–17.
Kotsantis P, Petermann E, Boulton SJ. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Disc. 2018;8:537–55.
Yap TA, O’Carrigan B, Penney MS, Lim JS, Brown JS, de Miguel Luken MJ, et al. Phase I Trial of First-in-Class ATR Inhibitor M6620 (VX-970) as Monotherapy or in Combination With Carboplatin in Patients With Advanced Solid Tumors. J Clin Oncol. 2020;38:3195–204.
Banerjee S, Stewart J, Porta N, Toms C, Leary A, Lheureux S, et al. ATARI trial: ATR inhibitor in combination with olaparib in gynecological cancers with ARID1A loss or no loss (ENGOT/GYN1/NCRI). International journal of gynecological cancer: official journal of the International Gynecological Cancer. Society. 2021;31:1471–5.
Roulston A, Zimmermann M, Papp R, Skeldon A, Pellerin C, Dumas-Berube E, et al. RP-3500: A Novel, Potent, and Selective ATR Inhibitor that is Effective in Preclinical Models as a Monotherapy and in Combination with PARP Inhibitors. Mol Cancer Therap. 2022;21:245–56.
Rafiei S, Fitzpatrick K, Liu D, Cai MY, Elmarakeby HA, Park J, et al. ATM Loss Confers Greater Sensitivity to ATR Inhibition Than PARP Inhibition in Prostate Cancer. Cancer Res. 2020;80:2094–100.
Lloyd RL, Urban V, Munoz-Martinez F, Ayestaran I, Thomas JC, de Renty C, et al. Loss of Cyclin C or CDK8 provides ATR inhibitor resistance by suppressing transcription-associated replication stress. Nucleic acids Res. 2021;49:8665–83.
Llorca-Cardenosa MJ, Aronson LI, Krastev DB, Nieminuszczy J, Alexander J, Song F. et al. SMG8/SMG9 Heterodimer Loss Modulates SMG1 Kinase to Drive ATR Inhibitor Resistance. Cancer Res. 2022;82:3962–73.
Hustedt N, Alvarez-Quilon A, McEwan A, Yuan JY, Cho T, Koob L, et al. A consensus set of genetic vulnerabilities to ATR inhibition. Open biology. 2019;9:190156.
Dogan S, Vasudevaraja V, Xu B, Serrano J, Ptashkin RN, Jung HJ, et al. DNA methylation-based classification of sinonasal undifferentiated carcinoma. Mod Pathol. 2019;32:1447–59.
García-Rubio M, Barroso SI, Aguilera A. Detection of DNA-RNA Hybrids In Vivo. Methods Mol Biol. 2018;1672:347–61.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Zhao C, Deng Y, Liu L, Yu K, Zhang L, Wang H, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. 2016;7:10883.
de Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 2019;8:1874.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–4.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinform. 2015;31:2382–3.
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010;11:237.
Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J Comput Biol. 2017;24:1138–43.
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008.
Meers MP, Tenenbaum D, Henikoff S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;12:42.
Chen Y, Lun AT, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438.
Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–5.
Acknowledgements
We thank Dr. Guang Peng (Department of Clinical Cancer Prevention, The University of Texas, M.D. Anderson Cancer Center) for discussion. This work was supported by General Program of National Natural Science Foundation of China (No. 81972667 to J.S., No. 81870187 to W.W.), National Key R&D Program of China (2021YFC2701103 to J.S., 2021YFF0703802 to W.W.), the Major Science and Technology R&D Project of the Science and Technology Department of Jiangxi Province (20213AAG01013 to W.W.), The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. TP2018046 to J.S.), Shanghai Municipal Education Commission-Two Hundred Talent (No. 20191817 to J.S.), Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZDCX20210902 to R.J.) and the Center for High Performance Computing at Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Contributions
CZ and JH performed the experiments, analyzed the results and wrote the manuscript. QL, YW and GZ assisted with the generation of expression constructs and stable cell lines. RH, XL, JY, and DC assisted with drug sensitivity assay, foci staining and imaging analysis. ZZ and LG analyzed the bioinformatic data and provide project consultation. RJ and WW provided research materials and project consultation. JS designed and supervised the experiments, analyzed the results and wrote the manuscript. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
The study was performed in accordance with World Medical Association Declaration of Helsinki and was approved by the Ethics Committee of Shanghai Jiao Tong University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Angelo Peschiaroli
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zong, C., Zhang, Z., Gao, L. et al. APOBEC3B coordinates R-loop to promote replication stress and sensitize cancer cells to ATR/Chk1 inhibitors. Cell Death Dis 14, 348 (2023). https://doi.org/10.1038/s41419-023-05867-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-05867-0
This article is cited by
-
Unscheduled excessive R-loops in immune response
Functional & Integrative Genomics (2024)