Abstract
Poly(ADP-ribose) polymerase 1 (PARP1) regulates gene transcription in addition to functioning as a DNA repair factor. Forkhead box O1 (FoxO1) is a transcription factor involved in extensive biological processes. Here, we report that PARP1 binds to two separate motifs on the FoxO1 promoter and represses its transcription in a polymerase-independent manner. Using PARP1-knock out (KO) cells, wild-type-PARP1-complemented cells and catalytic mutant PARP1E988K-reconstituted cells, we investigated transcriptional regulation by PARP1. PARP1 loss led to reduced DNA damage response and ~362-fold resistance to five PARP inhibitors (PARPis) in Ewing sarcoma cells. RNA sequencing showed 492 differentially expressed genes in a PARP1-KO subline, in which the FoxO1 mRNA levels increased up to more than five times. The change in the FoxO1 expression was confirmed at both mRNA and protein levels in different PARP1-KO and complemented cells. Moreover, exogenous PARP1 overexpression reduced the endogenous FoxO1 protein in RD-ES cells. Competitive EMSA and ChIP assays revealed that PARP1 specifically bound to the FoxO1 promoter. DNase I footprinting, mutation analyses, and DNA pulldown FREP assays showed that PARP1 bound to two particular nucleotide sequences separately located at −813 to −826 bp and −1805 to −1828 bp regions on the FoxO1 promoter. Either the PARPi olaparib or the PARP1 catalytic mutation (E988K) did not impair the repression of PARP1 on the FoxO1 expression. Exogenous FoxO1 overexpression did not impair cellular PARPi sensitivity. These findings demonstrate a new PARP1-gene promoter binding mode and a new transcriptional FoxO1 gene repressor.
Similar content being viewed by others
Introduction
Poly(ADP-ribose) polymerase 1 (PARP1) plays critical roles in DNA repair via catalyzing the transfer of the ADP-ribosyl group of NAD+ onto acceptor proteins (including PARP1 itself) to form poly(ADP-ribose) polymers, a process known as poly(ADP-ribosyl)ation (PARylation)1,2,3. PARP1 inhibitors (PARPis) have been shown to selectively kill homologous recombination repair (HRR) deficient cancer cells1,2,3 by increasing PARP1-DNA binding due to suppression of autoPARylation of PARP1 on DNA4. Four PARPis (olaparib, rucaparib, niraparib, and talazoparib) have been clinically used for cancer therapy, and more are undergoing clinical or preclinical tests3,5,6,7,8,9,10,11. Our recent studies have revealed that treatments of cancer cells with PARPis reduce the expression of 53BP1 or enhance the expression of COX-2, BIRC3, and SAMHD1, which contributes to cellular drug resistance12,13. These findings suggest that transcriptional regulation by PARP1 appears to affect the cellular sensitivity to PARPis or other anticancer drugs. Down-regulation of BRCA2 expression by PARP1 in an enzymatic activity dependent manner14 provides an additional supporting clue for this.
PARP1 has been reported to regulate gene transcription in several ways15,16,17,18. The transcriptional regulation by PARP1 is dependent on or independent of its polymerase activity and varies in gene-, cell type-, and context-specific manners15,16. All these indicate that the PARP1-mediated transcriptional regulation is complicated and unpredictable based on present knowledge. Therefore, further investigations, such as its DNA sequence dependency and its correlations with cellular PARPi sensitivity, are required.
We previously established PARP1-KO sublines of Ewing sarcoma RD-ES and SK-ES-1 cell lines, which were denoted as RD/KO1, RD/KO2, SK/KO1, and SK/KO2 separately4. Here, we first characterized these sublines about their responses to PARPis. Then, we conducted RNA profiling in both RD-ES and RD/KO1 cells to find changes in mRNA levels due to PARP1 loss. Following a series of analyses and verifications, Forkhead box O1 (FoxO1) was selected for further explorations because the PARP1 KO significantly increases its mRNA and protein levels in different cell lines, which was partly reversed by PARP1 complementation. Subsequently, we demonstrated by electrophoretic mobility shift (EMSA) and chromatin immunoprecipitation (ChIP) that PARP1 binds to the FoxO1 promoter. This binding was further confirmed to be DNA sequence specific by DNase I footprinting assays, EMSA, and flanking restriction enhanced pulldown (FREP) assays. Finally, the transcriptional inhibition of FoxO1 by PARP1 was shown to be independent of its enzymatic activity and cellular PARPi sensitivity.
Results
Characterization of PARP1-knockout variants of RD-ES and SK-ES-1 cells
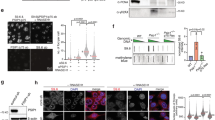
Ewing sarcoma is the fifth highest PARP1-expressing malignancy19. To investigate the transcriptional regulation by PARP1, we used cellular models generated from Ewing sarcoma RD-ES and SK-ES-1 cells by knocking out the PARP1 gene, denoted as RD/KO1, RD/KO2, SK/KO1, and SK/KO24. All these clones almost completely lost their PARP1 expression and PAR formation (Fig. 1a) and displayed ~362-fold resistance to five PARPis4. The treatment with PARPi olaparib led to apparently less increase in levels of γH2AX [a marker of DNA double-strand breaks (DSB)]20 in the PARP1-deficient cells than in their respective parental cells (Fig. 1b). The levels of central components involved in DNA damage response (DDR) such as RPA32, RAD51, CHK1, and CHK2 in both RD/KO1 cells and CRISPR-mediated PARP1 KO (Cri/KO) cells were similar to that of the parent RD-ES cells and the wild-type-PARP1-complemented RD/KO1 (RD/KO1-WT) cells4 (Fig. 1c). Notably, complementation with WT-PARP1 only partially restored PARPi sensitivity in RD/KO1-WT cells (Fig. 1d).
a Levels of PARP1 and PAR were detected by western blotting in different PARP1/KO variants (KO1 and KO2) of RD-ES and SK-ES-1 cells exposed to 200 μM H2O2 for 5 min. b Accumulation of γH2AX was reduced in PARP1/KO cells relative to parental cells treated with olaparib (0, 1, 3, or 10 μM). c Levels of DNA repair-related proteins in the RD-ES, RD/KO1, Cri/KO, and RD/KO1-WT cells were determined by western blotting. d Changes in PARPi sensitivity in response to PARP1 loss and PARP1 reconstitution. IC50 values from three independent experiments were expressed as mean ± SD. Error bars represent the SD. The resistance factor (RF) is the ratio of the averaged IC50 value of indicated PARPi in given cells to that of the same PARPi in RD-ES cells. e Volcano plots of the differentially expressed genes in RD/KO1 cells [log2 fold change >1 with statistical significance (p < 0.05)] detected by RNA-seq. Significantly upregulated and downregulated genes were colored in red and blue, respectively. X axis: log2 fold change of gene expression. Y axis: statistical significance of the differential expression in the scale of −log10 (p value). f Hierarchical clustered heatmap of differentially expressed genes in PARP1 loss cells: rows represent cell lines and columns represent genes. Genes with similar expression patterns are within the same cluster and close to each other, and they may have similar functions or participate in the same biological processes. In clustering analysis, high expression and low expression genes are colored in red and blue, respectively (Genes were shown in Supplementary Table S1 from top to bottom). g Differentially expressed genes in RD/KO1 cells involved in “pathways in cancer” and “regulation of sequence-specific DNA binding transcription factor activity” were plotted in a Venn diagram to display commonly affected genes (Genes were shown in Supplementary Table S2).
Depletion of PARP1 increases FoxO1 expression
To identify target genes transcriptionally regulated by PARP1, we conducted transcription profiling in RD/KO1 cells and parental cells by RNA-seq. The results showed that the expression of 492 genes changed significantly in PARP1/KO cells [log2 fold change >1 with statistical significance (p < 0.05)]. The volcano plot (Fig. 1e) and the hierarchical clustered heatmap (Fig. 1f) revealed that among these expression-changed genes, 277 genes were upregulated, and 215 genes were downregulated (These genes in hierarchical clustered heatmap were shown in Supplementary Table S1 from top to bottom.) Among them, KEGG analysis further demonstrated that 23 genes were involved in “pathways in cancer” while the GO analysis indicated that 20 genes participated in “regulation of sequence-specific DNA binding transcription factor activity.” Interestingly, 10 genes were common to both (Fig. 1g). Among the 10 genes, the expression of seven genes changed obviously (log2 fold change >2), including TNF alpha induced protein 3 (TNFAIP3; up), nuclear factor kappa B subunit 2 (NF-κB2; up), nuclear factor kappa B subunit 1 (NF-κB1; up), NF-κB inhibitor alpha (NF-κBIA, also known as IκBα; up), FoxO1 (up), inhibitor of nuclear factor kappa B kinase subunit beta (IκBKB; down), and androgen receptor (AR; down) (Supplementary Table S2).
PARP1 has been reported to support the transcriptional function of AR17. Additionally, the above results show that genes related to NF-κB1 signaling might be affected most due to PARP1 loss. Thus, to verify the results from the transcription profiling, we used parental cells (RD-ES and SK-ES-1), PARP1-KO cells (RD/KO1 and SK/KO1), and their WT-PARP1-complemented cells (RD/KO1-WT and SK/KO1-WT)4. RT-qPCR revealed that though the mRNA levels of TNFAIP3, IκBα, and NF-κB1 were strikingly elevated due to PARP1 loss, complementation with WT-PARP1 did not reduce their elevation (Fig. 2a). Moreover, western blotting further showed that PARP1 loss caused increased TNFAIP3 protein levels but no change in IκBα or NF-κB1 levels. In addition, PARP1 reconstitution led to no (IκBα or NF-κB1) or only weak (TNFAIP3) changes at the protein levels (Fig. 2b). These inconsistent results indicated that genes related to NF-κB1 signaling might not be regulated by PARP1, at least not in detected Ewing sarcoma cells.
a Confirmation of some results from RNA-seq by RT-qPCR in different cells. Log2 mRNA levels of TNFAIP3, IκBα, and NF-κB1 in PARP1-KO or complemented cells were normalized to that in corresponding parental cells. Error bars represent the SD. b Confirmation of some results from RNA-seq by western blotting in indicated cells. c Loss of PARP1 increased mRNA and protein levels of FoxO1, which was prevented, at least partially, by PARP1 complementation. The mRNA levels of FoxO1 were detected by RT-qPCR and normalized to that in the corresponding parental cells; Error bars represent the SD. *, p < 0.05. Protein levels of FoxO1 were detected by western blotting. d After RD-ES cells were transfected with GFP-PARP1 cDNA for 72 h, protein levels of FoxO1 were determined by western blotting. The relative FoxO1 levels were presented as the ratio of (FoxO1/GAPDH)GFP-PARP1/(FoxO1/GAPDH)CON when the value of (FoxO1/GAPDH)CON was normalized as 1.
Therefore, we turned to FoxO1, which encodes a transcription factor that regulates gene expression, controlling various cellular processes21. For changes in FoxO1 gene expression caused by either PARP1 loss or complementation in different cells, both RT-qPCR and western blotting provided consistent results to the transcription profiling (Fig. 2c and Supplementary Table S1). RNA-seq revealed increased FoxO1 mRNA levels up to more than five times in RD/KO1 cells (Supplementary Table S1), while RT-qPCR showed 2.7–6.8-fold increases in PARP1-KO Ewing sarcoma RD/KO1, Cri/KO and SK/KO1 cells and pancreatic CAPAN1/KO cells (Fig. 2c, left). Similar increases in FoxO1 protein levels were observed in these cells (Fig. 2c, middle and right). Notably, PARP1 complementation could reduce, though not eliminate, the PARP1 loss-mediated increase in either mRNA or protein levels of this gene (Fig. 2c). To further validate these results, we overexpressed PARP1 by transfecting GFP-PARP1 into RD-ES. The result showed that exogenous PARP1 overexpression could reduce endogenous FoxO1 protein levels by 44% (Fig. 2d). These results indicate that PARP1 negatively regulates FoxO1 gene transcription. Moreover, transcriptional regulation of FoxO1 by PARP1 is not just limited to Ewing sarcoma cells; similar changes were observed in CAPAN1 cells (Fig. 2c). Furthermore, though FoxO1 has been reported to be a direct target gene of EWS-FLI1 in Ewing sarcoma cells21, this regulation is independent of EWS-FLI1 because it does not exist in CAPAN1 cells.
PARP1 binding to the FoxO1 promoter
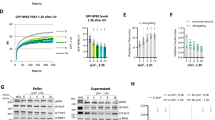
To demonstrate how PARP1 regulates FoxO1 transcription, we evaluated whether PARP1 binds to the FoxO1 promoter. To conduct the EMSA assay, we used three FAM-labeled fragments correspondingly located at −753 to −1032 (FoxO1-L), −1289 to −1565 (FoxO1-M), and −1678 to −1995 (FoxO1-R) upstream of the transcription start site (TSS) in the FoxO1 promoter region as probes (Fig. 3a; the 2 kb promoter region of the human FoxO1 gene and the location and sequence of FoxO1-L, FoxO1-M, and FoxO1-R were shown in Supplementary Table S3). The result showed a clear probe band in the control group (Lane 1, no PARP1 for each panel in Fig. 3b). As PARP1 was added by increasing amounts of 2 to 10 μg, the probe band for each group was progressively reduced in size and finally disappeared while more DNA-protein complexes formed (Lanes 2–4; 2, 5, and 10 μg PARP1 for each panel in Fig. 3b). Importantly, adding an excess of 40-fold of unlabeled cold probe DNA partially recovered the probe band and almost completely eliminated FAM-labeled DNA-protein complexes (Lane 5; 10 μg PARP1 for each panel in Fig. 3b). The results indicate that PARP1 specifically binds to DNA sequences that consist of the FoxO1 promoter region in the in vitro system.
a Schematic representation of the locations of particular nucleotide fragments (FoxO1-L, -M, and -R) on the FoxO1 promoter analyzed by EMSA. b The binding of purified PARP1 to particular nucleotide fragments (FoxO1-L, -M, and -R) were analyzed by EMSA. For the competition assays, 40-fold excess of unlabeled DNA fragments were added to the reaction mixture before adding FAM-labeled probes, and the labeled PARP1-FoxO1 complexes were almost completely displaced. c RD-ES, RD/KO1, and RD/KO1-WT cells were subjected to ChIP analyses using the antibody against PARP1 and an isotype-matched IgG as a negative control. The association of PARP1 with the FoxO1 gene promoter was quantified by RT-qPCR using primers targeting FoxO1-L, -M, and -R, respectively. Error bars represent the SD. d Identification of PARP1-protected regions on FoxO1-L and FoxO1-R by DNase I footprinting assays. Electropherograms showed the whole region of the FoxO1-L/R after digestion with DNase I following incubation in the presence (blue) or absence (red) of PARP1. The DNA sequences of the PARP1-protected regions were marked with dashed rectangles and denoted as FoxO1-L-B (−813TCACTGTATTCTT−826) and FoxO1-R-B (−1805TCTTGTGGTCTCTTCACGTTTAC−1828).
To confirm the in vitro data, we conducted a ChIP assay using RD-ES, RD/KO1 and RD/KO1-WT cells. The results showed that endogenous PARP1 protein bound to the FoxO1 promoter at the L, M, and R regions in RD-ES and RD/KO1-WT cells, while PARP1 binding in RD/KO1 cells was comparable to that of IgG in the same regions of the FoxO1 promoter in all tested cells (Fig. 3c). These data further strengthen the conclusion that PARP1 can specifically bind to the FoxO1 promoter.
To more accurately define the DNA sequences to which PARP1 binds in the FoxO1 promoter, we performed DNase I footprinting assays with purified PARP1 protein and DNA fragments (L, M, and R) corresponding to the FoxO1 promoter. The results showed that 30 μg PARP1 produced stronger protection against the DNase I-mediated degradation of L and R than M [Fig. 3d (the indicated sequences) and Supplementary Figure S1]. Two regions (denoted as FoxO1-L-B and FoxO1-R-B) that seemed to be protected were located at −813 bp to −826 bp and −1805 bp to −1828 bp on the FoxO1 promoter (Fig. 3d). The results indicate that PARP1 is likely to bind to the FoxO1 promoter via these two regions and regulate FoxO1 transcription.
The binding of PARP1 to the FoxO1 promoter is sequence-specific
As a DNA repair factor, PARP1 binds to single or double-strand broken DNA without any apparent sequence preference22,23. To demonstrate whether PARP1 binding to the FoxO1 promoter has sequence specificity, we used probes containing FoxO1-L-B and FoxO1-R-B and their respective mutants containing three or four point mutations (Fig. 4a) as probes in an EMSA assay. Data showed that these point mutations did not cause detectable changes in PARP1 binding to the probe DNA (Lane 3 vs Lane 2 and Lane 6 vs Lane 5 in Fig. 4b). In contrast, complete deletions of FoxO1-L-B and FoxO1-R-B (Fig. 4a) increased unbound probes (Lane 3 vs Lane 2 and Lane 6 vs Lane 5 in Fig. 4c) probably due to PARP1 binding to the probe DNA. However, these deletions did not fully restore unbound probes to control levels (Lane 3 vs Lane 1 and Lane 6 vs Lane 1 in Fig. 4c), revealing PARP1 non-specific binding, probably because the probes had 2 ends comparable to broken DNA. Nevertheless, the results reveal that PARP1 indeed binds to the FoxO1 promoter at the −813 to −826 bp (FoxO1-L-B) and −1805 to −1828 bp (FoxO1-R-B) regions in a sequence-specific manner.
a The FAM-labeled probes containing sequences of FoxO1-L-B (left) and FoxO1-R-B (right) and corresponding mutated probes (underlined; FoxO1-L-B-M and FoxO1-R-B-M), and the deleted sequences (line-through; FoxO1-L-B-D and FoxO1-R-B-D). b EMSA was carried out using normal or mutated FAM-labeled probes. The amount of DNA-protein complexes detected in FAM-labeled mutant probes was similar to that in FAM-labeled normal probes. c EMSA was carried out using FAM-labeled normal and deletion probes. Fewer DNA-protein complexes were detected with FAM-labeled deletion probes than the FAM-labeled normal probes followed by PARP1 incubation. d Schematic representation of the flanking restriction enhanced pulldown (FREP). A biotinylated DNA fragment is conjugated to streptavidin-coated magnetic Dynabeads (Invitrogen, Carlsbad, CA). This fragment is engineered to include the FoxO1-L-B or FoxO1-R-B specific (“bait”) sequence (black dashed box), flanked by restriction enzyme cleavage sites for BamH I proximally (gray dashed box) and EcoR I distally (gray box). DNA-beads are mixed with PARP1 protein. A free non-biotinylated FoxO1-L-B or FoxO1-R-B DNA fragment can be included in the control reaction at this stage as a specific competitor. Magnetic separation and wash remove non-DNA binding PARP1 protein. EcoR I digestion releases 3′ DNA end-binding PARP1, and BamH I digestion separates the sequence-specific FoxO1-L-B or FoxO1-R-B binding PARP1 from the 5′ DNA and Dynabeads. Western blotting identifies PARP1 binding to FoxO1-L-B or FoxO1-R-B. e The PARP1-DNA complexes cut with EcoR I and BamH I were detected by western blotting. The relative levels of FoxO1-bound PARP1 were presented as the ratio of FoxO1-bound PARP1 band intensity/PARP1 input band intensity when the value of PARP1 input band intensity was normalized as l. Lane 1: PARP1 inputs, lane 2: labeled FoxO1-L-B-beads, lane 3: PARP1 with labeled FoxO1-L-B-beads, lane 4: PARP1 with labeled FoxO1-L-B-beads and cold competitor (40-fold excess of free FoxO1-L-B-beads), lane 5: labeled FoxO1-R-B-beads, lane 6: PARP1 with labeled FoxO1-R-B-beads, lane 7: PARP1 with labeled FoxO1-R-B-beads and cold competitor (40-fold excess of free FoxO1-R-B-beads), lane 8: a labeled non-specific DNA sequence (NS-beads) and lane 9: PARP1 with labeled NS-beads.
Our conclusion was further supported by data from a novel DNA pulldown assay termed FREP that was done to minimize detectable non-specific PARP1 binding through restriction enzyme digestion24. To do this assay, the 3′ free end of the DNA was cleaved off by EcoR I and the single-stranded DNA that was not cleaved off with BamH I was therefore discarded along with the bead (Fig. 4d). FoxO1-L-B and FoxO1-R-B were used as corresponding competitors of the biotin-labeled FoxO1-L-B and FoxO1-R-B, while a non-specific 31 bp DNA sequence (NS) labeled with biotin was used as the control for non-specific PARP1 binding. The PARP1-DNA complexes digested with EcoR I and BamH I were detected by western blotting with an anti-PARP1 antibody. Results (Fig. 4e) revealed that PARP1 bound to the biotin-labeled FoxO1-L-B (Lane 3) and FoxO1-R-B (Lane 6) much more than the control (Lane 9). Importantly, PARP1 bound to biotin-labeled FoxO1-L-B and FoxO1-R-B was largely competed away by free FoxO1-L-B and FoxO1-R-B, respectively (Lane 4 and 7, Fig. 4e). Therefore, these data further indicate that PARP1 sequence-specific binding to the FoxO1 promoter inhibits FoxO1 transcription.
PARP1 transcriptional regulation of FoxO1 is independent of its catalytic activity
PARP1 has been shown to regulate gene transcription in two modes independent of or dependent on its catalytic activity15. To test whether its poly(ADP-ribose) polymerase activity is required for its transcriptional regulation on FoxO1, we treated RD-ES and SK-ES-1 cells with the PARPi olaparib. Olaparib inhibits the PARP1 enzymatic activity but does not affect PARP1 expression3,25. The treatments with olaparib did not cause obvious changes in mRNA or protein levels of FoxO1 (Fig. 5a and Supplementary Fig. S2). To verify this result, we used a catalytic mutant of PARP1 by replacing the glutamic acid residue at 988 with a lysine residue (E988K) to complement RD/KO1 (resulting cells, RD/KO1-E988K)4. E988K has no poly(ADP-ribose) polymerase activity but keeps the mono-ADP-ribosyl-transferase activity4. The expression of E988K in RD-ES/KO1 cells led to apparent decreases in FoxO1 mRNA and protein levels (Fig. 5b, c). These results further indicate that transcriptional regulation of FoxO1 by PARP1 is independent of its catalytic activity.
a RD-ES (upper) and SK-ES-1 (lower) cells were incubated in the indicated concentrations of olaparib for 24 h or 48 h. Then, mRNA levels of FoxO1 were detected by RT-qPCR. b mRNA levels of FoxO1 in RD/KO1 cells and their stably-transfected with mutated-PARP1 cDNA (E988K) variants were detected by RT-qPCR. c Protein levels of FoxO1 in RD/KO1 cells and their stably-transfected with mutated-PARP1 cDNA (E988K) variants were detected by western blotting. The relative FoxO1 levels were presented as the ratio of (FoxO1/GAPDH)KO1 or E988K/(FoxO1/GAPDH)RD-ES when the value of (FoxO1/GAPDH)RD-ES was normalized as l. Data were expressed as mean ± SD from three independent experiments. d Effects of PARP1 loss and PARP1 reconstitution on the expression of FoxO1 target genes. mRNA levels were detected by RT-qPCR. *, p < 0.05. e RD-ES cells were transfected with GFP-FoxO1 cDNA for 72 h and 96 h, and the mRNA (left) and protein (right) levels of FoxO1 were detected by RT-qPCR and western blotting, respectively. f Survival curves of olaparib, niraparib and talazoparib-treated RD-ES and FoxO1-overexpressed RD-ES (RD-ES FoxO1 OE) cells assessed by CCK-8 assays. Error bars represent the SD. g Changes of the mRNA levels of FoxO1 and its target genes. RD-ES cells were treated with cisplatin, carmustine and temozolomide for 12 h. Then, mRNA levels of FoxO1, p21, PUMA and Bim were detected by RT-qPCR and normalized to those in RD-ES cells without any treatments. *, p < 0.05. h FoxO1 overexpression by transfecting GFP-FoxO1 into RD-ES cells was determined by western blotting. i Sensitivity of RD-ES, RD/KO1 and RD-ES FoxO1 OE cells to cisplatin, carmustine and temozolomide. Cells were exposed to gradient concentrations of the tested agents for 72 h. IC50 values from three independent experiments were expressed as mean ± SD. Error bars represent the SD. *, p < 0.05.
FoxO1 does not contribute to the sensitivity of RD-ES cells to PARP inhibitors
We further evaluated the effects of PARP1 loss and reconstitution on the target genes of the transcription factor FoxO1. The data revealed that loss and exogenous re-expression of PARP1 in RD-ES cells caused similar significant changes in mRNA levels of FoxO1 and p21, one of the target genes of FoxO126. However, only marginal changes took place in mRNA levels of PUMA and Bim, the other two target genes of FoxO127 (Fig. 5d).
We demonstrate that PARP1 loss leads to FoxO1 overexpression and cellular resistance to PARP inhibitors. Therefore, we further investigated whether FoxO1 overexpression contributed to PARPi resistance. We established a FoxO1-overexpressed model by transfecting GFP-FoxO1 into RD-ES cells that normally express PARP1 protein. FoxO1 overexpression was verified in Fig. 5e. However, FoxO1 overexpression did not change the cellular sensitivity of PARPis olaparib, niraparib, or talazoparib (Fig. 5f), indicating that there is no correlation between FoxO1 expression and the sensitivity of RD-ES cells to PARPis.
FoxO1 has been shown to regulate cellular sensitivity to several DNA damaging agents28. We thus evaluated the changes in the mRNA levels of FoxO1 and its target genes in RD-ES cells exposed to cisplatin, carmustine, and temozolomide. As shown in Fig. 5g, only carmustine increased the mRNA levels of FoxO1 at its high concentration. Notably, changes in the mRNA levels of p21, PUMA, and Bim were different in response to different treatments.
Then we tested the sensitivity of RD-ES, RD/KO1, and FoxO1-overexpressed RD-ES (RD-ES FoxO1 OE) (Fig. 5h) cells to cisplatin, carmustine, and temozolomide. The results suggested that the decreased sensitivity of RD/KO1 cells to cisplatin was probably not caused by increased FoxO1 expression. In contrast, the sensitivity of both RD/KO1 and RD-ES FoxO1 OE cells to carmustine significantly increased while the sensitivity of these cells to temozolomide basically kept unchanged relative to that of RD-ES cells (Fig. 5i). Carmustine but not the other two alkylating agents showed some dependency on FoxO1 in its killing RD-ES, which was consistent with the fact that cisplatin and temozolomide did not cause significant changes in the mRNA levels of FoxO1 while carmustine could significantly increase the mRNA levels of FoxO1 at 50 μM (Fig. 5g).
Discussion
Recent studies on PARP1 have been largely focused on its functions in DNA repair, primarily due to the successful clinical uses of PARPis. This may be a reason why our understanding on PARP1 transcriptional regulation has been less established. In this study, we investigated PARP1-mediated gene transcription by using PARP1-KO cells which did not respond to PARPi treatments. FoxO1, encoding a transcription factor, was up-regulated in its mRNA and protein levels in the PARP1-KO Ewing sarcoma RD/KO1, Cri/KO, and SK/KO1 cells and pancreatic cancer CAPAN1/KO cells. Importantly, PARP1 complementation prevented PARP1 loss-mediated increase in its mRNA or protein levels, though partly in some cells. The results indicate that PARP1 represses FoxO1 gene transcription. We subsequently demonstrated that PARP1 specifically bound to the FoxO1 promoter. It was also revealed that regions of −813 to −826 bp (i.e., 5′-TCACTGTATTCTT-3′) and −1805 to −1828 bp (i.e., 5′-TCTTGTGGTCTCTTCACGTTTAC-3′) upstream of the TSS on the FoxO1 promoter were required for this binding, indicating its DNA sequence dependency or specificity. Moreover, negative transcriptional regulation of FoxO1 by PARP1 was independent of its enzymatic activity. This may be why FoxO1 expression is not correlated with cellular PARPi sensitivity because almost all the present PARPis are enzymatic inhibitors. Nevertheless, our data suggest a possible correlation of the transcriptional regulation of FoxO1 by PARP1 with the carmustine sensitivity in RD-ES cells. This provides a direction for our future exploration.
As a DNA repair factor, PARP1 can bind to DNA in a DNA sequence-independent manner, and inhibition of its enzymatic activity increases this binding4,23,29. In striking contrast, when functioning as a transcriptional regulation factor, the binding of PARP1 to gene promoter regions requires particular DNA sequences. For example, PARP1 was shown to bind to the 5′-GTTTCACAAT-3′ sequence in the BRCA2 promoter14, to the 5′-GCTGTGGGAA-3′ sequence in the Tcirg1 promoter29, to the 5′-ATGGTcttACCTA-3′ sequence in the HFE promoter30, to the 5′-GTTG-3′ sequence in the CXCL1 promoter31, and to the 5′-TGTTG-3′ sequence in the cTnT promoter32. The PARP1 binding results in negative transcriptional regulation of the former three genes but positive regulation of the latter two. Different from only a single-nucleotide sequence motif that is required for the PARP1 binding in these studies14,29,30,31,32, our data reveal that two specific motifs (i.e., 5′-TCACTGTATTCTT-3′ and 5′-TCTTGTGGTCTCTTCACGTTTAC-3′) are needed for FoxO1 transcriptional inhibition. The two motifs are separately located at the regions at a distance of 979 bp on the FoxO1 promoter. This is a new PARP1-gene promoter binding mode. Notably, the 5′-TG-3′ nucleotide sequence appears in the above mentioned PARP1 binding motifs (except BRCA2). However, this sequence might not be a critical consensus nucleotide sequence for the PARP1-gene promoter binding because our result revealed that the mutation of 5′-TG-3′ did not change the PARP1 binding. Therefore, previous studies and our own indicate that transcriptional regulation by PARP1, at least by way of nucleotide-sequence-dependent binding to the gene promoter, is gene specific in the aspects of PARP1-bound DNA sequence(s), transcriptional inhibition or stimulation and PARP1-polymerase dependency. At present, the issues on this specificity remain to be explored, particularly including what the determining factors are, whether any other cofactors are involved, and which domain(s) of PARP1 contribute to specific binding.
The transcription factor FoxO1 is a member of the FoxO family and regulates diverse gene expression in controlling various biological processes such as tumorigenesis and aging33. FoxO1 can function as a tumor suppressor. On the one hand, FoxO1 inhibits cancer cell proliferation by activating the transcription of p21, encoding a cell cycle inhibitor26; on the other, FoxO1 induces apoptosis by upregulating expression of several pro-apoptotic factors including PUMA and Bim26,34. Moreover, FoxO1 also affects cellular sensitivity or resistance to anticancer drugs26,28,34,35,36. Though its post-transcriptional modifications and its transcriptional control of target genes have been extensively investigated37, relatively little is known about transcriptional regulation of the FoxO1 gene itself. Our current study reveals that PARP1 binds to the FoxO1 promoter and represses FoxO1 expression, which is independent of PARP1 enzymatic activity. Knockout and complementation of PARP1 separately caused a consistent increase and reduction in both mRNA and protein levels of FoxO1. Notably, cells that normally express PARP1 and that are complemented with PARP1 (WT or mutated) have low levels of FoxO1 expression. Therefore, PARP1 binding appears not to affect the basal expression of the FoxO1 gene. This conclusion might be supported also by the finding that in HEK293T cells transfected with both FLAG-PARP1 and HA-FoxO1, both proteins can be detected at the same time38. Therefore, in addition to transcription factors E2F-139, FoxC140, FoxO341, and EWS-FLI121, PARP1 is another new transcriptional regulator of the FoxO1 gene by direct binding to its promoter.
Collectively, this study demonstrates a new PARP1-gene promoter binding mode evidenced by direct PARP1 binding to two separate motifs on the FoxO1 promoter. PARP1 is a new transcriptional repressor of FoxO1, encoding an important transcription factor with extensive biological functions. The regulation of FoxO1 expression by PARP1 is independent of its polymerase activity and cellular PARPi sensitivity. These findings provide new insights into both PARP1 functions and FoxO1 transcriptional regulation, helping to further understand the roles of PARP1 and FoxO1 in tumorigenesis and cancer therapy.
Materials and methods
Details about drugs and antibodies, cell culture, stable KO of PARP1 with the CRISPR/Cas9 technique, RNA sequencing (RNA-seq), quantitative real-time polymerase chain reaction (RT-qPCR), EMSA and competitive binding assays, ChIP, DNase I footprinting assays, and FREP are provided in Supplementary Materials and Methods.
Western blotting
Standard western blotting42 was used to detect the changes in protein levels caused by the indicated treatments.
Cytotoxicity assays
Cell Counting Kit 8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) assays were used to detect cytotoxicity as previously described43.
Plasmid construction, PARP1 protein purification, generation of cells expressing PARP1 or its mutants
Plasmid construction, PARP1 protein purification, generation of cells expressing PARP1, or its mutants were conducted as previously reported4.
Statistical analysis
All data were representative of three independent experiments. Data were presented as mean ± SD (standard deviation), and no data was excluded from analysis. A Student’s t test (two-tailed) was used to compare two groups. Only p < 0.05 was considered statistically significant.
References
Ding, J., Miao, Z. H., Meng, L. H. & Geng, M. Y. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol. Sci. 27, 338–344 (2006).
He, J. X., Yang, C. H. & Miao, Z. H. Poly(ADP-ribose) polymerase inhibitors as promising cancer therapeutics. Acta Pharmacol. Sin. 31, 1172–1180 (2010).
Wang, Y. Q. et al. An update on poly(ADP-ribose)polymerase-1 (PARP-1) inhibitors: opportunities and challenges in cancer therapy. J. Med. Chem. 59, 9575–9598 (2016).
Chen, H. D. et al. Increased PARP1-DNA binding due to autoPARylation inhibition of PARP1 on DNA rather than PARP1-DNA trapping is correlated with PARP1 inhibitor’s cytotoxicity. Int. J. Cancer 145, 714–727 (2019).
Yuan, B. et al. Poly(ADP-ribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett. 386, 47–56 (2017).
He, J. X. et al. Novel PARP1/2 inhibitor mefuparib hydrochloride elicits potent in vitro and in vivo anticancer activity, characteristic of high tissue distribution. Oncotarget 8, 4156–4168 (2017).
Ye, N. et al. Design, synthesis, and biological evaluation of a series of benzo[de][1,7]naphthyridin-7(8H)-ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J. Med. Chem. 56, 2885–2903 (2013).
Chen, J. Y. et al. Synthesis of isoquinolinone-based tricycles as novel poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. Bioorg. Med. Chem. Lett. 24, 2669–2673 (2014).
Chen, X. X. et al. Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors. Eur. J. Med. Chem. 145, 389–403 (2018).
Chen, W. H. et al. Discovery of potent 2,4-difluoro-linker poly(ADP-ribose) polymerase 1 inhibitors with enhanced water solubility and in vivo anticancer efficacy. Acta Pharmacol. Sin. 38, 1521–1532 (2017).
Chen, W. H. et al. Discovery, mechanism and metabolism studies of 2,3-difluorophenyl-linker-containing PARP1 inhibitors with enhanced in vivo efficacy for cancer therapy. Eur. J. Med. Chem. 138, 514–531 (2017).
Wang, Y. T. et al. Acquired resistance of phosphatase and tensin homolog-deficient cells to poly(ADP-ribose) polymerase inhibitor and Ara-C mediated by 53BP1 loss and SAMHD1 overexpression. Cancer Sci. 109, 821–831 (2018).
Yang, Z. M. et al. Combining 53BP1 with BRCA1 as a biomarker to predict the sensitivity of poly(ADP-ribose) polymerase (PARP) inhibitors. Acta Pharmacol. Sin. 38, 1038–1047 (2017).
Wang, J. H. et al. Poly(ADP-ribose) polymerase-1 down-regulates BRCA2 expression through the BRCA2 promoter. J. Biol. Chem. 283, 36249–36256 (2008).
Schiewer, M. J. & Knudsen, K. E. Transcriptional roles of PARP1 in cancer. Mol. Cancer Res. 12, 1069–1080 (2014).
Kraus, W. L. & Hottiger, M. O. PARP-1 and gene regulation: progress and puzzles. Mol. Asp. Med. 34, 1109–1123 (2013).
Schiewer, M. J. et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2, 1134–1149 (2012).
Frizzell, K. M. et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 284, 33926–33938 (2009).
Ohali, A. et al. Prediction of high risk Ewing’s sarcoma by gene expression profiling. Oncogene 23, 8997–9006 (2004).
Chao, O. S. & Goodman, O. B. Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol. Cancer Res. 12, 1755–1766 (2014).
Yang, L., Hu, H. M., Zielinska-Kwiatkowska, A. & Chansky, H. A. FOXO1 is a direct target of EWS-Fli1 oncogenic fusion protein in Ewing’s sarcoma cells. Biochem. Biophys. Res. Commun. 402, 129–134 (2010).
Gradwohl, G. et al. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA 87, 2990–2994 (1990).
Langelier, M. F. & Pascal, J. M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 23, 134–143 (2013).
Li, G. et al. The rheumatoid arthritis risk variant CCR6DNP regulates CCR6 via PARP-1. PLoS Genet. 12, e1006292 (2016).
Kim, G. et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 21, 4257–4261 (2015).
Farhan, M. et al. FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 13, 815–827 (2017).
Calnan, D. R. & Brunet, A. The FoxO code. Oncogene 27, 2276–2288 (2008).
Chen, C. et al. FOXO1 associated with sensitivity to chemotherapy drugs and glial-mesenchymal transition in glioma. J. Cell Biochem. 120, 882–893 (2019).
Beranger, G. E. et al. RANKL treatment releases the negative regulation of the poly(ADP-ribose) polymerase-1 on Tcirg1 gene expression during osteoclastogenesis. J. Bone Miner. Res. 21, 1757–1769 (2006).
Pelham, C. et al. Regulation of HFE expression by poly(ADP-ribose) polymerase-1 (PARP1) through an inverted repeat DNA sequence in the distal promoter. Biochim. Biophys. Acta 1829, 1257–1265 (2013).
Nirodi, C. et al. A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression. J. Biol. Chem. 276, 9366–9374 (2001).
Huang, K. et al. Analysis of nucleotide sequence-dependent DNA binding of poly(ADP-ribose) polymerase in a purified system. Biochemistry 43, 217–223 (2004).
Coomans de Brachene, A. & Demoulin, J. B. FOXO transcription factors in cancer development and therapy. Cell. Mol. Life Sci. 73, 1159–1172 (2016).
Hornsveld, M., Dansen, T. B., Derksen, P. W. & Burgering, B. M. T. Re-evaluating the role of FOXOs in cancer. Semin. Cancer Biol. 50, 90–100 (2018).
Gao, J. et al. The involvement of FoxO in cell survival and chemosensitivity mediated by Mirk/Dyrk1B in ovarian cancer. Int. J. Oncol. 40, 1203–1209 (2012).
Beretta, G. L., Corno, C., Zaffaroni, N. & Perego, P. Role of FoxO proteins in cellular response to antitumor agents. Cancers 11, 90 (2019).
Xing, Y. Q. et al. The regulation of FOXO1 and its role in disease progression. Life Sci. 193, 124–131 (2018).
Sakamaki, J., Daitoku, H., Yoshimochi, K., Miwa, M. & Fukamizu, A. Regulation of FOXO1-mediated transcription and cell proliferation by PARP-1. Biochem. Biophys. Res. Commun. 382, 497–502 (2009).
Nowak, K., Killmer, K., Gessner, C. & Lutz, W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim. Biophys. Acta 1769, 244–252 (2007).
Berry, F. B. et al. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum. Mol. Genet. 17, 490–505 (2008).
Essaghir, A., Dif, N., Marbehant, C. Y., Coffer, P. J. & Demoulin, J. B. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J. Bacteriol. 284, 10334–10342 (2009).
Yi, J. M. et al. Triptolide induces cell killing in multidrug-resistant tumor cells via CDK7/RPB1 rather than XPB or p44. Mol. Cancer Ther. 15, 1495–1503 (2016).
Wang, W. et al. MCL-1 degradation mediated by JNK activation via MEKK1/TAK1-MKK4 contributes to anticancer activity of new tubulin inhibitor MT189. Mol. Cancer Ther. 13, 1480–1491 (2014).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81573450 and 81773764), the Chinese Academy of Sciences (XDA12020104, XDA12020109 and CASIMM0120185003), the Nova Development Program of the Shanghai Institute of Materia Medica, the Science and Technology Commission of Shanghai Municipality (19ZR1467900), the State Key Laboratory of Drug Research and the Open Studio for Drugability Research of Marine Natural Products in the Qingdao National Laboratory for Marine Science and Technology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R. Mantovani
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tian, YN., Chen, HD., Tian, CQ. et al. Polymerase independent repression of FoxO1 transcription by sequence-specific PARP1 binding to FoxO1 promoter. Cell Death Dis 11, 71 (2020). https://doi.org/10.1038/s41419-020-2265-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-020-2265-y
This article is cited by
-
Poly(ADP-ribosyl)ating enzymes coordinate changes in the expression of metabolic genes with developmental progression
Scientific Reports (2023)
-
Poly(ADP-ribosyl)ating enzymes cooperate to coordinate development
Scientific Reports (2022)