Abstract
Despite past extensive studies, the pathoetiologies underlying tumor metastasis remain poorly understood, which renders its treatment largely unsuccessful. The methyl-CpG-binding domain 2 (MBD2), a “reader” to interpret DNA methylome-encoded information, has been noted to be involved in the development of certain types of tumors, while its exact impact on tumor metastasis remains elusive. Herein we demonstrated that patients with LUAD metastasis were highly correlated with enhanced MBD2 expression. Therefore, knockdown of MBD2 significantly attenuated the migration and invasion of LUAD cells (A549 and H1975 cell lines) coupled with attenuated epithelial–mesenchymal transition (EMT). Moreover, similar results were observed in other types of tumor cells (B16F10). Mechanistically, MBD2 selectively bound to the methylated CpG DNA within the DDB2 promoter, by which MBD2 repressed DDB2 expression to promote tumor metastasis. As a result, administration of MBD2 siRNA-loaded liposomes remarkably suppressed EMT along with attenuated tumor metastasis in the B16F10 tumor-bearing mice. Collectively, our study indicates that MBD2 could be a promising prognostic marker for tumor metastasis, while administration of MBD2 siRNA-loaded liposomes could be a viable therapeutic approach against tumor metastasis in clinical settings.
Similar content being viewed by others
Introduction
Tumor metastasis is featured by the dissemination of tumor cells to new sites, leading to the formation of secondary tumors, which has been under intensive investigations for preventing distant tumor spread and related mortality [1]. During the course of metastasis, malignant cells firstly become migratory and invasive to spread them from the primary tumor, by which they get into the circulation via blood vessel or lymphatic intravasation [2]. Approximately 0.2% of the circulating tumor cells (CTCs) could evade immune surveillance and infiltrate into the vascular system to initiate distant metastatic colonization. However, the disseminated tumor cells require a new microenvironment in favor of the establishment of a metastatic supportive niche at distant sites, where they convert from the migratory mode to the proliferative mode, thereby inducing the large-scale spread [3].
Accumulating evidence suggests that epithelial–mesenchymal transition (EMT) plays a central role in tumor metastasis [4]. EMT is an important morphological change that entails the transformation of epithelial cells into an elongated fibroblastic phenotype. The process is characterized by the downregulation of epithelial markers (e.g., E-cadherin) and upregulation of mesenchymal markers (e.g., N-cadherin and Vimentin) following the stimulation of specific growth factors, in which the transforming growth factor beta1 (TGF-β1) is the prototype [5, 6]. Cancer cells undergone EMT display increased motility and invasiveness to promote their dissemination to the remote spots [6]. Indeed, TGF-β1 activates its downstream signaling pathways and several transcription factors that are essential for EMT process [7]. Therefore, targeting TGF-β1 has been proved to be an efficient therapeutic strategy for repressing tumor metastasis [8]. However, the adverse effects are even worse than the therapeutic impacts, which limited its clinical application.
DNA methylation is a chemical epigenetic modification essential for cell viability and embryonic development in mammals. Recent studies demonstrated evidence supporting that DNA methylation also plays a crucial role in EMT process [9]. Given that global inhibition of DNA methylation is always coupled with severe side effects, strategies aimed at suppressing DNA methylation cannot be applied in clinical settings. However, studies including our own revealed that the information encoded by DNA methylome is read by a family of methyl-CpG–binding domain (MBD) proteins such as MBD1 to 4, and the methyl CpG binding protein 2 (MeCP2) [10]. MBD1, MBD2, MBD4, and MeCP2 selectively bind to the methylated CpG DNA to suppress target gene transcription via a transcriptional repressor domain (TRD). In particular, MBD2 possesses the highest binding activity [11], but mice deficient in MBD2 is viable and fertile without perceptible defects in physiological activities [12]. Therefore, MBD2 could be a viable target against tumor metastasis without discernable side effect. Indeed, altered MBD2 expression has been consistently noted to impact tumorigenesis on mouse cancer cell lines and on human cancer cell xenograft models [13], but its impact on tumor metastasis is yet to be elucidated.
Herein in this report, we demonstrated convincing evidence that patients with lung adenocarcinoma (LUAD) at the stage of lymphatic or distant metastasis were featured by the enhanced MBD2 expression. Moreover, knockdown of MBD2 significantly attenuated TGF-β1-induced EMT in LUAD cell lines (A549 and H1975) as evidenced by the repressed migration and invasion. Interestingly, consistent results were also observed in murine melanoma B16F10 cells, suggesting that altered MBD2 expression is a common feature adopted by tumor cells for metastasis. Mechanistically, MBD2 selectively bound to the methylated CpG DNA within the damage-specific DNA binding protein 2 (DDB2) promoter, by which it repressed DDB2 expression to enhance EMT process, thereby leading to tumor metastasis. As a result, liposomes loaded with MBD2 siRNA suppressed tumor metastasis in a B16F10 tumor-bearing mouse model. Together, our data support that MBD2 could be a promising prognostic marker for tumor metastasis, while strategies aimed at silencing MBD2 with siRNA-loaded liposomes could be a viable approach against tumor metastasis in clinical settings.
Methods
Tissue samples
Fresh tissue samples (138 in total) including lung adenocarcinoma (LUAD) (n = 69) and paired adjacent normal lung (n = 69, >5 cm from the tumor edge) tissues, were collected from Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. All patients had received no chemotherapy or radiotherapy before surgery and signed a written informed consent before sample collection. Clinical data of the patients are provided in Supplementary Table 1. The studies were approved by the Human Assurance Committee of Tongji Hospital (IRB: TJ-IRB20160601).
Data mining from TCGA and Human Protein Atlas database
We assessed the association between DDB2 expression and melanoma using the cohorts from TCGA (The Cancer Genome Atlas) database (TCGA-melanoma cohort). The survival curves for MBD2 and DDB2 in LUAD and melanoma patients were evaluated in the data from the Human Protein Atlas database (https://www.proteinatlas.org/). The UCSC Xena browser (https://xenabrowser.net) was employed to access and analyze the data.
Animals
The C57BL/6 male mice (8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All animal experiments were conducted in accordance with the guidelines approved by the Institutional Animal Care and Use Committee at Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China).
Cell culture
The A549 cells, NCI-H1975 and murine melanoma B16F10 cells were purchased from the American Type Culture Collection (Rockville, MD, USA). A Mycoplasma contamination test was carried out, which was negative. Those cells were cultured at 37 °C in a 5% CO2 incubator. To stimulate EMT, each type of cells was induced by TGF-β1 (10 ng/ml, 24 h) after transfected with MBD2 siRNA or a scramble siRNA for 48 h. The scramble siRNA transfected cells without TGF-β1 were served as the controls.
Reagents
Recombinant human TGF-β1 (Cat: 100-21) was purchased from the PeproTech Corporation (Cranbury, NJ, USA). Antibodies against E-cadherin (Cat: Sc-7870), N-cadherin (Cat: Sc-8424), Vimentin (sc-58899) and GAPDH(Sc-47724) were obtained from the Santa Cruz Biotechnology (Dallas, TX, USA). The crystal violet staining solution (Cat: C0121) and the ChIP assay kit (Cat: P2078) were purchased from the Beyotime Biotechnology (Shanghai, China). The lipidoid C12-200 was purchased from the Xinjiahecheng Medical Chemistry Corporation (Wuhan, Hubei, China). 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (mPEG-DMG) was ordered from the NOF Corporation (Tokyo, Japan). The CCK-8 assay kit (Cat: 40203ES60) was originated from Yeasen Biotechnology (Wuhan, China), and the Lipofectamine 3000 (Cat: L3000015) was obtained from the Invitrogen Corporation (Shanghai, China). All other reagents were obtained from Sigma (St. Louis, MO, USA).
RNA interference and preparation of siRNA-loaded liposomes
The MBD2 siRNAs and a corresponding scramble siRNA were synthesized by the RiboBio (Guangzhou, China), and were then transfected into A549 cells, B16F10 cells, and H1975 cells using the Lipofectamine 3000 according to the manufacturer’s protocol or a liposome-based siRNA transfection method as previously described [14]. The following two siRNAs were used for MBD2: siRNA1 5’-GCA AGA GCG AUG UCU ACU A-3’, and siRNA2 5’-GCG AAA CGA UCC UCU CAA U-3’. The siRNA-loaded nanoparticles were prepared as reported [15].
MBD2 overexpression
A549 cells and B16F10 cells were seeded on 12-well plates and transduced with Adenovirus-MBD2 and a Vector (Abcam, MA, USA) at the multiplicity of infection (MOI) 1:4 for 48 h. The transduced cells were then stimulated with TGF-β1 (10 ng/ml) for 24 h and analyzed by Western blotting.
Western blot analysis
Total proteins were extracted from tissues and cells using the Lysis buffer (Beyotime, Shanghai, China), and then subjected to Western blot analysis using the established techniques [16]. The primary antibodies against MBD2; E-cadherin, N-cadherin and Vimentin were employed for the analyses, and GAPDH band was used as the internal control.
Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 15 min. The samples were washed with PBS and blocked with 5% BSA in PBS for 1 h. Subsequently, the cells were incubated firstly with indicated primary antibodies overnight at 4 °C, and then with an Alexa Fluor 488-conjugated antibody or an Alexa Fluor 594-conjugated antibody (Abbkine, Redlands, CA, USA, 1:200) for 1 h at room temperature, respectively. Nuclei were counterstained with DAPI for 8 min. Immunofluorescence images were acquired under a microscope (Olympus, Tokyo, Japan) in a blinded manner.
Quantitative real-time PCR
Quantitative real-time PCR was conducted using the SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan) as previously reported [17]. The primer sequences were provided in Supplementary Table 2.
Cell migration and invasion assays
The cell migration and invasion were evaluated by using a Transwell assays (Corning, MA, USA) using the established techniques [18].
Cell Counting Kit-8 Assay
The cell viability was examined by a Cell Counting Kit-8 kit based on the manufacturer’s protocol. The absorbance was measured at 450 nm using a microplate reader (ELx800, BioTek Instruments, Winooski, VT, USA).
Apoptosis assay
Apoptosis assay was conducted using an Annexin V/PI detection kit (Beyotime, Shanghai, China) and analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA) as reported previously [18].
RNA deep sequencing (RNA-seq)
RNA from A549 cells activated by TGF-β1 was extracted using an RNA isolation kit (TaKaRa, Tokyo, Japan). The prepared mRNA libraries were sequenced on an Illumina HISEQ 2500 platform, and the HISAT2 v2.0.4 software (Center for Computational Biology, Baltimore, Maryland) was used to map the clean reads to the mm10 reference genome. Fragments per kilobase of exon per million mapped fragments (FPKM) values were calculated using the CuffNorm version 2.2.1 software (University of Washington, Seattle, Washington, USA). The genes with a calculated normalized FPKM value greater than 5.0 were considered to be expressed. Significantly differentially expressed genes were defined as those with a |log2 (fold change)| ≥ 0 and a p value ≤ 0.05. Heatmaps were generated using the R package heatmap.
GO term and KEGG pathway enrichment analysis
The biological significance of the differentially expressed genes (DEGs) was explored by the Gene Ontology (GO) term enrichment analysis covering 3 functional groups (biological process, cellular component and molecular function), and the Genomes (KEGG) pathway enrichment analysis of the DEGs was performed with the Bioconductor package “GeneAnswers” to identify vital pathways related to EMT in A549 cells.
Global DNA methylation assay and bisulfite DNA sequencing
Global DNA methylation was evaluated by a MethylFlashTM Methylated DNA Quantification Kit (Epigentek, NY, USA) according to the instructions and bisulfite DNA sequencing was carried by Sequenom MassARRAY platform (BGI.write, Beijing, China) as previously reported [19].
Chromatin immunoprecipitation (ChIP) assay and DDB2 promoter reporter assay
ChIP assays were conducted using a ChIP assay kit (Beyotime, Shanghai, China) as reported [20]. The primers for DDB2 used in the ChIP PCR were F: 5’ ACT CCC CAA CTA CAC CCT GT 3’ and R: 5’ CCG GCT AAT TTC TCT CTC TCT 3’. A dual luciferase reporter system (Promega, Madison, WI) was used for DNA methylation dependent DDB2 promoter luciferase reporter assays, in which the MBD2 binding sites within the DDB2 promoter were disrupted using the established techniques [21].
Flow cytometric analysis
A549 and B16F10 cells were transfected with DiI-labeled liposomes for 2 or 4 h. After washing, the cells were analyzed using a FACSCanto II (BD Biosciences, San Jose, CA, USA) as reported [22]. All data were analyzed using the FACSExpress V3 software (De Novo Software) based on the manufacturer’s protocol.
Anticancer activity in the B16F10 lung metastasis model
The melanoma tumor-bearing mouse model was established through the subcutaneous injection of B16F10 melanoma cells into the right flanks of C57BL/6 mice (6 ×105 cells/mouse). Once the tumor volume reached approximately 80 mm3, the mice were randomly intratumorally injected with PBS, L-scramble siRNA, empty liposomes or L-MBD2 siRNA (1 mg/kg) for a total of three times with a three-day of interval, eight mice were included in each study group. Tumor progression was assessed by monitoring the tumor volume with the following equation: V = (length × width2)/2. Two days after the last injection, the mice were sacrificed, and the tumor tissues were collected and weighed. The tumors were cryosectioned into pieces. The sections were co-stained with a FITC-labeled anti-N-cadherin antibody and an Alexa Fluor 594-labeled anti-E-cadherin antibody at 37 °C for 30 min. After rinsing the sections with PBS. the distribution of N-cadherin and E-cadherin was observed by confocal microscopy, and the relative fluorescence intensity was determined using the ImageJ software.
To further investigate the inhibitory capability of L-MBD2 siRNA in tumor metastasis, B16F10 melanoma cells were incubated with PBS, empty liposomes, 50 nM L-scramble siRNA or 50 nM L-MBD2 siRNA for 6 h, respectively. The pretreated B16-F10 tumor cells were then intravenously injected into the C57BL/6 mice (2 × 105 cells/mouse). After 20 days of injection, metastatic tumor-bearing lung tissues were collected from sacrificed mice, and the pulmonary metastatic nodules were counted. Tumor-bearing lung sections were also prepared for H&E staining.
Statistical analysis
All experiments were conducted with at least three independent replicates, and statistical analyses were performed using the GraphPad Prism 5.0 software (San Diego, CA, USA). Data are presented as the mean ± SEM, and an independent Student’s t test was applied to analyze the statistical significance of differences between two groups. As for comparisons among multiple groups, one-way ANOVA followed by Tukey’s post hoc test was performed. In all cases p < 0.05 were considered with statistical significance.
Results
LUAD metastasis is coupled with MBD2 upregulation
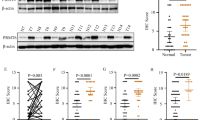
Since lung cancers ((non-small cell lung cancers (NSCLCs) in particular) are one of the highest-ranking causes of cancer-related death worldwide and LUAD is the most prevalent subtype of NSCLCs [23], we thus first examined MBD2 expression in the lung samples obtained from 69 LUAD patients. No significant difference in terms of MBD2 mRNA was observed between the adjacent normal tissue samples and the LUAD tissue samples (Fig. 1A). A similar expression pattern was also found in LUAD tumor tissues (n = 58) versus matched adjacent nontumor tissues (n = 58) [24] derived from the Oncomine database (https://www.oncomine.org/resource/login.html) (Supplementary Fig. S1A, B), a publicly accessible online cancer microarray database.
A The mRNA level of MBD2 was compared between 69 pairs of lung adenocarcinoma tissue samples (LUAD) and adjacent nontumor tissue samples (Normal). The expression of MBD2 was associated with sex (B), smoking history (C), degree of tumor differentiation (D), tumor stage (E), N stage (F), and M stage (G) in lung adenocarcinoma patients (n = 69). H Survival curves for MBD2 high group and MBD2 low group (The cutoff value is 15.02, n = 269). The data were originated from the Human Protein Atlas database. Each dot expresses one patient sample. N node, M metastasis. The data are presented as the mean ± SD. *p < 0.05.
To explore the functional impact of MBD2 on LUAD, we evaluated the associations between MBD2 expression and the clinicopathological features of LUAD patients. First, we observed that MBD2 expression in tumors was not correlated with sex (Fig. 1B), smoking history (Fig. 1C), or the degree of tumor differentiation (Fig. 1D). Similarly, we failed to detect a significant difference of MBD2 expression between tumors at different tumor stages (Fig. 1E). In sharp contrast, tumors with lymphatic or distant metastasis exhibited significantly higher MBD2 expression than that of tumors in situ (Fig. 1F, G). Moreover, high levels of MBD2 expression were found to be associated with poor outcome in patients with LUAD derived from the Human Protein Atlas database (Fig. 1H). To further confirm the above results, we examined our collected LUAD samples. Collectively, these data support that MBD2 is likely involved in tumor metastasis during the course of LUAD progression.
MBD2 regulates tumor metastasis in vitro
To confirm the above observations in LUAD patients, we next conducted Transwell assays to assess the impact of MBD2 on the migration and invasion of multiple cancer cell lines. To this end, A549 cells, a NSCLC cell line, were transfected with a scramble siRNA or an MBD2 siRNA for 48 h with Lipofectamine 3000 and then stimulated with TGF-β1 for 24 h, respectively. Notably, Western blot analysis revealed that TGF-β1 induced a significant increase of MBD2 expression, which was robustly blocked by the MBD2 siRNA (Fig. 2A), indicating that the MBD2 siRNA possesses high potency to knock down MBD2 expression. Importantly, TGF-β1 induced a dramatic increase in the number of migrated and invaded cells as compared to those of control cells, while knockdown of MBD2 potently reversed TGF-β1-induced migration and invasion (Fig. 2B). These results prompted us to examine the expression of E-cadherin, N-cadherin and Vimentin, the critical EMT markers relevant to tumor metastasis [25]. As expected, TGF-β1 robustly repressed E-cadherin expression, but promoted N-cadherin and Vimentin expression (Fig. 2A). Remarkably, the MBD2 siRNA significantly blocked the effect of TGF-β1 on the induction of EMT process (Fig. 2A and Supplementary Fig. S2A). Next, we checked morphological changes of the above cells by FITC-phalloidin staining. It was noted that A549 cells treated with TGF-β1 displayed an elongated spindle-like morphology, while significantly less amount of spindle-shaped cells was noted in MBD2 siRNA transfected cells (Supplementary Fig. S2B), suggesting that knockdown of MBD2 may attenuate TGF-β1-induced EMT in A549 cells.
A Western blot analysis of E-cadherin, N-cadherin, Vimentin and MBD2 in A549 cells after MBD2 siRNA transfected. B Transwell assays were performed to investigate the migration ability (left panel) and invasion ability (right panel) of A549 cells. C Western blot analysis of E-cadherin, N-cadherin and MBD2 in A549 cells after AdMBD2 transduced. D Western blot analysis of E-cadherin, N-cadherin, Vimentin and MBD2 in B16F10 cells after MBD2 siRNA transfected. E Transwell assays were performed to investigate the migration ability (left panel) and invasion ability (right panel) of B16F10 cells. F Western blot analysis of E-cadherin, N-cadherin and MBD2 in B16F10 cells after AdMBD2 transduced. All images were acquired at ×200 magnification. The data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
To address whether the above findings were limited to A549 cells, we conducted studies in murine melanoma B16F10 cells, which is generally considered as a potent metastatic tumor cell line. Similar results were noted as above in B16F10 cells, and MBD2 siRNA substantially repressed TGF-β1-induced MBD2 expression (Fig. 2D), and the MBD2 siRNA exhibited high potency to suppress TGF-β1-induced migration, invasion (Fig. 2E) coupled with attenuated EMT, as evidenced by the changes of expression for EMT related makers (Fig. 2D and Supplementary Fig. S2C) and cellular morphology (Supplementary Fig. S2D). Consistent results were obtained from in parallel experiments conducted in LUAD NCI-H1975 cells (Supplementary Fig. S2E). The next key question is whether MBD2 overexpression would promote EMT. To address this issue, adenoviruses carrying MBD2 cDNA were transduced into A549 and B16F10 cells. Indeed, the AdMBD2-transduced cells manifested a significantly increased EMT program (Fig. 2C, F). Taken together, our data support that MBD2 enhances EMT, by which it promotes cancer cell migration and invasion predisposing to tumor metastasis.
MBD2 represses DDB2 transcription to enhance EMT
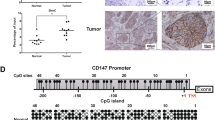
To dissect the mechanisms by which MBD2 facilitates tumor metastasis, RNA deep sequencing was conducted in scramble and MBD2 siRNA transfected A549 cells following TGF-β1 stimulation. Comparative analysis revealed that the MBD2 siRNA upregulated the expression of 4386 genes but downregulated the expression of 4559 genes (Fig. 3A). GO enrichment analysis characterized top 10 terms enriched with the differential expressed genes (DEGs) which involved in cell morphogenesis, adhesion and migration, indicating a pivotal role of MBD2 in tumor metastasis (Supplementary Fig. S3). KEGG pathway analysis revealed 10 pathways enriched with the upregulated genes (Fig. 3B), among which P53 signaling pathway was the most significant one under the comprehensive consideration of the enrichment ratio and p-value. Given that P53 is one of the most important tumor suppressor genes frequently mutated in human cancers [26], we then picked out P53-related genes from DEG and listed them in the heat map (Fig. 3C). Indeed, RT-PCR confirmed that MBD2 siRNA enhanced TP53 expression following TGF-β1 induction (Fig. 3D). Interestingly, the damaged DNA binding 2 (DDB2), a well-known and potent inhibitor for tumor metastasis [27], was characterized in the cluster of genes whose expression was upregulated in the MBD2 siRNA transfected cells (Fig. 3D). Specifically, the MBD2 siRNA rendered the TGF-β1 stimulated cells with a 3-fold increase of DDB2 expression as compared to the scramble siRNA transfected cells (Fig. 3D), suggesting that DDB2 could be the critical MBD2 target gene. Since MBD2 serves as a reader to interpret the information encoded by DNA methylation [13], we analyzed the DNA methylation status of genomic DNA by a global DNA methylation assay. Remarkably, TGF-β1 induced a global genomic DNA hypermethylation in A549 cells (Fig. 3E), while bioinformatic analysis using EMBOSS Cpgplot (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/) revealed that the DDB2 promoter sequence contained a CpG island (Supplementary Fig. S4A). This result prompted us to assess the DNA methylation status of the DDB2 promoter by bisulfite DNA sequencing. Indeed, TGF-β1 induced the CpG DNA within the DDB2 promoter (Supplementary Fig. S4B, from −555 to −557 bp, the transcription start site as +1) to undergo a DNA hypermethylation. However, no perceptible difference in terms of DNA methylation within the DDB2 promoter was noted between the scramble siRNA and MBD2 siRNA transfected A549 cells, indicating that MBD2 itself does not DNA methylation (Fig. 3F).
A Scatter plot showing the average gene expression level (log10) in A549 cells upon TGF-β1 stimulation vs the fold change (log2). The blue and red dots represent downregulated and upregulated genes, respectively. B KEGG pathway enrichment analysis of the upregulated DEGs. C Heatmap showing the upregulated P53-related genes in TGF-β1-treated A549 cells transfected with MBD2 siRNA. D RT-PCR analysis of MBD2, TP53 and DDB2 expression in TGF-β1 induced A549 cells after MBD2 siRNA transfected. E Global DNA methylation level in A549 cells following TGF-β1 treatment. F Results of the bisulfite DNA sequencing analysis of the DDB2 promoter in TGF-β1 induced A549 cells following MBD2 siRNA transfected. G ChIP-PCR analysis of MBD2 binding to the DDB2 promoter. H Relative luciferase activity in A549 cells after WT or MU plasmid transfected. I RT-PCR analysis of CDH1, CDH2 and VIM expression in A549 cells. J Comparison of DDB2 mRNA expression between lung adenocarcinoma tissue samples and adjacent nontumor tissue samples in the TCGA-LUAD cohort. K Comparison of DDB2 mRNA expression between premetastatic and metastatic tissues in the TCGA melanoma cohort. The data are presented as the mean ± SEM. GO Gene Ontology, DEG differentially expressed gene, KEGG Kyoto Encyclopedia of Genes and Genomes. *p < 0.05, **p < 0.01, ***p < 0.001.
We next sought to address whether MBD2 selectively binds to the above hypermethylated CpG DNA within the DDB2 promoter. Chromatin immunoprecipitation (ChIP) assays were employed to determine the MBD2 binding activity to the DDB2 promoter in A549 cells. Indeed, MBD2 could bind to the methylated CpG DNA within the DDB2 promoter following TGF-β1 stimulation (Fig. 3G). To dissect the effect of MBD2 on DDB2 transcription following binding to its promoter, a pGL3-Basic luciferase reporter plasmid, in which the cytosine within the CpG DNA of DDB2 promoter (from −555 to −557 bp) were mutated to adenine, was constructed (Mutant plasmid). A pGL3-Basic plasmid containing the wild-type DDB2 promoter sequence was also prepared (WT plasmid). The A549 cells were co-transfected with either the mutant or WT luciferase reporter plasmid along with an MBD2 plasmid, and the cells were harvested for assays of reporter activity following TGF-β1 stimulation. In line with the above results, cells transfected with the mutant plasmid displayed a 2.5-fold higher reporter activities (Fig. 3H), indicating that MBD2 represses DDB2 expression following binding to the methylated DNA within its promoter. We then embarked on the effect of DDB2 on TGF-β1 induced EMT program. A549 cells were transfected with an MBD2 siRNA or a DDB2 siRNA alone or in the presence of TGF-β1 stimulation. Similar as our previous data, knockdown of MBD2 significantly increased CDH1 expression while repressed CDH2 and VIM expression (Fig. 3I). However, knockdown of DDB2 completely abolished the effect of MBD2 siRNA, and particularly, no perceptible difference was noted in terms of the expression levels for the above genes between DDB2 siRNA and MBD2 + DDB2 siRNA transfected cells (Fig. 3I). These results support that MBD2 enhances TGF-β1-induced EMT dependent on its suppressive effect on DDB2 expression.
Based on the above data, we next examined the association between MBD2 and DDB2 expression in lung tissues from LUAD patients. We first compared DDB2 mRNA levels between Normal and LUAD in the TCGA-LUAD cohort. Analysis of the paired samples indicated that LUAD was featured by the decreased DDB2 mRNA levels (Fig. 3J). More importantly, the DDB2 mRNA levels were substantially lower in samples derived from metastatic tissue than that of samples originated from premetastatic tissue in the TCGA-melanoma cohort (Fig. 3K). To investigate the prognostic values of DDB2 in patients with LAUD or melanoma, the Human Protein Atlas was employed to assess the relationship between DDB2 and prognosis of tumor patients. A total of 267 LUAD patients and 47 melanoma patients were available for the analysis of overall survival. The survival curves indicated that lower levels of DDB2 expression were associated with shorter overall survival (Supplementary Fig. S5A, B), and this association is consistent with what we found for MBD2 (Fig. 1H). Collectively, our data support that MBD2 promotes tumor metastasis by enhancing EMT via attenuating DDB2 expression.
Generation of MBD2 siRNA-loaded liposomes
To translate the above discoveries into a clinical strategy against tumor metastasis, we generated cationic lipid-based liposomes carrying MBD2 siRNA as previously reported [21] (Supplementary Fig. S6A). The prepared liposomes dispersed well in PBS (polydispersity index 0.15) and possessed an MBD2 siRNA entrapment efficiency above 95% with a zeta potential of 2.3 mV (Supplementary Fig. S6B). The hydrodynamic diameter of these liposomes was approximately 98 nm (Supplementary Fig. S6B), which was smaller than that of the empty liposomes likely due to the electronegativity of siRNA. Transmission electron microscopy demonstrated that the liposomes exhibited a uniform distribution of spherical particles (Supplementary Fig. S6C, D) and could maintain its intact structure in PBS for at least 24 h (Supplementary Fig. S6E).
Next, we assessed the toxicity of liposome-loaded siRNA by CCK-8 assay. As expected, the safety profile of those liposomes was promising, as we failed to detect any toxicity of liposome-loaded scramble siRNA in both A549 and B16F10 cells, in terms of viability, at the different concentrations tested (Supplementary Fig. S6F). In contrast, administration of MBD2 siRNA-loaded liposomes strikingly decreased the viability of A549 and B16F10 cells once the dose of liposomes was higher than 200 nM (Supplementary Fig. S6G), while 200 nM of MBD2 siRNA-loaded liposomes only rendered less than 10% cells to undergo apoptosis (Supplementary Fig. S6H), and therefore, this concentration was chosen for the subsequent studies. Flow cytometry analysis revealed that those liposomes could be efficiently taken up by A549 (Fig. 4A) and B16F10 cells in a time-dependent manner (Fig. 4B). A549 and B16F10 cells were then co-cultured with siRNA-loaded liposomes for 48 h and then stimulated with TGF-β1 for 24 h. The MBD2 siRNA-loaded liposomes (L-MBD2 siRNA) significantly repressed TGF-β1-induced MBD2 expression (Fig. 4C, D) along with attenuated migration (Fig. 4E, F) and invasion (Fig. 4G, H). Wound healing assay further confirmed the inhibitory effect of L-MBD2 siRNA on the TGF-β1-induced motility in A549 and B16F10 cells (Supplementary Fig. S7A, B).
MIF in A549 (A) and B16F10 (B) cells transfected with DiI-labeled liposomes. C, D Western blot analysis of MBD2 expression in A549 (C) and B16F10 (D) cells after L-Scr siRNA or L-MBD2 siRNA transfected. E, F The migration ability of A549 (E) and B16F10 (F) cells transfected with L-Scr siRNA or L-MBD2 siRNA. G-H The invasion ability of A549 (G) and B16F10 (H) cells transfected with L-Scr siRNA or L-MBD2 siRNA. I, J Coimmunostaining of liposomes (Red) and E-cadherin (Green) in A549 (I) and B16F10 (J) cells after TGF-β1 stimulation. K, L Coimmunostaining of liposomes (Red) and Vimentin (Green) in A549 (K) and B16F10 (L) cells after TGF-β1 stimulation. All images were acquired at ×200 magnification. Nuclei were stained with DAPI (blue). M, N RT-PCR analysis of CDH1, CDH2 and VIM in TGF-β1 induced A549 (M) and B16F10 (N) cells following L-Scr siRNA or L-MBD2 siRNA treatment. MFI mean fluorescence intensity, L-Scr siRNA liposome-based Scrambled siRNA, L-MBD2 siRNA liposome-based MBD2 siRNA. The data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
To determine the impact of L-MBD2 siRNA on the progression of EMT, A549 and B16F10 cells following TGF-β1 stimulation were immunostained with green fluorescence labeled antibodies against E-cadherin or Vimentin, and liposomes were labeled with Dil (red). It was noted that cells transfected with L-MBD2 siRNA were coupled with enhanced E-cadherin (Fig. 4I, J) and attenuated Vimentin expression (Fig. 4K, L). Moreover, RT-PCR analysis of EMT markers (CDH1, CDH2 and VIM) in above cells obtained consistent results (Fig. 4M, N). Taken together, these results support that the liposome-based MBD2 siRNA antagonizes TGF-β1-induced cancer cell migration, invasion coupled with attenuated EMT, and therefore, it could be a viable therapeutic approach against tumor metastasis.
Administration of L-MBD2 siRNA attenuates tumor metastasis
Finally, we sought to assess the therapeutic potential of L-MBD2 siRNA in tumor metastasis. The mice bearing subcutaneous B16F10 melanoma tumors with a volume of approximately 50 mm3 were intratumorally injected with PBS, empty liposomes, liposome-based scramble siRNA (L-Scrambled siRNA), and L-MBD2 siRNA, respectively. Neither the empty liposomes nor the L-scramble siRNA exhibited perceptible impact on tumor growth inhibition (Fig. 5A, B). However, the L-MBD2 siRNA treated mice were featured by a marked reduction in terms of tumor volume (Fig. 5A) or size (Fig. 5B) along with a decreased tumor mass (Fig. 5C). Likewise, no body mass difference was noted (Supplementary Fig. S8), demonstrating the outstanding biocompatibility of L-MBD2 siRNA in vivo. Injection of L-MBD2 siRNA significantly suppressed MBD2 expression in the tumors (Fig. 5D), which were accompanied by the robust increase of E-cadherin and reduction of N-cadherin expression, as determined by RT-PCR analysis (Fig. 5E, F) and immunostaining (Fig. 5G).
A The B16F10 melanoma tumor volume in each group was determined at the indicated time points. B Images of tumor nodules at the endpoint of the experiment. C The weight of the tumor nodules from each group at the endpoint of the experiment. D Western blot analysis of MBD2 in each group. E, F RT-PCR analysis of Cdh1 (E) and Cdh2 (F) in the indicated groups. G Representative images of E-cadherin and N-cadherin coimmunostaining in the tumor nodule sections. H Images of pulmonary metastatic nodules from the indicated groups. I Representative H&E staining of pulmonary metastatic nodules from the indicated groups. J MBD2 binds to methylated CpG DNA within the DDB2 promoter and thus represses DDB2 expression, resulting in tumor metastasis. Intravenous administration of liposomes carrying MBD2 siRNA suppressed tumor growth in an animal model bearing tumor metastasis. L-Scr siRNA: liposome-based Scrambled siRNA; L-MBD2 siRNA: liposome-based MBD2 siRNA. The data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
To further evaluate the anti-metastatic capability of L-MBD2 siRNA, a metastasis model was established by intravenous injection of B16F10 melanoma cells. Pulmonary metastatic nodules were noted following 20 days of injection. Remarkably, L-MBD2 siRNA strikingly suppressed the metastasis of B16F10 cells as compared to that in the other three groups, as manifested by the reduction for the number of tumor nodules in the lungs (Fig. 5H). Consistent with the above results, H&E staining also revealed alleviation of metastasis in the L-MBD2 siRNA-treated group (Fig. 5I). Collectively, our data support the notion that L-MBD2 siRNA could be a promising therapeutic approach against tumor metastasis and in situ tumor growth.
Discussions
In the present study, we performed studies in human patients and animal models to elaborate the impact of MBD2, a reader for interpretation of DNA methylome-encoded information, on tumor metastasis. We demonstrated evidence suggesting an association between MBD2 expression and tumor metastasis in LUAD patients. Moreover, in vitro studies corroborated that MBD2 is involved in the regulation of TGF-β1-induced migration, invasion and EMT in multiple types of tumor cells. Mechanistically, MBD2 selectively binds to the methylated CpG DNA within the DDB2 promoter, by which it represses DDB2 expression predisposing to tumor metastasis. Therefore, administration of liposomes carrying MBD2 siRNA suppressed tumor metastasis in the lung (Fig. 5J). Together, our data not only provide novel insights into the pathogenesis underlying tumor metastasis, but also demonstrate evidence supporting that MBD2 siRNA liposomes could be a viable approach against tumor metastasis in clinical settings.
Cancer is a class of diseases referring to a series of molecular changes to prefer uncontrolled cell growth and ultimately metastasize to other anatomical locations. Metastasis, a life-threatening event in cancer progression, contributes substantially to the current ranking of cancer as the second leading cause of death worldwide [28]. Although a great effort has recently been devoted to clarifying the pathoetiology underlying tumor metastasis, the exact molecular mechanisms, however, remain poorly understood. Aberrant DNA methylation has long been recognized in tumor development, and recently becomes more attractive due to its therapeutic potential [29]. Nevertheless, the severe side effect following global inhibition of DNA methylation limited the development of therapeutic strategies in clinical settings [30]. The DNA methylome-encoded information is read by a family of MBD proteins, through which they interpret the impact of DNA methylation on gene transcription while without affecting DNA methylation. Those properties rendered MBD proteins with feasibility to be therapeutic targets for tumor metastasis with acceptable side effects [31]. Particularly, MBD2 could be the most promising one, as deletion of MBD2 in mice does not generate any major deleterious effects [13, 32], supporting that MBD2 might be dispensable under physiological conditions.
Indeed, MBD2 has been shown to inhibit aberrantly methylated tumor suppressive genes by binding to the methylated CpG DNA within the promoter regions [33]. More recently, studies indicate that MBD2 probably modulates tumorigenesis in different types of cancer [34, 35]. In the past few years, our laboratory has concentrated on the effect of MBD2 on cancer progression. In Notch1-driven T-cell acute lymphoblastic leukemia (T-ALL), we first noted that MBD2 is required for the progression and maintenance of leukemia [13]. We showed evidence that MBD2 ablation under “steady-state” conditions not only causes a profound decrease in lymphocytes but also substantially impedes the progression of Notch1-driven T-ALL. In the current report, we focused on the role of MBD2 in pathogenesis of tumor metastasis, particularly in the metastasis of LUAD, as metastasis is one of the most important hallmarks of LUAD. Intriguingly, we demonstrated that higher expression of MBD2 was observed in tumors of LUAD patients with either lymphatic or distant tumor metastasis than in tumors in situ, but no significant difference in terms of MBD2 expression was noted between normal lung tissues and LUAD tumor tissues by analysis of paired samples from 73 LUAD patients (Fig. 1) and the data from 58 pairs of LUAD patients from the Oncomine database (Supplementary Fig. 1). These observations strongly suggest that MBD2 might be a critical prognostic marker for tumor metastasis. Indeed, the data from the Human Protein Atlas illustrated that higher MBD2 levels were correlated with poorer outcome in patients with LUAD.
EMT has been reported to increase the motility and invasiveness of cancer cells by repressing the expression of epithelial markers and promoting the expression of mesenchymal markers, which in turn facilitates the formation of metastases [6]. Abundant evidence suggests that TGF-β1 can promote the migration and invasion of various cancer cells [7, 36]. We thus stimulated A549, H1975 and B16F10 cells with TGF-β1 to assess the impact of MBD2 on cancer cell EMT process. We demonstrated convincing evidence that knockdown of MBD2 markedly inhibited TGF-β1-induced cell migration and invasion coupled with attenuated EMT. However, in contrast to our observation, Pei and colleagues reported that lower MBD2 expression was significantly correlated with higher tumor stage and metastasis via deteriorating the EMT process [37]. This discrepancy is most likely caused by the differences of experimental approaches employed. Specifically, they assessed the EMT process in A549 cells without TGF-β1 induction, while we stimulated the cancer cells with TGF-β1, as TGF-β1 is highly enriched in the tumor microenvironment and essential for tumor metastasis. Nevertheless, regardless of which point of view is more persuasive, all of these findings suggest the implication of MBD2 in modulating tumor metastasis, thereby contributing to the pathogenesis of cancer progression.
Extensive studies support the therapeutic potential for cancer metastasis by inhibiting EMT program, which highlights an urgent need for developing effective anticancer drugs to target the TGF-β1 signaling. Generally, TGF-β1 activates the ALK5 type I receptor (which phosphorylates SMAD2/3) as well as noncanonical (e.g., Src kinase, EGFR, JAK/STAT, P53) pathways that jointly drive cancer progression [38, 39]. In nucleotide excision repair, P53 functions as a transcription factor to regulate both the basal and DNA damage-inducible expression of a set of genes involved in DNA repair, including DDB2 [40], which has been proposed to act as a suppressor of tumorigenesis by transcriptionally repressing the expression of genes essential for EMT program, such as Snail and Zeb1 [41]. Indeed, our RNA-seq data revealed that TGF-β1-treated A549 cells transfected with MBD2 siRNA were characterized by a gain of the DDB2 signature, which was further confirmed by RT-PCR. In fact, TGF-β1 induced A549 cells to undergo a DNA hypermethylation within the DDB2 promoter along with the induction of MBD2 overexpression, which in turn bound to the methylated CpG DNA within the DDB2 promoter to suppress its expression, thereby activating and sustaining the EMT program.
Over the past few decades, remarkable scientific progress has been made in cancer treatments, such as surgical therapy, chemotherapy and radiotherapy. Improvements in the use of these therapies have improved the disease-free survival of cancer patients [42]. However, there are still many challenges in cancer treatment unsettled. Regarding the discovery of various proto-oncogenes and tumor suppressor genes, gene therapy has become a popular research direction in the treatment of metastatic cancers [43]. Nanovehicles have been extensively applied for the effective delivery of siRNA or chemotherapeutics [44]. In particular, positively charged cationic liposomes have been widely used as siRNA and chemotherapeutic carriers due to their high loading capacity and significantly enhanced cellular internalization through adsorptive interactions with the tumor cell membrane [45]. For example, patisiran (Onpattro) has been approved by the Food and Drug Administration (FDA) in 2018 as the first lipid nanoparticle-based siRNA drug for the treatment of hereditary transthyretin-mediated amyloidosis, which translates such an idea into the clinical practice [46]. The most exciting discovery in this study was that administration of MBD2 siRNA-loaded liposomes significantly attenuated xenograft metastatic potential in animal models, highlighting a promising prospect and important application value of MBD2 siRNA-loaded liposomes in clinical settings. However, our study still has some limitations. First, the cohort of patients with metastatic LUAD was relatively small. More patients are needed to verify the association of MBD2 and tumor metastasis. Second, MBD2 was upregulated in A549, H1975 and B16F10 cells after TGF-β1 stimulation. However, the mechanism underlying MBD2 overexpression is far from clear. In our recent study, TGF-β1 could upregulate the expression of MBD2 via the canonical SMAD pathway in fibroblasts [47]. We speculated that the expression of MBD2 induced by TGF-β1 might be depended on the canonical SMAD pathway in tumor cells as well, but it needs to be confirmed. Third, the expression of MBD2 may facilitate tumor growth, as evidenced by the significantly lower tumor volume, size and mass in the L-MBD2 siRNA treated mice. We believe that this phenotype caused by the enhancing effect of MBD2 siRNA on inducing cancer cell apoptosis (Supplementary Fig. 8H). However, the detailed mechanism needs to be further explored.
In summary, this study indicated that MBD2 facilitates cancer metastasis, and therefore, knockdown of MBD2 inhibited TGF-β1-induced EMT, migration and invasion of cancer cells. Mechanistically, MBD2 selectively binds to the methylated CpG DNA within the DDB2 promoter region, by which MBD2 suppresses DDB2 expression to motivate the EMT program. Given that MBD2 itself does not influence DNA methylation and seems to be dispensable for the biological processes under physiological conditions [32], MBD2 is likely to be a viable target to suppress cancer metastasis. Indeed, intratracheal instillation of liposomes carrying MBD2 siRNA substantially inhibited tumor growth and metastasis in a B16F10 tumor-bearing animal model. Together, our data provide important proof of principle for MBD2 as a useful metastasis biomarker and describe liposome-based MBD2 siRNA as a candidate anticancer agent in clinical settings.
Data availability
The data used in the current study are all available from the corresponding author upon reasonable request.
Materials availability
The materials used in the current study are all available from the corresponding author upon reasonable request.
References
Zeeshan R, Mutahir Z. Cancer metastasis - tricks of the trade. Bosn J Basic Med Sci. 2017;17:172–82.
Elgundi Z, Papanicolaou M, Major G, Cox TR, Melrose J, Whitelock JM, et al. Cancer metastasis: the role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front Oncol. 2019;9:1482.
Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–26.
Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39.
Marcucci F, Stassi G, De Maria R. Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15:311–25.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Foroutan M, Cursons J, Hediyeh-Zadeh S, Thompson EW, Davis MJ. A transcriptional program for detecting TGFbeta-induced EMT in cancer. Mol Cancer Res. 2017;15:619–31.
Zhou Q, Li Y, Zhu Y, Yu C, Jia H, Bao B, et al. Co-delivery nanoparticle to overcome metastasis promoted by insufficient chemotherapy. J Control Release. 2018;275:67–77.
Casalino L, Verde P. Multifaceted roles of DNA methylation in neoplastic transformation, from tumor suppressors to EMT and metastasis. Genes. 2020;11:922.
Cheng J, Song J, He X, Zhang M, Hu S, Zhang S, et al. Loss of Mbd2 protects mice against high-fat diet-induced obesity and insulin resistance by regulating the homeostasis of energy storage and expenditure. Diabetes. 2016;65:3384–95.
Dhasarathy A, Wade PA. The MBD protein family-reading an epigenetic mark? Mutat Res. 2008;647:39–43.
Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23.
Zhou M, Zhou K, Cheng L, Chen X, Wang J, Wang XM, et al. MBD2 ablation impairs lymphopoiesis and impedes progression and maintenance of T-ALL. Cancer Res. 2018;78:1632–42.
Asik E, Akpinar Y, Caner A, Kahraman N, Guray T, Volkan M, et al. EF2-kinase targeted cobalt-ferrite siRNA-nanotherapy suppresses BRCA1-mutated breast cancer. Nanomedicine. 2019;14:2315–38.
Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277.
Zhang L, Wang Y, Wu G, Rao L, Wei Y, Yue H, et al. Blockade of JAK2 protects mice against hypoxia-induced pulmonary arterial hypertension by repressing pulmonary arterial smooth muscle cell proliferation. Cell Prolif. 2020;53:e12742.
Yao Y, Wang Y, Zhang Z, He L, Zhu J, Zhang M, et al. Chop deficiency protects mice against bleomycin-induced pulmonary fibrosis by attenuating M2 macrophage production. Mol Ther. 2016;24:915–25.
Hu Y, Yu J, Wang Q, Zhang L, Chen X, Cao Y, et al. Tartrate-resistant acid phosphatase 5/ACP5 interacts with p53 to control the expression of SMAD3 in lung adenocarcinoma. Mol Ther Oncolytics. 2020;16:272–88.
Aldamiz-Echevarria T, Resino S, Bellon JM, Jimenez-Sousa MA, Miralles P, Medrano LM, et al. European mitochondrial haplogroups predict liver-related outcomes in patients coinfected with HIV and HCV: a retrospective study. J Transl Med. 2019;17:244.
Wang F, Sun F, Luo J, Yue T, Chen L, Zhou H, et al. Loss of ubiquitin-conjugating enzyme E2 (Ubc9) in macrophages exacerbates multiple low-dose streptozotocin-induced diabetes by attenuating M2 macrophage polarization. Cell Death Dis. 2019;10:892.
Wang Y, Zhang L, Wu GR, Zhou Q, Yue H, Rao LZ, et al. MBD2 serves as a viable target against pulmonary fibrosis by inhibiting macrophage M2 program. Sci Adv. 2021;7:eabb6075.
Wang Y, Zhu J, Zhang L, Zhang Z, He L, Mou Y, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor alpha positive feedback loop in M2 macrophages. J Allergy Clin Immunol. 2017;140:1550.e8–61.e8.
Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:e342–51.
Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–211.
Brabletz T, Hlubek F, Spaderna S, Schmalhofer O, Hiendlmeyer E, Jung A, et al. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 2005;179:56–65.
Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets. 2014;15:80–9.
Barbieux C, Bacharouche J, Soussen C, Hupont S, Razafitianamaharavo A, Klotz R, et al. DDB2 (damaged-DNA binding 2) protein: a new modulator of nanomechanical properties and cell adhesion of breast cancer cells. Nanoscale. 2016;8:5268–79.
Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, et al. Tissue invasion and metastasis: molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35:S244–75.
Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607.
Schulze KV, Swaminathan S, Howell S, Jajoo A, Lie NC, Brown O, et al. Edematous severe acute malnutrition is characterized by hypomethylation of DNA. Nat Commun. 2019;10:5791.
Ginder GD, Williams DC Jr. Readers of DNA methylation, the MBD family as potential therapeutic targets. Pharmacol Ther. 2018;184:98–111.
Rao X, Zhong J, Zhang S, Zhang Y, Yu Q, Yang P, et al. Loss of methyl-CpG-binding domain protein 2 enhances endothelial angiogenesis and protects mice against hind-limb ischemic injury. Circulation. 2011;123:2964–74.
Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, et al. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145:853.e9–64.e9.
Martin V, Jorgensen HF, Chaubert AS, Berger J, Barr H, Shaw P, et al. MBD2-mediated transcriptional repression of the p14ARF tumor suppressor gene in human colon cancer cells. Pathobiology. 2008;75:281–7.
Mian OY, Wang SZ, Zhu SZ, Gnanapragasam MN, Graham L, Bear HD, et al. Methyl-binding domain protein 2-dependent proliferation and survival of breast cancer cells. Mol Cancer Res. 2011;9:1152–62.
Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30.
Pei YF, Xu XN, Wang ZF, Wang FW, Wu WD, Geng JF, et al. Methyl-CpG binding domain protein 2 inhibits the malignant characteristic of lung adenocarcinoma through the epigenetic modulation of 10 to 11 translocation 1 and miR-200s. Am J Pathol. 2019;189:1065–76.
Oh IR, Raymundo B, Kim M, Kim CW. Mesenchymal stem cells co-cultured with colorectal cancer cells showed increased invasive and proliferative abilities due to its altered p53/TGF-beta1 levels. Biosci Biotechnol Biochem. 2020;84:256–67.
Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321.e10–37.e10.
Burger K, Ketley RF, Gullerova M. Beyond the trinity of ATM, ATR, and DNA-PK: multiple kinases shape the DNA damage response in concert with RNA metabolism. Front Mol Biosci. 2019;6:61.
Roy N, Bommi PV, Bhat UG, Bhattacharjee S, Elangovan I, Li J, et al. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer Res. 2013;73:3771–82.
Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–19.
Xu C, Liu W, Hu Y, Li W, Di W. Bioinspired tumor-homing nanoplatform for co-delivery of paclitaxel and siRNA-E7 to HPV-related cervical malignancies for synergistic therapy. Theranostics. 2020;10:3325–39.
Qian J, Xu M, Suo A, Xu W, Liu T, Liu X, et al. Folate-decorated hydrophilic three-arm star-block terpolymer as a novel nanovehicle for targeted co-delivery of doxorubicin and Bcl-2 siRNA in breast cancer therapy. Acta Biomater. 2015;15:102–16.
Yang ZZ, Li JQ, Wang ZZ, Dong DW, Qi XR. Tumor-targeting dual peptides-modified cationic liposomes for delivery of siRNA and docetaxel to gliomas. Biomaterials. 2014;35:5226–39.
Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21.
Wang Y, Zhang L, Huang T, Wu GR, Zhou Q, Wang FX, et al. The methyl-CpG-binding domain 2 facilitates pulmonary fibrosis by orchestrating fibroblast to myofibroblast differentiation. Eur Respir J. 2022;60:2003697.
Funding
Our study was supported by the National Natural Science Foundation of China (8213000540, 91749207, 81920108009, 82070808, 81770823, 81974456, and 82100079), the Ministry of Science and Technology (2017YFC1309603), NHC Diabetes Drug Discovery Program (2017ZX09304022-07), the Department of Science and Technology of Hubei State (2020DCD014), the Integrated Innovative Team for Major Human Disease Programs of Tongji Medical College, Huazhong University of Science and Technology, and the Innovative Funding for Translational Research from Tongji Hospital.
Author information
Authors and Affiliations
Contributions
LZ and SYW were responsible for conducting all experiments and data analysis and wrote the manuscript. G-RW, HHY, and SZ were involved in Western blot, immunostaining, and animal breeding. RHD was responsible for nanoparticle preparation. QLY and PY provided help for flow cytometry. HLZ, JPZ, and JY were involved in study design and review of the manuscript. WNX and XLY contributed to discussion and review of the manuscript, study design, and manuscript preparation. XLY, TYY, and C-YW contributed to the study design and manuscript preparation. All authors were involved in drafting the article or revising it critically, and all authors gave their approval for the final manuscript to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of Tongji Hospital. All procedures were conducted in accordance with the Declaration of Helsinki and International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS).
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Stephen Tait
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, L., Wang, S., Wu, GR. et al. MBD2 facilitates tumor metastasis by mitigating DDB2 expression. Cell Death Dis 14, 303 (2023). https://doi.org/10.1038/s41419-023-05804-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-023-05804-1
This article is cited by
-
Methyl-CpG binding domain protein 2 (Mbd2) drives breast cancer progression through the modulation of epithelial-to-mesenchymal transition
Experimental & Molecular Medicine (2024)