Abstract
Malignant characteristics of cancers, represented by rapid cell proliferation and high metastatic potential, are a major cause of high cancer-related mortality. As a multifunctional RNA-binding protein, heterogeneous nuclear ribonucleoprotein K (hnRNPK) is closely associated with cancer progression in various types of cancers. In this study, we sought to identify hnRNPK-regulated long intergenic non-coding RNAs (lincRNAs) that play a critical role in the regulation of cancer malignancy. We found that hnRNPK controlled malignant phenotypes including invasiveness, proliferation, and clonogenicity. RNA sequencing and functional studies revealed that LINC00263, a novel target of hnRNPK, is involved in the oncogenic functions of hnRNPK. Knockdown of LINC00263 mitigated the malignant capabilities. Conversely, increased malignant phenotypes were observed in LINC00263-overexpressing cells. Since LINC00263 was mainly localized in the cytosol and highly enriched in Argonaute 2-immunoprecipitation (Ago2-IP), we hypothesized that LINC00263 acts as a competitive endogenous RNA (ceRNA), and thus sought to identify LINC00263-associated microRNAs. Using small RNA sequencing followed by antisense oligonucleotide pull-down, miR-147a was selected for further study. We found that miR-147a negatively regulates LINC00263 via direct interaction, thus suppressing malignant capabilities. Moreover, knockdown of hnRNPK and LINC00263 upregulated miR-147a, indicating that LINC00263 serves as a ceRNA for miR-147a. By analyzing RNA sequencing data and miRNA target prediction, calpain 2 (CAPN2) was identified as a putative target of miR-147a. Ago2-IP and luciferase reporter assay revealed that miR-147a suppressed CAPN2 expression by directly binding to the 3′UTR of CAPN2 mRNA. In addition, we found that the weakened malignant capabilities following knockdown of hnRNPK or LINC00263 were restored by miR-147a inhibition or CAPN2 overexpression. Furthermore, our findings were validated in various other types of cancer cells including lung cancer, colorectal cancer, neuroblastoma, and melanoma. Collectively, we demonstrate that hnRNPK-regulated LINC00263 plays an important role in cancer malignancy by acting as a miR-147a decoy and thus upregulating CAPN2.
Similar content being viewed by others
Introduction
Malignant properties of cancer cells, including their highly aggressive characteristics, are major obstacles in the successful treatment of cancer. In fact, the degree of malignancy is a major factor that affects cancer mortality1. The limitless growth of cancer cells is a result of the abnormal activation of oncogenic signals that enhance the proliferation rate and inhibit apoptotic processes2. Cancer metastasis involves multiple steps in which cancer cells are disseminated from the primary site to other tissues or organs far from where they first occurred. Through the control of metastasis-associated genes, primary tumor cells prepare for metastasis by acquiring invasive capacity and proliferative properties to disseminate and survive in the secondary sites3. Rapid proliferation and metastatic potential are the main phenotypes associated with cancer malignancy. Therefore, the control of proliferation and metastasis is considered a promising approach for the development of cancer therapeutics.
Heterogeneous nuclear ribonucleoprotein K (hnRNPK) is a DNA- and RNA-binding protein (RBP) that contains three K homology (KH) domains, a nuclear shuttling (KNS) domain, and a nuclear localization signal (NLS)4. HnRNPK controls the expression of target genes mainly by directly binding to the untranslated region (UTR) of the target mRNAs. Through a wide range of regulatory mechanisms, including post-transcriptional gene regulation, hnRNPK is known to induce genes involved in the extracellular matrix, cell motility, and angiogenesis5,6. Furthermore, a loss-of-function screening using randomized intracellular antibodies has revealed that hnRNPK is a potential target for metastasis therapy and its cytoplasmic accumulation is crucial for its role in metastasis7. We previously reported that hnRNPK regulates the proliferation of cancer cells by targeting polo-like kinase 1 (PLK1) and heme oxygenase-1 (HO-1). Further, we demonstrated that mechanically, hnRNPK competes for interaction with PLK1 mRNA8 and increases the expression of HO-1 through PTOV1-miR-1207-5p9.
MicroRNAs (miRNAs) typically regulate gene expression at the post-transcriptional level by recognizing miRNA-recognition elements (MREs) within their target mRNA. Noncoding RNAs (ncRNAs) may share MREs with target mRNA of coding genes and therefore be targeted by miRNAs. This interaction and sequestering of miRNA by ncRNAs constitutes the basis for the competitive endogenous RNA (ceRNA) theory10. Recently, long ncRNAs (lncRNAs) have received increasing attention for their key roles in cancer progression as oncogenes and tumor suppressors11,12. Emerging evidence suggests that the ceRNA-mediated gene regulatory network is closely associated with cancer progression in various types of cancers.
In this study, we screened hnRNPK-regulated lncRNAs that are responsible for the oncogenic function of hnRNPK. LINC00263 was identified as a novel target of hnRNPK and potentiates malignant properties including proliferation and invasiveness by functioning as a decoy for miR-147a and thus upregulating calpain 2 (CAPN2) expression.
Materials and methods
Cell culture and transfection
Human cervical cancer (HeLa) cells were maintained in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA). Human non-small cell lung cancer (H460 and H1299), human colon cancer (DLD1 and LoVo), human melanoma (A375P), and human neuroblastoma (T98G and A172) cells were maintained with RPMI 1640 medium (GIBCO-BRL, Grand Island, NY, USA). Both culture media were supplemented with 10% fetal bovine serum (GIBCO-BRL, Grand Island, NY) and 1% antibiotic–antimycotic solution (GIBCO-BRL). All cell lines were recently authenticated by the STR profiling and regularly tested for mycoplasma contamination. For siRNA transfection, cells were plated at 60% density and transfected with the indicated siRNAs using Lipofectamine2000 (Invitrogen, Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. The siRNAs for HNRNPK and LINC00263 were synthesized by Bioneer (Daejeon, Republic of Korea; sequences are shown in Supplementary Table 1). CAPN2-targeting siRNA was purchased from Santa Cruz Biotechnology (sc-41459: Santa Cruz, CA). Precursor miR-147a (pre-miR-147a: PM10020) and antisense miR-147a (anti-miR-147a: AM10020) were purchased from Ambion (Ambion, Thermo Fisher Scientific, Waltham, MA) and used for overexpression or inhibition of miR-147a, respectively, using Lipofectamine2000 (Invitrogen).
Western blot analysis
Cells were lysed using a radioimmunoprecipitation (RIPA) buffer containing protease and phosphatase inhibitors (Roche, Basel, Switzerland). Equal amounts of the cell lysate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking with 5% skim milk, the membranes were incubated with the indicated primary antibody (Supplementary Table 2), washed with tris-buffered saline containing tween-20, and incubated with the appropriate secondary antibody. The protein bands were detected using an enhanced chemiluminescent reagent. GAPDH was used as a loading control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions and used as a template to synthesize cDNA, using the SuperScript III First-Strand Synthesis System (Invitrogen). The expression levels of mRNAs were quantified by RT-qPCR analysis with appropriate primers (sequences are shown in Supplementary Table 3) using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). To determine the stability of LINC00263, cells were transfected with control and HNRNPK siRNA. Following treatment of actinomycin D (0.5 μg/ml), cells were harvested at the indicated times and the levels of LINC00263 and GAPDH mRNA were determined by RT-qPCR analysis.
Determination of malignant phenotypes
The invasive ability of the cells was determined using the BD Biocoat™ Matrigel invasion chamber (BD Bioscience, San Jose, CA). Equal number of transfected cells in serum-free media were added into the upper chamber. The invasion was triggered by adding the same medium supplemented with 10% FBS to the bottom chambers as a chemoattractant. After incubation for 24 h, the invaded cells were fixed with 95% MeOH for 5 min and stained with 0.1% hematoxylin and eosin. Invasiveness was determined by counting the number of invaded cells in at least ten randomly selected fields. For analysis of cell proliferation rate, the transfected cells were plated in 6-well plates at a density of 5 × 104–1 × 105 cells/well. Cells were trypsinized and the number of viable cells was assessed under a microscope at the indicated time points. For clonogenicity assay, the transfected cells were plated in triplicate in 6-well plates and cultured for 2 weeks. Cells were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. The stained colonies were counted using the Image J program.
Cellular fractionation
A cellular fractionation assay was performed to determine the subcellular localization of LINC002638. Briefly, HeLa cells were lysed with RSB buffer (10 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 100 mM NaCl) containing 4 mg/ml digitonin (BN2006, Thermo Fisher Scientific). After centrifugation, the supernatant was collected as the cytosolic extract. The remaining nuclear pellet was washed five times with RSB buffer and then lysed with RIPA buffer. The protein levels of α-tubulin and lamin B served as markers for the cytosolic and nuclear fraction, respectively.
Ribonucleoprotein immunoprecipitation
The association of hnRNPK with LINC00263 was assessed by ribonucleoprotein immunoprecipitation (RNP-IP) using a hnRNPK-specific antibody (ChIP grade) as described in our previous report8. In case of direct interaction between miRNA and its targets, we used antibody recognizing Argonaute 2 (Ago2). Dynabeads™ Protein G (Invitrogen) was coupled with the indicated antibody followed by incubation of cytoplasmic lysate prepared using polysome extraction buffer with the antibody-conjugated beads. Following treatment with DNase I and protease K, RNAs were isolated from beads and the enrichment of target RNA level was determined by RT-qPCR analysis. The level of 18S was used for normalization in all RNP-IP experiments. Representative results are put into the figures and three independent results are presented in Supplementary figures. Details of the antibodies and primers used are provided in Supplementary Tables 2 and 3.
Antisense oligonucleotide (ASO) pull-down assay
To identify LINC00263-associated miRNAs, ASO pull-down was performed using nonoverlapping biotinylated ASOs recognizing LacZ (four ASOs) and LINC00263 (eight ASOs). Incubation of the whole-cell lysates with the biotinylated ASO was followed by coupling with Streptavidin-coupled Dynabeads™ (Invitrogen). RNAs were isolated from the pull-down materials and small RNA sequencing was performed.
Stoichiometric quantitation of LINC00263 and miR-147a by RT-qPCR and droplet digital PCR analysis (ddPCR)
We employed ddPCR to quantify the absolute RNA copy numbers of LINC00263 and the miR-147a in HeLa cells. Briefly, 1.875 μg of total RNA was reversely transcribed with either a First-Strand cDNA synthesis kit or a Mir-X miRNA First-Strand Synthesis kit. The PCR reaction using the droplets was generated using EvaGreen Supermix (1864033, Bio-Rad) containing 1 μl of cDNA for quantifying the copy numbers of LINC00263 and miR-147a. In both cases, we employed 250 nM primers of the respective RNA. The droplets were generated using the QX200™ AutoDG™ Droplet Digital™ PCR System. The PCR amplification setup was: 5 min at 95 °C, 30 s at 95 °C followed by 60 s at 60 °C for 40 cycles, 5 min at 95 °C, and then held at 4 °C. The absolute RNA copy numbers were assessed using QX200 Droplet Digital PCR System and calculated as described13. We further estimated the copy numbers per cell using a reference mRNA of known abundance as described14. Following primer sets were used: (F) ATTGGCAAGATGTTCCTTGC and (R) CAAAGCCTGTTTGGTGGTTT for LINC00263¸ and GTGTGTGGAAATGCTTCTGC for miR-147a.
Luciferase reporter assay
To verify the direct interaction between miR-147a and MRE in its target, pmirGLO dual-luciferase vectors (E133A, Promega, Madison, WI) containing wild-type or mutant MRE sequences from LINC00263 or CAPN2 mRNA were constructed. Following transfection with control or pre-miR-147a, an equal number of HeLa cells were plated into 24-well plates. Then the cells were transfected with either wild-type or mutant luciferase vector. Luciferase expression was assessed using a Dual-GLO™ Luciferase Assay System (E2940, Promega).
Results
hnRNPK is responsible for the malignant phenotypes of cancer cells
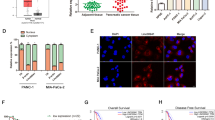
We investigated the role of hnRNPK in malignant phenotypes including metastatic potential and proliferation in HeLa cells. Individual and mixture of HNRNPK siRNAs efficiently decreased hnRNPK expression (Fig. 1a). Conversely, the introduction of Flag-hnRNPK8 resulted in a significant increase of hnRNPK in a dose-dependent manner (Fig. 1b). The invasive ability was reduced by hnRNPK knockdown (Fig. 1c); conversely, overexpression of hnRNPK enhanced the invasive ability (Fig. 1d), indicating that hnRNPK is closely associated with the invasiveness of cancer cells.
To investigate the role of hnRNPK in malignant phenotype, two individual and mixed HNRNPK siRNAs were transfected into HeLa cells (a, c, e, and g). In contrast, the Flag-hnRNPK vector was constructed and used for hnRNPK overexpression (b, d, f, and h). a, b The efficiency of hnRNPK knockdown (a) and overexpression (b) was determined by analyzing the level of hnRNPK by Western blot analysis. c, d Invasive ability was assessed by Transwell invasion assay in vitro. Representative images of the invaded cells are shown. Invasiveness was determined by counting the number of invaded cells from more than ten fields. e, f To determine the proliferation rate, an equal number of transfected HeLa cells were plated in 12-well plates and the number of viable cells was counted under a microscope at the indicated times. g, h Equal number of transfected cells were plated in six-well plates, and clonogenicity was determined by counting the number of colonies. Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t-test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
Two other distinctive features of malignancy, the proliferation rate, and clonogenicity were also examined. Knockdown of hnRNPK resulted in a decrease in the proliferation rate (Fig. 1e). On the other hand, hnRNPK-overexpressing cells showed a higher proliferation rate than the blank vector control cells (Fig. 1f). Knockdown of hnRNPK abrogated the colony-forming ability (Fig. 1g). In contrast, the number of colonies was dose-dependently increased following hnRNPK overexpression (Fig. 1h). Collectively, our findings demonstrate that hnRNPK is responsible for the malignant characteristics including high invasiveness and rapid proliferation.
LINC00263 is identified as a novel hnRNPK-regulated lincRNA
To identify hnRNPK-regulated lincRNAs, we performed RNA sequencing using hnRNPK-silenced HeLa cells (whole sequencing data were shown in Supplementary materials). The various plots representing RNA sequencing data and gene ontology (GO) analysis are shown in Supplementary Fig. 1a, c. Based on the data analysis and processing, five lincRNAs were identified to be significantly regulated by hnRNPK: two lincRNAs were upregulated and three lincRNAs were downregulated (Fig. 2a, b). Since LINC00263 showed the most significant effect on the metastatic potential (data not shown), we chose to investigate its role in the control of cancer malignancy through hnRNPK.
a, b To identify hnRNPK-regulated lincRNAs, RNA sequencing was performed using total RNAs isolated from hnRNPK- and LINC00263-silenced HeLa cells. a Volcano plots were generated by analyzing the sequencing data. b Information about the five hnRNPK-regulated lincRNAs is summarized. c, d RNA sequencing results were verified by assessing the level of LINC00263 in hnRNPK-silenced cells. Knockdown of hnRNPK by two independent siRNAs was verified by Western blot analysis (c) and the levels of HNRNPK mRNA and LINC00263 were determined by RT-qPCR analysis (d). To check whether knockdown of LINC00263 affects hnRNPK expression, the levels of hnRNPK protein (e) and HNRNPK mRNA (f) were determined by Western blot and RT-qPCR analyses, respectively. g, h To confirm that hnRNPK regulates LINC00263, a specific siRNA targeting the 3′UTR of HNRNPK mRNA was used. HeLa cells were cotransfected with the 3′UTR-specific HNRNPK siRNAs and Flag-hnRNPK vector. The level of endogenous and ectopic hnRNPK (Flag-hnRNPK) was determined by Western blot analysis. GAPDH was used as a loading control (g). The level of LINC00263 in transfected cells as described above was determined by RT-qPCR analysis (h). i Direct association of hnRNPK with LINC00263 was tested by RNP-IP experiment using control IgG and hnRNPK antibody. The enrichment of LINC00263 was calculated by comparing the level of LINC00263 in IgG and hnRNPK IP materials. The level of LINC00263 was determined by RT-qPCR analysis and 18S was used for normalization. j Following transfection of HeLa cells with wild-type (full length, FL) or four deletion mutant vectors, RNP-IP was performed using an anti-Flag antibody. The level of LINC00263 in the Flag IP was quantified by RT-qPCR analysis. The Schematic represents wild-type and four mutants of hnRNPK (ΔKH1, ΔKH1/2, ΔKH2, and ΔKH3) used in this study. k The effect of hnRNPK on the stability of LINC00263 was examined. Following treatment of actinomycin D (0.5 μg/ml), cells were harvested at the indicated times and the levels of LINC00263 and GAPDH mRNA were determined by RT-qPCR analysis. Statistical analyses were performed using the Student’s t-test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
To verify the RNA sequencing data, we performed transient knockdown of hnRNPK and observed a substantial decrease in hnRNPK expression with two individual siRNAs (Fig. 2c). Further, knockdown of hnRNPK also reduced the level of LINC00263 significantly (Fig. 2d). Although LINC00263-targeting siRNAs caused a substantial decrease in LINC00263 level, they did not affect the levels of hnRNPK protein and mRNA (Fig. 2e, f, respectively). In addition, knockdown of hnRNPK by 3′UTR-targeting siRNA efficiently decreased the expression of hnRNPK without significant change in the ectopic hnRNPK (Flag-hnRNPK) (Fig. 2g) and resulted in decreased expression of LINC00263. However, the level of LINC00263 was restored to the control level following the ectopic expression of hnRNPK (Fig. 2h).
Next, we investigated the detailed molecular mechanism by which hnRNPK regulates the expression of LINC00263. Since five hnRNPK motives are predicted in the sequence of LINC00263 (http://rbpmap.technion.ac.il/) (Supplementary Fig. 2a, b), the direct interaction between hnRNPK and LINC00263 was examined through the RNP-IP experiment. LINC00263 was found to be highly enriched in hnRNPK IP material compare to control IgG (Fig. 2i). In addition, RNP-IP using full-length Flag-hnRNPK and its various deletion mutants (ΔKH1, ΔKH1/2, ΔKH2, and ΔKH3) revealed that interaction of hnRNPK with LINC00263 was dependent on its K homology 1 (KH1) and KH2 domains (Fig. 2j). We also examined whether hnRNPK influences the stability of LINC00263 (Fig. 2k). Knockdown of hnRNPK induced a more rapid decrease in LINC00263 to compare to control. However, the level of GAPDH mRNA was barely affected by the knockdown of hnRNPK.
LINC00263 promotes malignant phenotypes including invasiveness, proliferation, and clonogenicity
To investigate whether LINC00263 is responsible for hnRNPK-mediated invasiveness, we assessed invasive ability. Knockdown of LINC00263 in HeLa cells showed a decrease in the number of invading cells (Fig. 3a). Conversely, overexpression of LINC00263 potentiated the invasive ability of HeLa cells (Fig. 3b). The level of LINC00263 in the overexpressing cells was verified by RT-qPCR analysis (Supplementary Fig. 3a). We also observed decreased proliferation rate in LINC00263-silenced cells (Fig. 3c). In contrast, the proliferation rate tended to increase in LINC00263-overexpressing cells (Fig. 3d). The colony-forming assay revealed that approximately 40% decrease in the number of colonies in LINC00263-silenced cells (Fig. 3e), while the colony-forming ability was increased following overexpression of LINC00263 (Fig. 3f). These results indicate that LINC00263 is associated with the oncogenic function of hnRNPK.
To investigate whether LINC00263 is required for hnRNPK-mediated malignant phenotypes, two independent siRNAs (a, c, and e) and overexpression vector (pcDNA/LINC00263: b, d, and f) were used. Invasiveness was determined using Transwell invasion assay (a, b). Cellular proliferation (c, d) and clonogenicity (e, f) were assessed by counting the number of viable cells and colonies, respectively, as described in “Materials and methods”. Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
miR-147a is involved in the regulation of cancer malignancy by hnRNPK/LINC00263
To determine the molecular mechanism through which LINC00263 positively regulates malignant properties, we first examined the subcellular localization of LINC00263. For verification of the appropriate cellular fractions, the levels of α-tubulin (cytosolic marker) and lamin B (nuclear marker) were analyzed in each fraction (Fig. 4a). The levels of NEAT1, MALAT1, 18S, GAPDH, and ACTB were assessed for reference (Fig. 4b). The cellular fractionation assay revealed that LINC00263 was mainly localized in the cytosol, whereas NEAT1 and MALAT1 were dominantly expressed in the nucleus. In addition, we performed Argonaute 2 immunoprecipitation (Ago2-IP) assay to examine whether LINC00263 was associated with the function of miRNAs (Fig. 4c). LINC00263 was more enriched in Ago2-IP compared to control IgG-IP, indicating that LINC00263 is involved in the regulatory pathway of miRNAs (Supplementary Fig. 5a).
a, b Cellular fractionation assay was performed to check the localization of LINC00263. To ensure the purity of the fractions, the levels of α-tubulin (cytosolic marker) and lamin B (nuclear marker) were analyzed by Western blot analysis (a). The levels of LINC00263, 18S, GAPDH, and ACTB mRNA in each fraction were determined by RT-qPCR analysis (b). c To check whether LINC00263 is involved in miRISC, Ago2 RNP-IP was performed using a specific antibody. The level of LINC00263 in control IgG and Ago2 IP materials was determined by RT-qPCR analysis and normalized to the level of GAPDH mRNA. d–f To screen for LINC00263-associated miRNAs, antisense oligonucleotide pull-down (ASO PD) was performed. A schematic of the experimental design is shown (d). Detailed information of the ASO sequences for LacZ (control) and LINC00263 was provided in Supplementary Fig. 4a. The efficiency of ASO PD was examined by comparing the level of LINC00263 in ASO PD materials (e). Small RNA sequencing was performed with RNAs isolated from ASO PD materials. miRNAs with higher expression in LINC00263 ASO PD are listed (f and Supplementary fig. 4b). g Quantification of copy numbers of LINC00263 and miR-147a (copy number per cell) was performed by RT-qPCR and ddPCR analyses. h–k HeLa cells were transfected with pre-miR-147a (for overexpression, h and j) or anti-miR-147a (for inhibition, i and k). Direct association of LINC00263 with miR-147a-involved miRISC was analyzed by Ago2 RNP-IP (h, i) and the level of LINC00263 was determined by RT-qPCR analysis (j, k). l Following the knockdown of hnRNPK and LINC00263, the level of miR-147a was determined by RT-qPCR analysis. m Bioinformatic analyses revealed that two MREs of miR-147a exist in the LINC00263 sequence (Supplementary Fig. 7a, b). To examine the sequence-specific interaction, luciferase reporter vectors containing wild-type or mutant sequences of miR-147a MREs were constructed. Following overexpression of miR-147a, the luciferase activity was assessed as described in “Materials and methods”. n–s To investigate the effect of miR-147a on malignant capabilities, pre-miR-147a (n, p, and r) or anti-miR-147a (o, q, and s) were introduced into HeLa cells. Malignant phenotypes including invasiveness (n, o), proliferation rate (p, q), and clonogenicity (r, s) were examined as described in “Materials and methods”. Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
From the above results, we hypothesized that LINC00263 may function as a competitive endogenous RNA (ceRNA) for tumor-suppressing miRNA. To identify LINC00263-associated miRNAs, ASOs for LINC00263 and LacZ were designed to perform ASO pull-down experiments (Fig. 4d and Supplementary Fig. 4a). The efficiency of the ASO pull-down was verified by determining the levels of LINC00263 and ACTB mRNA in ASO pull-down materials. Whereas ACTB mRNA was not enriched, LINC00263 was selectively enriched in the pull-down materials using the corresponding ASOs as compared to LacZ ASO (Fig. 4e). To screen LINC00263-bound miRNAs, small RNA sequencing was performed using the RNA isolated from the ASO pull-down. Analysis of sequencing data revealed that 24 miRNAs showed higher enrichment in LINC00263 ASO pull-down material than in LacZ ASO pull-down (Fig. 4f and Supplementary Fig. 4b). Next, we predicted the potential miRNA binding sites within the LINC00263 sequence using a miRNA target discovery tool RNA22 (https://cm.jefferson.edu/rna22). This bioinformatics tool revealed that LINC00263 possessed MREs for only four miRNAs (miR-147a, miR-492, miR-601, and miR-1268a) out of the 24 miRNAs found by the ASO pull-down analyses (Supplementary Fig. 4c). Since miR-147a showed the most significant folding energy, we chose to further investigate whether miR-147a was responsible for the oncogenic function of hnRNPK/LINC00263.
Under basal conditions, LINC00263 was present in 20–23 copies per cell, whereas miR-147a was present in 17–31 copies per cell as assessed by RT-qPCR and ddPCR experiments (Fig. 4g). The fact that the copy numbers are comparable in cells lends support to their stoichiometric interaction. In addition, miR-147a was found to be highly enriched in LINC00263 ASO pull-down materials (Supplementary Fig. 4d). Ago2-IP assay indicated that overexpression of miR-147a resulted in an increase in LINC00263 in Ago2 IP materials, indicating that miR-147a guided the interaction of LINC00263 with Ago2 to form miRNA-induced silencing complex (miRISC) (Fig. 4h). In contrast, inhibition of miR-147a using anti-miR-147a decreased the level of LINC00263 in Ago2-IP (Fig. 4i). Pre- and anti-miR-147a significantly increased and decreased the level of miR-147a (Supplementary Fig. 3b, c). These results indicate that LINC00263 is associated with miR-147a-guided miRISC. Decreased level of LINC00263 was observed in miR-147a-overexpressing cells compared to that in the control (Fig. 4j); conversely, LINC00263 was highly expressed following miR-147a knockdown (Fig. 4k). Accordingly, we assessed the level of miR-147a in hnRNPK- and LINC00263-silenced cells (Fig. 4l). Knockdown of both hnRNPK and LINC00263 resulted in an increase of miR-147a, indicating that LINC00263 acts as a ceRNA for miR-147a. Two MREs of miR-147a in LINC00263 were predicted by the bioinformatic tool (Supplementary Fig. 7a, b). Consequently, we constructed luciferase reporter vectors containing wild-type or mutant sequence of miR-147a MREs. In both reporter vectors, overexpression of miR-147a suppressed the expression of luciferase in the wild-type reporter vector but not in the mutant (Fig. 4m). The results of the Ago2-IP and luciferase reporter assay revealed that miR-147a directly binds to LINC00263.
Next, we tested whether miR-147a influences malignant phenotypes. Invasiveness was reduced by overexpression of miR-147a; conversely, inhibition of miR-147a resulted in increased invasive ability (Fig. 4n, o, respectively). In addition to invasiveness, proliferation rate and colony-forming ability were also regulated by miR-147a. Under conditions of high miR-147a levels, the proliferation rate and clonogenicity were diminished (Fig. 4p, r, respectively). Conversely, a decrease in miR-147a level resulted in higher proliferative and clonogenic abilities compared to those of the control (Fig. 4q, s, respectively). Collectively, we concluded that LINC00263 controls malignant properties by functioning as a ceRNA of miR-147a.
CAPN2 is a target of hnRNPK/LINC00263/miR-147a axis
To search for target genes responsible for the oncogenic function of hnRNPK/LINC00263/miR-147a, we conducted RNA sequencing using total RNA isolated from hnRNPK- and LINC00263-silenced HeLa cells (Supplementary Fig. 1a, b). TargetScan (http://www.targetscan.org) was used to predict miR-147a target genes. Eight genes (CAPN2, CCND1, CDKN1A, CSDC2, L1CAM, PAQR4, PARP12, and TRIM47) were identified as common target genes that are simultaneously regulated by hnRNPK, LINC00263, and miR-147a (Fig. 5a). RT-qPCR analysis indicated that knockdown of either hnRNPK or LINC00263 significantly decreased the level of CAPN2 mRNA, suggesting that it may be a putative target of hnRNPK/LINC00263/miR-147a (Supplementary Fig. 6). Whereas LINC00263 was highly enriched in hnRNPK IP material, CAPN2 mRNA was barely bound to hnRNPK (Fig. 5b). Overexpression of miR-147a resulted in decreased CAPN2 protein expression without any change in hnRNPK (Fig. 5c). Conversely, inhibition of miR-147a resulted in increased CAPN2 protein and mRNA expression (Fig. 5d). To determine the direct binding between CAPN2 mRNA and miR-147a, the level of CAPN2 mRNA in Ago2-IP material was assessed. Ago2-IP revealed that miR-147a increased the enrichment of CAPN2 mRNA in miRISC (Fig. 5e); conversely, knockdown of miR-147a decreased the level of CAPN2 mRNA in the Ago2-IP material (Fig. 5f). In addition to Ago2-IP, luciferase reporter vectors containing the wild-type and mutant MRE of miR-147a were constructed to confirm the direct binding of miR-147a to the 3′UTR of CAPN2 mRNA. Overexpression of miR-147a inhibited luciferase activity in wild-type vector, whereas it did not affect the expression of luciferase in mutant vector (Fig. 5g). In addition to luciferase assay, knockdown of hnRNPK and LINC00263 significantly decreased the level of CAPN2 protein and mRNA (Fig. 5h). CAPN2 mRNA was enriched in Ago2-IP following knockdown of hnRNPK or LINC00263 (Fig. 5i), suggesting that a decrease in hnRNPK strengthens the function of miR-147a by reducing LINC00263.
a By comparing RNA sequencing data and miR-147a predicted target genes (TargetScan v7), eight genes were selected as putative targets of hnRNPK/LINC00263/miR-147a. b To check whether the interaction of hnRNPK with CAPN2 mRNA is required for the regulation of its expression, an RNP-IP experiment was performed. The levels of CAPN2 mRNA and LINC00263 in each IP material were determined by RT-qPCR analysis. c–g Following transfection of HeLa cells with pre-miR-147a (for overexpression, c, e, and g) or anti-miR-147a (for inhibition, d and f), the levels of CAPN2 protein and mRNA were determined by Western blot and RT-qPCR analyses, respectively. To examine whether miR-147a directly binds to the 3′UTR of CAPN2 mRNA, Ago2 RNP-IP (e, f) and luciferase reporter assay (g) were performed. Detailed information of luciferase reporter vector is presented in Supplementary Fig. 7c. h Protein and mRNA expression of CAPN2 in hnRNPK- and LINC00263-silenced cells were determined by Western blot and RT-qPCR analyses, respectively. i To examine whether knockdown of hnRNPK or LINC00263 influences the interaction between miR-147a and CAPN2 mRNA, Ago2 RNP-IP assay was performed as described in “Materials and methods”. j–m The effect of CAPN2 silencing on malignant phenotypes including invasiveness (k), proliferation (l), and clonogenicity (m) were investigated. The efficiency of CAPN2 silencing was determined by Western blot analysis (j). Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
To validate whether CAPN2 is involved in the regulation of malignant phenotypes by hnRNPK/LINC00263/miR-147a, the effect of CAPN2 silencing on invasiveness, proliferation, and clonogenicity was examined. Introduction of CAPN2-specific siRNA into HeLa cells markedly decreased CAPN2 expression (Fig. 5j). As expected, knockdown of CAPN2 decreased the number of invading cells (Fig. 5k), inhibited cell proliferation (Fig. 5l), and suppressed colony-forming ability (Fig. 5m).
In the proteome profiler human p-kinase array, phosphorylation of ERK and p70S6K was found to be diminished in HNRNPK- or LINC00263-silenced cells compared to the controls (Supplementary Fig. 8a, b). Western blot analysis verified that knockdown of hnRNPK or LINC00263 reduced phosphorylated ERK and p70S6K. Further, decreased expression of CAPN2 using miR-147a or siRNA inhibits the activation of ERK and p70S6K (Supplementary Fig. 8c). Collectively, we demonstrated that CAPN2 was responsible for the oncogenic function as a target of hnRNPK/LINC00263/miR-147a and that ERK and p70S6K pathways are partly involved.
Repression of malignant capabilities is restored by miR-147a inhibition or CAPN2 overexpression
From the above results, we found that hnRNPK-regulated LINC00263 decoys miR-147a and thus increases CAPN2 expression. To verify our findings, we performed rescue experiments by downregulating miR147-a. The level of miR-147a was significantly decreased not only in the control but also in hnRNPK- or LINC00263-silenced cells by anti-miR-147a where miR-147a is upregulated by lowering its ceRNA, LINC00263 (Fig. 6a). Whereas knockdown of hnRNPK or LINC00263 increased the level of CAPN2 mRNA in Ago2 IP, the inhibition of miR-147a by anti-miRNA lowered the enrichment of CAPN2 mRNA in miRISC, indicating that miR-147a is responsible for the repression of CAPN2 in hnRNPK- and LINC00263-silenced cells (Fig. 6b). Inhibition of miR-147a reversed the decrease in CAPN2 protein and mRNA caused by the knockdown of hnRNPK and LINC00263 as well (Fig. 6c). Consistent with the recovery of reduced CAPN2 expression, invasiveness, and colony-forming abilities were restored by anti-miR-147a (Fig. 6d, e, respectively). These results demonstrate that miR-147a is closely involved in the regulation of CAPN2 expression, and thus plays an important role in the gain of malignant phenotypes by hnRNPK/LINC00263.
a–e To examine whether inhibition of miR-147a restores the malignant capabilities, siRNAs for hnRNPK or LINC00263 were introduced into HeLa cells with control miRNA or anti-miR-147a. Following isolation of total RNA, the level of miR-147a was determined by RT-qPCR analysis (a). Ago2 RNP-IP experiment was performed using the cytoplasmic lysates. The level of CAPN2 mRNA in Ago2 IP material was determined by RT-qPCR analysis (b). The expression levels of CAPN2 protein and mRNA were determined by Western blot and RT-qPCR analyses, respectively (c). Invasiveness (d) and colony-forming ability (e) were examined as described in “Materials and methods”. f–h For the rescue experiments, CAPN2 was ectopically overexpressed in hnRNPK- or LINC00263-silenced HeLa cells. The protein level of CAPN2 was determined by Western blot analysis (f). Invasiveness (g) and colony-forming ability (h) were examined as described in “Materials and methods”. Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
In addition to inhibition of miR-147a, we examined whether the ectopic expression of CAPN2 reverses the lowered malignant capabilities resulting from the knockdown of hnRNPK and LINC00263. The appropriate concentration of overexpression vector was determined by introducing various concentrations of Flag-CAPN2 vector (Supplementary Fig. 3d). Western blot analysis showed that knockdown of hnRNPK and LINC00263 decreased CAPN2 expression and that ectopic CAPN2 did not affect the expression of hnRNPK (Fig. 6f). As observed earlier, invasiveness was significantly decreased following the knockdown of hnRNPK and LINC00263. However, following ectopic overexpression of CAPN2, the invasive ability was restored (Fig. 6g). Consistent with the results of the invasion assay, the colony-forming assay revealed that ectopic CAPN2 restored the clonogenic ability that was reduced in the hnRNPK- and LINC00263-silenced cells (Fig. 6h). Collectively, we concluded that CAPN2 is a major effector of the oncogenic function of hnRNPK/LINC00263/miR-147a.
The ability to sponge miR-147a is required for the oncogenic potential of LINC00263
To test whether hnRNPK is required for the regulation of CAPN2 expression by LINC00263, the expression levels of CAPN2 protein and mRNA were examined in the conditions of hnRNPK presence or absence (Fig. 7a, b). Overexpression of LINC00263 showed an increase in the expression of CAPN2, regardless of the level of hnRNPK. These results suggest that the control of CAPN2 by LINC00263 resulted from inhibiting the function of miR-147a.
a, b HeLa cells were transfected with HNRNPK siRNA and/or a LINC00263 expression vector. The expression levels of CAPN2 protein (a) and CAPN2 mRNA (b) were determined by Western blot and RT-qPCR analyses, respectively. c–e LINC00263 expression vectors harboring mutant sequences of both miR-147a MREs were constructed. As with constructing the luciferase reporter vectors, four nucleotides of each miR-147a MRE in LINC00263 were changed to block the binding of miR-147a. c The invasive and clonogenic effects of three mutated LINC00263 (miR-147a MRE mutant #1, #2, and #1/#2) were determined as described in “Material and methods”. d The expression levels of CAPN2 protein and CAPN2 mRNA were determined by Western blot and RT-qPCR analyses, respectively. e Effect of mutated LINC00263 on miR-147a expression was assessed by RT-qPCR. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD). f Schematic of the proposed mechanism of action of the oncogenic hnRNPK/LINC00263/miR-147a/CAPN2-axis. A detailed description is shown in the main text.
In addition, we newly constructed LINC00263 expression vector harboring mutant sequences of two miR-147a MREs to block the binding of miR-147a (detailed information in Supplementary Fig. 10). As observed earlier, LINC00263 enhanced the invasive ability and increased the number of colonies (Fig. 7c). However, the mutants of LINC00263 did not show an increase in invasive and clonogenic abilities. Rather, they showed some inhibitory effects (Fig. 7d). It is assumed that ectopic expression of mutated LINC00263 possibly interrupts the interaction between endogenous LINC00263 and miR-147a, thus resulting in an increasing amount of working miR-147a (Fig. 7e). Collectively, we concluded that the decoying ability for miR-147a is responsible for the oncogenic functions of LINC00263.
Based on our findings, the regulatory role of hnRNPK/LINC00263/miR-147a/CAPN2 in cancer malignancy is schematically summarized in Fig. 7f. Briefly, LINC00263 is regulated by hnRNPK and functions as a ceRNA for CAPN2-targeting miR-147a. Under conditions of high hnRNPK, LINC00263 is highly expressed thereby reducing the amount of CAPN2-targeting miR-147a. Conversely, low hnRNPK results in the decreased LINC00263, which potentiates miR-147a-mediated suppression of CAPN2. Therefore, the malignant capabilities are diminished. Taken together, our data suggest that hnRNPK/LINC00263/miR-147a/CAPN2 represents a promising target for the development of cancer therapeutics.
hnRNPK/LINC00263/miR-147a/CAPN2 axis is applicable to various types of cancer cells
To generalize our findings to various types of cancer cells, the regulatory action of hnRNPK/LINC00263/miR-147a/CAPN2 was examined in two lung cancer cells (H460 and H1299). We compared the level of LINC00263 in two GSE datasets (Supplementary Fig. 9a, b)15. LINC00263 was highly expressed in non-small cell lung cancer tissues compared to nonmalignant tissues (GSE81089) and in tumor tissues compared to normal (GSE40419) tissues. In addition, we compared the level of HNRNPK mRNA and LINC00263 in two lung cancer cells with those in non-cancerous WI-38 cells (Fig. 8a). Compared to that in WI-38 cells, the expression of HNRNPK and LINC00263 was significantly increased in both lung cancer cells. Further, the expression level of HNRNPK mRNA and LINC00263 was positively correlated (Fig. 8b). Moreover, H1299 cells showed higher invasive ability than H460 cells (Supplementary Fig. 9c), indicating that the higher the invasiveness, the greater the increase of HNRNPK mRNA and LINC00263. Consistent with the previous results, knockdown of hnRNPK and LINC00263 induced a decrease of CAPN2 mRNA in both the lung cancer cells (Fig. 8c). Moreover, the introduction of pre-miR-147a and CAPN2 siRNA also decreased the expression of CAPN2 (Fig. 8d). As expected, the invasive and clonogenic abilities were diminished following the knockdown of hnRNPK and LINC00263 (Fig. 8e). We found that LINC00263 increased the number of invading cells in H460 cells (Supplementary Fig. 9d). The number of colonies was also decreased in HNRNPK- and LINC00263-silenced cells (Fig. 8f). Overexpression of miR-147a lowered invasive and colony-forming abilities of both the lung cancer cells (Fig. 8g, h, respectively). Conversely, inhibition of miR-147a induced an increase in invasiveness and clonogenicity. From these results, we confirmed that hnRNPK/LINC00263/miR-147a/CAPN2 regulatory axis is very closely related to the malignant phenotype of lung cancer cells.
To expand our findings to various types of cancer, we checked whether the regulatory mechanism of hnRNPK/LINC00263/miR-147a/CAPN2 is applicable to various types of cancers. a The relationship between the expression of HNRNPK mRNA and LINC00263 was examined in lung cancer cells by comparing them to the levels in WI-38 cells. The levels of HNRNPK mRNA and LINC00263 in WI-38, H460, and H1299 were determined by RT-qPCR analysis. b To determine whether hnRNPK regulates LINC00263, the level of LINC00263 was analyzed by RT-qPCR analysis in hnRNPK-silenced lung cancer cells. c Regulation of CAPN2 by hnRNPK and LINC00263 was verified by assessing the level of CAPN2 mRNA in hnRNPK- or LINC00263-silenced lung cancer cells. d–h To examine whether hnRNPK/LINC00263/miR-147a/CAPN2 axis regulates the invasive and clonogenic abilities, H460 and H1299 cells were transfected with siRNA targeting HNRNPK mRNA or LINC00263, or pre-miR-147a. The level of CAPN2 protein was determined by Western blot analysis (d). Invasiveness (e, g) and colony-forming ability (f, h) were examined as described in “Materials and methods”. i The effect of hnRNPK/LINC00263/miR-147a on CAPN2 expression was evaluated in various cancer cells including DLD1, LoVo, A375P, T98G, and A172. The expression level of CAPN2 and hnRNPK were determined by Western blot analysis. Bars on microscopic images represent 100 μm. Statistical analyses were performed using the Student’s t test using three independent experiments (*p < 0.05). All data represent mean ± standard variation (SD).
Next, the role of hnRNPK/LINC00263/miR-147a in the regulation of CAPN2 expression was verified in various cancer cells including DLD1, LoVo, A375, T98G, and A172 cells (Fig. 8i). All the cells tested showed suppression of CAPN2 expression as observed in HeLa and lung cancer cells. From the above results, we confirmed that our findings are applicable to various types of cancers.
Discussion
Cancer malignancy, the main cause of high cancer-related mortality, is controlled by strict and precise control of gene expression. Accumulating evidence indicates that RBPs and ncRNAs are key players in post-transcriptional gene regulation by affecting multiple steps of gene expression16. RBPs and ncRNAs are also known to modulate multiple cancer traits related to cancer progression, for instance, rapid proliferation and high metastatic potential. HnRNPK is known to be one of the most promising RBP targets for the treatment of various cancers. Typically, hnRNPK accelerates cellular proliferation and potentiates metastatic potential by upregulating a wide range of oncogenes that trigger malignant phenotypes. Recently, the interaction of hnRNPK and ncRNAs, especially lncRNAs, was reported to play a critical role in gene regulation at transcriptional and post-transcriptional levels17.
lncRNAs function as critical regulators of cancer metastasis, and their abnormal expression has been reported in many malignant tumors18,19. Accumulating evidence suggests that lncRNAs govern many cellular processes related to cancer malignancy partly by associating with various RBPs20. Recently, several lncRNAs have been reported to directly interact with hnRNPK in the cytoplasm. The elevated level of hnRNPK-associated lincRNA correlates with metastasis and poor prognosis of cancer patients, for example, c-Myc upregulated lncRNA (MYU), cancer susceptibility candidate 11 (CASC11), Ets-1 promoter-associated noncoding RNA (pancEts-1)21,22,23,24. In addition, hnRNPK-regulated lincRNAs including LINC01413 and LINC00460 potentiates the metastatic potential by inducing epithelial–mesenchymal transition (EMT)24,25.
Salmena et al.10 established the concept of ceRNA as a group of RNA transcripts that can quantitatively regulate miRNA through the sequence called MRE in ceRNA. Because miRNAs are partially complementary to the 3′UTR of the target mRNA, each miRNA has few hundreds of target genes. So far, several ncRNAs are reported to function as ceRNAs including pseudogenes, antisense transcripts, and lncRNAs26. Many ceRNAs are highly expressed in cancer cells compared to normal cells and thus miRNA is more susceptible to degradation in cancer cells. Since the functional efficacy of ceRNA is primarily dependent on its relative level, understanding how ceRNAs are controlled may be useful for the development of cancer treatments. As a ceRNA, lncRNAs compete with miRNAs for binding to their MRE in the endogenous target mRNA, thus causing a reduction and impairment of miRNA. For these reasons, ceRNAs are also termed endogenous miRNA sponges. Several lncRNAs such as MEG3 and TUG1, sequester multiple miRNAs from their target mRNA, thus leading to the derepression of target genes27,28,29,30. In addition, lncRNA H19 harboring MREs for miR-138 and miR-200a positively regulates the expression of VIM, ZEB1/2, and TWIST2, which are known EMT regulators. Therefore, lncRNA H19 potentiates metastasis by abolishing the function of EMT-suppressing miRNAs31,32.
Here, we identify LINC00263, also called oligodendrocyte maturation-associated long intergenic ncRNA (OLMALINC), as a novel hnRNPK-regulated lincRNA. And we also find that hnRNPK directly interacts with LINC00263 and stabilizes LINC00263. ASO pull-down assays and small RNA sequencing revealed that miR-147a interacts strongly with LINC00263, indicating that LINC00263 controls the repressive function of miR-147a. LINC00263 is known to be upregulated in a wide range of cancer types including lung adenocarcinoma, colorectal cancer, and renal carcinoma15. Consistent with our observation, LINC00263 was recently reported to be abnormally regulated in lung cancer15.
Comparison of RNA sequencing data and miR-147a target genes revealed that CAPN2 is a downstream effector of hnRNPK/LINC00263/miR-147a. Mechanistically, LINC00263 increases CAPN2 expression by functioning as a ceRNA of miR-147a, and thus potentiates malignant capabilities. CAPN2 is a calcium-dependent protease and known to play an important role in the proliferation and metastasis of cancer cells.33,34. It was also reported to function as an oncogene by inducing EMT and increasing expression of matrix metalloproteinase 9 (MMP9)33,35. Abnormal expression of CAPN2 is closely associated with poor prognosis of ovarian cancer patients36. Moreover, CAPN2 is also related to metastatic prostate cancer by potentiating proliferative and invasive capabilities37,38.
Our results demonstrate that LINC00263 is a novel target of hnRNPK and functions as a ceRNA for miR-147a, a CAPN2-targeting miRNA. Therefore, increased expression of LINC00263 in cancer cells suppresses the repressive effects of miR-147a, thereby increasing CAPN2 expression. CAPN2 controls the malignant properties of cancer cells partly through ERK and p70S6K. Taken together, the hnRNPK/LINC00263/miR-147a/CAPN2 axis identified in this study represents a promising target for the treatment of human cancer.
References
Demicheli, R., Dillekas, H., Straume, O. & Biganzoli, E. Distant metastasis dynamics following subsequent surgeries after primary breast cancer removal. Breast Cancer Res. 21, 57 (2019).
Feitelson, M. A. et al. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol. 35, S25–S54 (2015).
Zhuang, X., Zhang, H. & Hu, G. Cancer and microenvironment plasticity: double-edged swords in metastasis. Trends Pharm. Sci. 40, 419–429 (2019).
Siomi, H., Matunis, M. J., Michael, W. M. & Dreyfuss, G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21, 1193–1198 (1993).
Gao, R. et al. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 288, 15046–15056 (2013).
Chung, I. C. et al. Matrix metalloproteinase 12 is induced by heterogeneous nuclear ribonucleoprotein K and promotes migration and invasion in nasopharyngeal carcinoma. BMC Cancer 14, 348 (2014).
Inoue, A., Sawata, S. Y., Taira, K. & Wadhwa, R. Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl Acad. Sci. USA 104, 8983–8988 (2007).
Shin, C. H. et al. Regulation of PLK1 through competition between hnRNPK, miR-149-3p and miR-193b-5p. Cell Death Differ. 24, 1861–1871 (2017).
Shin, C. H., Ryu, S. & Kim, H. H. hnRNPK-regulated PTOV1-AS1 modulates heme oxygenase-1 expression via miR-1207-5p. BMB Rep. 50, 220–225 (2017).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Cao, M. X., Jiang, Y. P., Tang, Y. L. & Liang, X. H. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget 8, 12472–12483 (2017).
Yuan, J. H. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014).
Wojciechowska, M. et al. Quantitative methods to monitor RNA biomarkers in myotonic dystrophy. Sci. Rep. 8, 5885 (2018).
Kanno, J. et al. “Per cell” normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics 7, 64 (2006).
Liu, S. et al. Annotation and cluster analysis of long noncoding RNA linked to male sex and estrogen in cancers. NPJ Precis. Oncol. 4, 5 (2020).
Wang, Z. et al. The emerging roles of hnRNPK. J. Cell Physiol. 235, 1995–2008 (2020).
Xu, Y. et al. New Insights into the interplay between non-coding RNAs and RNA-binding protein HnRNPK in regulating cellular functions. Cells 8, 62 (2019).
Xu, Q. et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 7, e2254 (2016).
Dhamija, S. & Diederichs, S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer 139, 269–280 (2016).
Dangelmaier, E. & Lal, A. Adaptor proteins in long noncoding RNA biology. Biochim. Biophys. Acta 1863, 194370 (2020).
Kawasaki, Y. et al. MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep. 16, 2554–2564 (2016).
Zhang, Z. et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 376, 62–73 (2016).
Li, D. et al. Long noncoding RNA pancEts-1 promotes neuroblastoma progression through hnRNPK-mediated beta-catenin stabilization. Cancer Res. 78, 1169–1183 (2018).
Ji, L. et al. LINC01413/hnRNP-K/ZEB1 axis accelerates cell proliferation and EMT in colorectal cancer via inducing YAP1/TAZ1 translocation. Mol. Ther. Nucleic Acids 19, 546–561 (2020).
Li, K. et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 420, 80–90 (2018).
Karreth, F. A. & Pandolfi, P. P. ceRNA cross-talk in cancer: when ce-bling Rivalries go Awry. Cancer Discov. 3, 1113–1121 (2013).
Moradi, M. T., Fallahi, H. & Rahimi, Z. Interaction of long noncoding RNA MEG3 with miRNAs: a reciprocal regulation. J. Cell Biochem. 120, 3339–3352 (2019).
Peng, W. et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J. Exp. Clin. Cancer Res. 34, 79 (2015).
Zhou, H., Sun, L. & Wan, F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol. Lett. 18, 4393–4402 (2019).
Tang, T. et al. Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed. Pharmacother. 107, 338–346 (2018).
Liang, W. C. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525 (2015).
Yang, Q., Wang, X., Tang, C., Chen, X. & He, J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int. J. Oncol. 50, 1801–1809 (2017).
Miao, C. et al. Overexpression of CAPN2 promotes cell metastasis and proliferation via AKT/mTOR signaling in renal cell carcinoma. Oncotarget 8, 97811–97821 (2017).
Li, P. et al. Silencing CAPN2 expression inhibited castration-resistant prostate cancer cells proliferation and invasion via AKT/mTOR signal pathway. Biomed. Res. Int. 2017, 2593674 (2017).
Chen, B. et al. Calpains are required for invasive and metastatic potentials of human HCC cells. Cell Biol. Int. 37, 643–652 (2013).
Storr, S. J. et al. Calpain-2 expression is associated with response to platinum based chemotherapy, progression-free and overall survival in ovarian cancer. J. Cell Mol. Med. 16, 2422–2428 (2012).
Rios-Doria, J., Kuefer, R., Ethier, S. P. & Day, M. L. Cleavage of beta-catenin by calpain in prostate and mammary tumor cells. Cancer Res. 64, 7237–7240 (2004).
Rios-Doria, J. et al. The role of calpain in the proteolytic cleavage of E-cadherin in prostate and mammary epithelial cells. J. Biol. Chem. 278, 1372–1379 (2003).
Acknowledgements
We appreciate the support of NIA IRP, NIH for P.R.P., D.T. and M.G.
Funding
This study was supported by a grant from the Mid-career Research Program (HHK, NRF-2017R1A2A2A05069691) and the Basic Research Program (HHK, NRF-2020R1F1A1068888), through the National Research Foundation (NRF) funded by Ministry of Science and ICT, and Global Ph.D. Fellowship Program (W.J.L., NRF-2017H1A2A1045644) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education. P.R.P. and M.G. were supported by the NIA IRP, NIH.
Author information
Authors and Affiliations
Contributions
W.J.L., M.G. and H.H.K. contributed to the concept and design of the study. W.J.L., C.H.S., H.J., S.D.J., M.-S.P., H.-H.W., P.R.P. and D.T. contributed to the experimental and statistical data acquisition. W.J.L., H.-H.W., M.G. and H.H.K. contributed to the analysis and interpretation of results; W.J.L., M.G. and H.H.K. contributed to the drafting and revising of the paper. All authors carefully read the paper and agreed to the submission.
Corresponding author
Ethics declarations
Conflict of interest
This study did not require ethical approval.The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by B. Rotblat
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, W.J., Shin, C.H., Ji, H. et al. hnRNPK-regulated LINC00263 promotes malignant phenotypes through miR-147a/CAPN2. Cell Death Dis 12, 290 (2021). https://doi.org/10.1038/s41419-021-03575-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03575-1
This article is cited by
-
The role of ncRNAs in neuroblastoma: mechanisms, biomarkers and therapeutic targets
Biomarker Research (2022)
-
Exosomes derived from stem cells of human deciduous exfoliated teeth inhibit angiogenesis in vivo and in vitro via the transfer of miR-100-5p and miR-1246
Stem Cell Research & Therapy (2022)
-
Identification of a novel Calpain-2-SRC feed-back loop as necessity for β-Catenin accumulation and signaling activation in hepatocellular carcinoma
Oncogene (2022)