Abstract
Angiotensin II (Ang II) is known to promote proliferation of vascular smooth muscle cells (VSMCs) in vascular remodeling, but whether it has an anti-apoptotic effect needs to be explored. Neuregulin-1 (NRG-1) as a member of the epidermal growth factor family was reported to suppress the proliferation of VSMCs by activating ErbB receptors, and therefore we hypothesized that there might be a cross talk between the anti-apoptotic effect of Ang II and the anti-proliferative effect of NRG-1 in VSMCs. The aim of the present study was to observe the expression and role of NRG-1 underlying the inhibitory effect of Ang II on apoptosis of mouse aortic smooth muscle cells (MASMCs). It was found that NRG-1 expression was down-regulated via the circNRG-1/miR-193b-5p-mediated post-transcriptional mechanism in response to Ang II. In addition, NRG-1 overexpression reversed the inhibitory effect of Ang II on apoptosis in MASMCs. Our data may provide a molecular basis for further understanding the mechanism of Ang II in suppressing the apoptosis of MASMCs by decreasing NRG-1 expression at circular RNA and micro RNA levels. The circNRG-1/miR-193b-5p/NRG-1 axis may prove to be a potential target for Ang II to inhibit the apoptosis of VSMCs and lead to vascular remodeling.
Similar content being viewed by others
Introduction
Vascular remodeling is a pathophysiological process in many cardiovascular diseases, such as atherosclerosis and hypertension1. Increasing evidence has demonstrated that proliferation and apoptosis of vascular smooth muscle cells (VSMCs) are key events in vascular remodeling2. Changes of the renin-angiotensin-aldosterone system (RAAS) may alter the balance between proliferation and apoptosis of VSMCs3,4. Angiotensin II (Ang II) is an effector peptide of the RAAS, and also a modulator of VSMC growth with proliferation/apoptosis effects mediated by activation of Ang II type 1 receptor (AT1R) or Ang II type 2 receptor (AT2R)5. In the early stages of vascular remodeling, Ang II promoted proliferation6,7 but inhibited apoptosis8,9 of VSMCs. A fundamental strategy for the treatment of these cardiovascular diseases is to accelerate apoptosis of VSMCs10. It is therefore important to gain insights into details of the molecular mechanism of Ang II in inhibiting apoptosis of VSMCs.
Neuregulin-1 (NRG-1) is a member of the epidermal growth factor (EGF) family, whose isoforms can be produced from the NRG-1 gene by alternative splicing11. Its transmembrane isoform includes an extracellular domain with an EGF-like sequence (NRG-1-ECD) and a highly conserved intracellular domain (NRG-1-ICD)12. NRG-1-ECD is a bioactive fragment, which can bind to ErbB family receptor tyrosine kinases to activate ErbB signaling in target cells13. NRG-1-ICD can translocate into the nucleus to regulate the gene expression, which has been confirmed by our previous and other studies14,15. Several lines of evidence have demonstrated that NRG-1 plays important roles in vascular physiopathology16. First, NRG-1 is expressed in vascular endothelial cells and smooth muscle cells, and its receptors are localized to the underlying smooth muscle cells17. Second, treatment of cultured VSMCs with NRG-1-ECD significantly decreases platelet-derived growth factor (PDGF)-BB-stimulated proliferation and migration18. Third, NRG-1-ICD is stimulated by transforming growth factor-β1 (TGF-β1), and translocates into the nucleus to regulate the α-SMA gene expression in human aortic smooth muscle cells (HASMCs)15. Previous studies showed that Ang II decreased NRG-1-ECD expression in endothelial cells19. However, the role of NRG-1 in the regulation of VSMC apoptosis in the context of Ang II signaling remains unclear.
Circular RNAs (circRNAs), which are shown to be a kind of critical gene regulator, are a novel class of non-coding RNAs with the characteristic of covalent bond linking the 3′ and 5′ ends generated by back splicing20. CircRNAs are known to play roles in the regulatory networks governing gene expression with multi-functions, such as cytoplasmic miRNA sponges21, RNA-binding protein participants22, and nuclear transcriptional regulators23. For example, one abundant circHIPK3, which regulates cell growth in cancerous tissues, acts as miRNA sponges to inhibit miR-124 activity by directly binding to miR-12424. The exon-intron circular RNAs circEIF3J and circPAIP2 interact with U1 snRNP and enhance transcription of their parental genes in the nucleus via specific RNA-RNA interaction25. However, the function of circRNAs under the treatment of Ang II in VSMC apoptosis remains unknown.
In this study, we showed that Ang II inhibited apoptosis through decreasing the expression of NRG-1 in mouse aortic smooth muscle cells (MASMCs), and this inhibitory effect could be reversed by NRG-1. In addition, circNRG-1 targeted NRG-1 for degradation by binding to miR-193b-5p. The results of the present study revealed a novel anti-apoptosis mechanism of Ang II via the circNRG-1/miR-193b-5p/NRG-1 axis, which may provide a potential therapeutic strategy for the prevention and treatment of vascular remodeling diseases.
Materials and methods
Cell culture and treatment
MASMCs (ATCC, No.CRL-2797TM) were cultured in low-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing penicillin, streptomycin and 10% fetal bovine serum (FBS). Human embryonic kidney 293A cells were obtained from ATCC (Manassas, VA) and maintained in high-glucose DMEM supplemented with 10% FBS. The cells were all in a humidified incubator at 37 °C with 5% CO2. Before stimuli and infection with adenovirus vectors or plasmids, MASMCs were incubated in serum-free medium for 24 h. Angiotensin II (A9525), Losartan (6188) and PD123319 (P186) were purchased from Sigma-Aldrich.
Cell apoptosis assay
MASMC apoptosis assays were performed with the Annexin V-FITC Apoptosis Detection Kit (KeyGEN BioTECH) according to the manufacturer’s recommendations. Cells were trypsinized, washed with cold PBS twice, re-suspended in 500 μl binding buffer containing 5 μl annexin V-FITC and 5 μl PI, and cultured at room temperature in the dark for 15 min. Finally, apoptosis was detected with a flow cytometer. All groups were evaluated in a minimum of three separate wells per experiment.
Western blot analysis
Proteins from cultured MASMCs were prepared as previously described15. Proteins were separated on 8%, 10% or 12% SDS-PAGE gels, and transferred to PVDF membranes. The membranes were blocked with 5% milk in TTBS for 2 h at room temperature and incubated with primary antibodies overnight at 4 °C. Antibodies are as follows: anti-NRG-1 (1:500, sc-348, santa), anti-NRG-1-ICD (1:500, sc-393009, santa), anti-Cleaved-caspase-3 (1:5000, ab214430, abcam), anti-caspase-3 (1:2000, ab184787, abcam), bax (1:1000, ab32503, abcam), bcl-2 (1:2000, ab182858, abcam), p-ErbB2 (1:500, ab47262, abcam), ErbB2 (1:500, ab131490, abcam), p-ErbB3 (1:1000, ab133459, abcam), ErbB3 (1:100, ab5470, abcam), p-ErbB4 (1:1000, ab76132, abcam), ErbB4 (1:1000, ab76303, abcam) and anti-β-actin (1:1000, sc-47778, Santa Cruz). The membranes were washed, incubated with the HRP-conjugated secondary antibodies for 1 h at room temperature, treated with the Immobilon™ Western (Millipore) and detected by ECL (enhanced chemiluminescence) BIO-RAD (721BR10829). All experiments were replicated three times.
Isolation of RNA and PCR
MASMCs were lysed by using the QIAzol Lysis Reagent (QIAGEN, Catalog no.79306). RNA was extracted from the above sample using a miRNeasy Mini Kit (QIAGEN, Catalog no.217004) according to the manufacturer’s instructions. The quality of the RNA was determined using a NanoPhotometer P-Class (IMPLEN). For large mRNA and circRNA, cDNA was synthesized using a M-MLV First Strand Kit (Life Technologies) with random hexamer primers. Quantitative real-time PCR (qRT-PCR) of mRNAs or circRNAs was performed using Platinum SYBR Green qPCR Super Mix UDG Kit (Invitrogen). For microRNA, reverse transcription and qRT-PCR were performed using the miRNA Detection Kit (Genepharma, Shanghai, China) and internal control U6 according to the manufacturer’s protocol. qRT-PCR was carried on an ABI 7500 FAST system (Life Technologies). Relative amounts of transcripts were normalized with GAPDH and calculated using the 2−ΔΔCt formula as previously described26. For reverse transcriptional PCR (RT-PCR) of mRNA and circRNA, 5 μl 1:5 diluted cDNA or gDNA was amplified (22–37cycles, depending on the template) in a 25 μl PCR reaction using the KOD XtremeTM HotStart Polymerase Kit (71975-3, Novagen). The experiments were carried on a Thermal Cycler Block 5020 (Thermo Fisher). The primer sequences were as follows: mouse GAPDH, 5′-AAGGTGAAGGTCGGAGTC-3′ and 5′- GATTTTGGAGGGATCTCG-3′; mouse NRG-1, 5′- GGAGATGCGAGCATAGACCG-3′ and 5′-GTGTCTCGGGGCTACTCTTG-3′; mouse circNRG-1, 5′-AACCCCTGACTCCTACAGAGACT-3′ and 5′- CTGGTCCCAGTCGTGGATGT-3′.
Microarray analysis
Circular RNA expression profiling was performed using Arraystar Mouse circRNA Array V2 analysis (Arraystar, USA). Circular RNAs of MASMCs were extracted using QIAzol Lysis Reagent (QIAGEN, Catalog no.79306) according to the manufacturer’s instructions. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols. Briefly, total RNAs were digested with Rnase R (Epicentre, Inc.) to remove linear RNAs and enrich circular RNAs. Then, the enriched circular RNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were hybridized onto the Arraystar Mouse circRNA Array V2 (8 × 15K, Arraystar). After washing slides, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images.
Fluorescence in situ hybridization
For circRNA FISH, cells were fixed in 4% paraformaldehyde for 5 min at room temperature, then permeabilized with 0.5% Triton X-100 and washed with PBS. The process used a RiboTM Fluorescent In Situ Hybridization Kit (RiboBio, China). In brief, MASMCs were blocked with pre-hybridization buffer for 30 min at 37 °C and then incubated with a circNRG-1 FISH Probe Mix and hybridization buffer overnight at 37 °C. Cell slides were washed with hybridization washing buffer I, II, and III for 15 min at 42 °C and 0.2 × SSC at 53 °C.
The double FISH assay was performed as described previously15. The procedures were conducted with a miR-193b-5p double-fluorescein (both 5′- and 3′- were labeled with FITC) FISH probe (Genepharma, China) and a cy3-circNRG-1 FISH probe. Hybridization was performed using fluorescence-labeled probes in hybridization buffer by incubation at 55 °C for 1.5 h. After stringent washing with SSC buffer, then DAPI (157574, MB biomedical) was used for nuclear counter staining. Images were captured by confocal microscopy and processed by LAS AF software.
Luciferase assay
Human embryonic kidney 293A cells were maintained as previously described27. For luciferase assays, HEK 293A cells were transfected with miR-193b-5p mimic (Gene pharma; Shanghai) and NC mimic combined with luciferase reporter plasmid of circNRG-1, circNRG-1 mut, NRG-1 3′-UTR, NRG-1 3′-UTR mut or an empty vector. Cells were harvested and luciferase activity was measured using a dual luciferase assay kit (Promega) after 24 h. The specific target activity was expressed as the relative activity ratio of firefly luciferase to renilla luciferase. All constructs were evaluated in a minimum of three separate wells per experiment.
Transfection of siRNA, plasmid, miRNA, and adenovirus vectors
Small interfering RNAs (siRNAs) targeting mouse circNRG-1 (si-circNRG-1) was designed and synthesized by RiboBio (Guangzhou, China). The siRNA sequence was as follows: circNRG-1 siRNA (si-circNRG-1), 5′-ATAGTGAAAGCCACATCTA-3′. Non-specific siRNA (si-Control) was purchased from RiboBio. The plasmid of circNRG-1 (pLVX-circNRG-1) was entrusted to Likely Biotechnology (Beijing, China). Sequences of circNRG-1 and NRG-1 gene 3′-UTR contain miR-193b-5p target sites or its mutant sequences were inserted into pmir-GLO Dual-Luciferase miRNA Target Expression Vector. miR-193b-5p mimic, inhibitor and control RNAs were designed and synthesized by Gene Pharma (Shanghai, China). Adenoviruses encoding NRG-1 (Ad-GFP-NRG-1) and GFP control (Ad-GFP) were entrusted to Hanbio (Shanghai, China). Transfection was performed using Lipofectamine 2000 following the manufacturer’s instructions. Twenty-four hours after transfection, MASMCs were treated with Ang II, miR-193b-5p-mimic, or anti-miR-193b-5p, harvested and lysed for Western blot, PCR and luciferase assay.
Statistical analysis
All the data are presented as the means ± S.E.M. Differences between two groups were assessed using analysis of variance followed by a Student’s t-test. A value of P < 0.05 was considered statistically significant. Statistical analysis was performed using Graphpad Prism 5 software (GraphPad Software, San Diego, CA, USA).
Results
Ang II decreases NRG-1 protein expression in MASMCs
To examine the effect of Ang II on NRG-1, we firstly analyzed the expression of NRG-1 in MASMCs treated with Ang II. Knowing that NRG-1 included an extracellular domain (ECD) and an intracellular domain (ICD), we first detected the expression levels of full-length NRG-1, NRG-1-ICD, and NRG-1-ECD, respectively. Western blot showed that Ang II significantly decreased both full-length NRG-1 and NRG-1-ICD protein levels in a dose- and time-dependent manner (Fig. 1a, c). Furthermore, there was a reduction in NRG-1 from cell membrane lysates and accumulation of NRG-1-ECD in the media from MASMCs in response to Ang II (Fig. 1b, d). Next, we sought to determine whether Ang II decreased the transcription of NRG-1 gene. MASMCs were treated with Ang II 10−7 M for different times and then NRG-1 mRNA was detected by qRT-PCR. Interestingly, we found that NRG-1 mRNA level was not affected by Ang II treatment (Fig. 1e). Based on these findings, we hypothesized that Ang II reduced protein expression of NRG-1 through post-transcriptional regulation.
a NRG-1 and NRG-1-ICD expression levels in MASMCs treated with Ang II with different doses were analyzed by Western blot (left). The right panel shows densitometric analyses from three independent experiments. ***P < 0.001 vs. Ang II for 0 M. b Levels of NRG-1-ECD expression in the culture medium of MASMCs treated with different doses of Ang II were determined by ELISA. *P < 0.05, ***P < 0.001 vs. Ang II for 0 M. c Western blot was used to assess the NRG-1 and NRG-1-ICD expression levels in MASMCs treated with Ang II (10−7 M) for the indicated times (left). The right panel shows densitometric analyses from three independent experiments. ***P < 0.001 vs. Ang II for 0 h. d Levels of NRG-1-ECD expression in medium treated with Ang II (10−7 M) for the indicated times were analyzed by ELISA. ***P < 0.001 vs. Ang II for 0 h. e MASMCs were treated with Ang II (10−7 M) for the indicated times, and NRG-1 mRNA was detected by qRT-PCR. Data represent the means ± SEM of three independent experiments
Ang II suppresses circNRG-1 expression in MASMCs
The finding that Ang II downregulated NRG-1 protein level without affecting mRNA level suggested that Ang II regulated NRG-1 expression post-transcriptionally via miRNAs or circRNAs. An Arraystar mouse circRNA Microarray was accomplished with MASMCs to detect whether circRNAs were involved in the regulation of NRG-1. The differential expression levels of circRNAs between Ang II and control groups were identified through scatter plots (Fig. 2a). In the scatter plots, circRNAs above the top green line and below the bottom green line showed >1.5-fold change between the two compared groups. A heat map of 382 circRNAs was differentially expressed between Ang II and control groups. Among the 382 circRNAs, 235 were upregulated and 147 were downregulated by >2-fold change in Ang II group. The results showed that mmu-circRNA-42742, whose parental gene is NRG-1, was significantly down-regulated in the MASMCs treated with Ang II as compared with the control group (Fig. 2b). To further validate the results, we used convergent and divergent primers, respectively, to amplify total RNA and circular RNA transcripts derived from the NRG-1 gene by RT-PCR (Fig. 2c). Its PCR products were confirmed by DNA sequencing (Fig. 2d). The result of fluorescence in situ hybridization (FISH) showed that circNRG-1 mostly located in the cytoplasm of MASMCs (Fig. 2e). A general consistency was shown between the results of qRT-PCR and microarray analysis. The results showed that circNRG-1 was decreased in a time-dependent manner in the group treated with Ang II in MASMCs (Fig. 2f). These data suggested that Ang II reduced circNRG-1 expression.
a Scatter plots were used to evaluate the difference in the expression of circRNAs between Ang II and control groups. The values plotted on X and Y axes are the averaged normalized signal values of each group (log2 scaled). The circRNAs above the top green line and below the bottom green line indicate >1.5-fold change between the two groups. b Hierarchical clustering analysis showed the differentially expressed circRNAs over 2.0-fold change. Red color indicates high expression level, and blue color indicates low expression level. c Divergent and convergent primers were used to verify whether circNRG-1 was a circRNA. Convergent primers were used to detect NRG-1 mRNA. Divergent primers amplified circNRG-1 in cDNA but not gDNA. GAPDH served as linear control and size marker in base pairs. d Sanger sequencing confirmed head-to-tail junction of circNRG-1. e RNA fluorescence in situ hybridization for circNRG-1 was detected. Nuclei were stained with DAPI. Scale bars = 50 μm. f qRT-PCR detected circNRG-1 expression in MASMCs treated with Ang II (10−7 M) for the different times. Data represent the means ± SEM of three independent experiments. *P < 0.05, ***P < 0.001 vs. Ang II for 0 h
circNRG-1 upregulates NRG-1 expression by acting as miR-193b-5p sponge
Knowing that Ang II decreased the expression levels of NRG-1 and circNRG-1, we sought to determine the relationship between them by overexpressing or knocking down circNRG-1 in MASMCs. Knowing that the exogenous circNRG-1 was effectively overexpressed in pLVX-circNRG-1-transfected MASMCs, we designed a siRNA targeting the backsplice sequence for circNRG-1 (si-circNRG-1) as shown by qRT-PCR (Fig. 3a). Western blot analyses revealed that circNRG-1 overexpression or knockdown markedly increased or decreased NRG-1 protein level, respectively (Fig. 3b). Knowing that circRNAs can function as miRNA sponges, we predicted miRNAs, whose nucleotide sequences are known to be complementary to a region of circNRG-1, and found out their interactions. The circRNA/miRNA interaction was predicted with Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda. The result showed that circNRG-1 contained sequences complementary to miR-193b-5p seed sequence (Fig. 3c). In addition, Ang II significantly increased the miR-193b-5p level (Fig. 3d). Localization of miR-193b-5p and circNRG-1 was showed by RNA in situ hybridization in MASMCs, and their co-localization was predominantly in cytoplasm (Fig. 3e). To further clarify whether the sponge effect of circNRG-1 on miR-193b-5p affected gene expression, circNRG-1 sequence was inserted immediately in downstream of the luciferase reporter gene. Luciferase assay revealed that transfection with miR-193b-5p mimic significantly decreased the luciferase activity via mediation of wild-type circNRG-1 sequence but not via its mutant (Fig. 3f). These findings suggested that circNRG-1 regulated NRG-1 expression and bound miR-193b-5p to prevent it from binding to the target genes.
a MASMCs were transfected with pLVX-circNRG-1 or si-circNRG-1, as well as with their corresponding controls. circNRG-1 expression was analyzed by qRT-PCR. ***P < 0.001 vs. pLVX-Ctl, ###P < 0.001 vs. si-Con. b MASMCs were transfected with pLVX-Ctl, pLVX-circNRG-1, si-Con and si-circNRG-1 for 24 h. NRG-1 was quantitated by densitometry and values were normalized to total β-actin (Right panel). Data were means ± SEM, n = 3. ***P < 0.001 vs. pLVX-Ctl, ###P < 0.001 vs. si-Con. c miR-193b-5p binding sites in the circNRG-1 sequence were predicted. d miR-193b-5p level in MASMCs treated with Ang II (10−7 M) for the indicated times was examined by qRT-PCR. Data are presented as means ± SEM, n = 3. **P < 0.01, ***P < 0.001 vs. 0 h. e Co-localization between circNRG-1 and miR-193b-5p was detected by RNA in situ hybridization in MASMCs. Nuclei were stained with DAPI. Scale bars = 25 μm. f HEK 293A cells were co-transfected with wild-type pmirGLO-circNRG-1 or its mutant pmirGLO-circNRG-1 mut and miR-193b-5p mimic or miR-Ctl. Luciferase activity was measured. Data are the means ± SEM of three independent experiments. **P < 0.01 vs. pmirGLO, pmirGLO-circNRG-1 mut
miR-193b-5p suppresses NRG-1 expression by targeting its 3′-UTR in MASMCs
NRG-1 was regarded as a miRNA target based on a network in which circRNA-miRNA-mRNA was constructed using cytoscape. The predicted ceRNA mechanism was finally described: circNRG-1 acted as a sponge or ceRNA for miR-193b-5p, while NRG-1 was the target gene (Fig. 4). To see whether NRG-1 was a direct target of miR-193b-5p, we used a bioinformatics approach to search the potential matching site of miR-193b-5p in the NRG-1 3′-UTR, and found that mouse NRG-1 3′-UTR contained two miR-193b-5p-binding sites at nucleotides 544–550 and 3195–3201 (Fig. 5a). To evaluate the effect of miR-193b-5p on NRG-1 expression, we constructed the wild-type pmirGLO-NRG-1 3'-UTR and its mutant pmirGLO-NRG-1 3′-UTR mut and co-transfected HEK 293A cells with them and miR-193b-5p mimic or miR-Ctl. The luciferase assay showed that miR-193b-5p mimic, but not control oligonucleotide (miR-Ctl), decreased the luciferase activity by 50%, whereas the mutation of the miR-193b-5p-binding site in the NRG-1 3′-UTR completely restored luciferase activity in the presence of the miR-193b-5p mimic (Fig. 5b). In the subsequent experiment, we transfected MASMCs with miR-193b-5p mimic and inhibitor anti-miR-193b-5p (Fig. 5c) and confirmed that the mimic and its antagomir markedly reduced or increased the NRG-1 protein expression, respectively, as compared with their corresponding controls (Fig. 5d). In addition, circNRG-1 overexpression and miR-193b-5p knockdown alone, or in combination in particular, significantly increased the NRG-1 expression level (Fig. 5e). In contrast, knockdown of circNRG-1 plus miR-193b-5p mimic worked together to decrease the NRG-1 expression level (Fig. 5f). These findings indicated that miR-193b-5p inhibited the NRG-1 expression in MASMCs by targeting its 3′-UTR, and that circNRG-1 facilitated the NRG-1 expression by absorbing miR-193b-5p.
a The miR-193b-5p-binding sites in the 3'-UTR of NRG-1 mRNA are shown in red. b HEK 293A cells were co-transfected with miR-193b-5p mimic and the wild-type pmirGLO-NRG-1 3′-UTR or its mutant pmirGLO-NRG-1 3′-UTR mut. After 24 h, luciferase activities were measured. Data are the means ± SEM of three independent experiments. **P < 0.01 vs. pmirGLO or pmirGLO-NRG-1 3′-UTR mut. c qRT-PCR detection of miR-193b-5p expression in MASMCs transfected with miR-193b-5p mimic or anti-miR-193b-5p and their corresponding controls. Data represent the means ± SEM of three independent experiments. ***P < 0.001 vs. miR-Ctl, ###P < 0.001 vs. anti-miR-Ctl. d MASMCs were transfected with miR-193b-5p mimic or anti-miR-193b-5p, as well as with their corresponding controls. NRG-1 expression was analyzed by Western blot. The right panel shows densitometric analyses from three independent experiments. **P < 0.01 vs. miR-Ctl, ##P < 0.01 vs. Anti-miR-Ctl. e Western blot detected NRG-1 expression in MASMCs transfected with pLVX-circNRG-1, anti-miR-193b-5p, or both of them. NRG-1 expression was quantitated as described above (Down panel). ***P < 0.001 vs. pLVX-Ctl+Anti-miR-193b-5p-untreated group. f MASMCs were transfected with si-circNRG-1, miR-193b-5p, or both of them for 24 h. NRG-1 expression was analyzed by Western blot and was quantitated as described above (Down panel). **P < 0.01, ***P < 0.001 vs. si-Con+miR-193b-5p-untreated group
Ang II inhibits apoptosis via down-regulating NRG-1 expression in MASMCs
To examine the role of Ang II in MASMC apoptosis, we used Western blot to examine proteins related to apoptosis and anti-apoptosis. The apoptosis proteins Cleaved-caspase-3 and bax were decreased and the anti-apoptosis protein bcl-2 was increased in a time-dependent manner with Ang II (10−7 M), but the caspase-3 protein level remained unchanged significantly (Fig. 6a). To further confirm the ability of Ang II to confer resistance to apoptosis, flow cytometry combined with annexin V/propidium iodide (PI) staining was used to assess apoptosis. As suggested in Fig. 6b, the apoptotic rate of MASMCs induced by Ang II (10−7 M) decreased from 12% to 6%. These results indicated that Ang II attenuated apoptosis in MASMCs. To verify whether NRG-1 participated in Ang II-reduced apoptosis in MASMCs, we overexpressed NRG-1 through transfecting adenovirus vectors with or without Ang II. The results showed that overexpression of NRG-1 could reverse the expression levels of Cleaved-caspase-3, bax and bcl-2 (Fig. 6c). In addition, the apoptotic rate of overexpression NRG-1 was reversed from 9.7% to 17.4% (Fig. 6d). In conclusion, up-regulation of NRG-1 abrogated the effect of Ang II on the inhibition of MASMC apoptosis.
a MASMCs were treated with Ang II (10−7 M) for indicated times. Cleaved-caspase-3, caspase-3, bax and bcl-2 expression levels were detected by Western blot. The right panel shows densitometric analysis from three independent experiments. ***P < 0.001 vs. 0 h. b MASMCs were treated with different times of Ang II (10−7 M) and then the apoptotic rate was determined by flow cytometry analysis using annexin V/propidium iodide double staining. The right panel shows the apoptotic ratio from three independent experiments. ***P < 0.001 vs. 0 h. c MASMCs were infected with Ad-GFP-NRG-1 or Ad-GFP for 24 h and then treated with or without Ang II (10−7 M) for an additional 12 h, Western blot detected NRG-1, Cleaved-caspase-3, caspase-3, bax and bcl-2 levels. Data represent the means ± SEM of three independent experiments. ***P < 0.001 vs. Ad-GFP+Ang II-untreated group. d The apoptotic rate was determined and quantitative analysis for the flow cytometry experiment is presented on the right panel. Data represent the means ± SEM, n = 3. ***P < 0.001 vs. Ad-GFP+Ang II-untreated group
Discussion
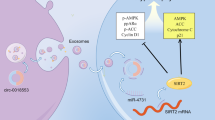
This study revealed that (1) Ang II decreased NRG-1 expression, including NRG-1-ECD and NRG-1-ICD in MASMCs; (2) Ang II down-regulated circNRG-1 expression, and circNRG-1 acted as a sponge binding miR-193b-5p, which targeted the 3′-UTR of NRG-1 mRNA, to regulate NRG-1 expression; and (3) Ang II inhibited apoptosis of MASMCs by reducing the NRG-1 expression. Based on the above findings, we concluded that the circNRG-1/miR-193b-5p/NRG-1 axis regulated the anti-apoptotic effect of MASMCs caused by Ang II.
It has been shown that endocardial endothelia and small subendocardial vessels are the primary cell source of NRG-1, and NRG-1 and its receptor ErbB play crucial roles in cardiac development, structural maintenance, and functional integrity of the heart28. Although our previous study found that TGF-β1 and PDGF-BB increased and decreased NRG-1 expression in HASMCs15, respectively, whether and how Ang II regulated NRG-1 in MASMCs remained unknown. The present data support the idea that Ang II decreases the NRG-1 protein expression in MASMCs and down-regulates it post-transcriptionally. NRG-1/ErbB signaling is an important potential mechanism for regulating vascular remodeling29. Hedhli et al. found that ErbB2, ErbB3 and ErbB4 were expressed in HASMCs30. Interestingly, we found that full-length NRG-1, NRG-1-ECD and NRG-1-ICD were all down-regulated by Ang II, p-ErbB2 and p-ErbB4 expression levels were also decreased in a time-dependent manner after stimulation by Ang II (Supplementary Fig. S1). This means that reduced phosphorylation of ErbB receptors was due to the low expression of NRG-1-ECD.
Notably, we found that Ang II down-regulated NRG-1 protein expression without affecting the mRNA level in MASMCs, which urged us to make clear whether some miRNAs or circRNAs regulated the NRG-1 expression at the post-transcriptional level. According to the circRNA microarray, we found that circNRG-1 was also down-regulated by Ang II, and this circRNA was not on the list of circBase, meaning that circNRG-1 is a new circRNA, so we used RT-PCR and Sanger sequencing to confirm whether circNRG-1 was derived from NRG-1 gene. In addition, we found that circNRG-1 overexpression or knockdown could increase or decrease the NRG-1 expression, so we wondered whether this mechanism of circNRG-1 was involved in the regulation of NRG-1 protein level. To simplify this problem, we tried to use the ceRNA theory31 to explain how circNRG-1 exerted its effect through bioinformatic analysis by establishing a circRNA-miRNA-mRNA regulatory model based on the ceRNA theory. As expected, we found that circNRG-1 acted as a miR-193b-5p sponge and interacted with miR-193b-5p to target the 3'-UTR of NRG-1 mRNA, thus increasing the NRG-1 expression in MASMCs.
In general, Ang II has been shown to induce or prevent apoptosis in several cell types32,33 but the role of Ang II in regulating apoptosis of VSMCs remains controversial. Here, we firstly demonstrated a novel mechanism of Ang II in inducing MASMCs to exhibit apoptosis resistance. The results showed that Ang II (10−7 M) decreased the apoptosis gene Cleaved-caspase-3 and bax, and increased the anti-apoptosis gene bcl-2 in MASMCs. In addition, the apoptotic rate of MASMCs was reduced by about 50%. This is similar to the finding in a previous study that Ang II inhibited VSMC apoptosis8, but different from the result of other studies that Ang II promoted apoptosis of VSMCs34. This may be due to different concentrations of Ang II used, or the different receptors activated by Ang II. In the early stages of vascular remodeling, a low Ang II environment may inhibit VSMC apoptosis, but with the phase advancing, the high Ang II environment stimulated VSMC apoptosis. In addition, Ang II exerted its biological effect through AT1R and AT2R, which are known to be associated with proliferation and apoptosis35,36. Several studies demonstrated that AT1R activation resulted in proliferation and anti-apoptosis whereas AT2R activation induced apoptosis in VSMCs37. In the present study, we blocked AT1R or AT2R to observe the apoptotic rate of MASMCs, and found that AT1R blockage could reverse the apoptotic rate of Ang II, but not the AT2R inhibitor (Supplementary Fig. S2). Finally, Ang II-induced anti-apoptosis was shown to be dose-dependent, and AT1R signaling was found to be a decisive response.
To further investigate the role of NRG-1 in Ang II-induced apoptosis resistance, we overexpressed NRG-1 and found that higher expression of NRG-1 reversed the inhibitory effect of Ang II on apoptosis in MASMCs. Together, these data suggest NRG-1 as a critical factor for mediating the resistance to apoptosis conferred by Ang II in MASMCs, and NRG-1 may be a potential therapeutic target for vascular remodeling associated with RAAS overactivation in atherosclerosis, hypertension and vascular diseases.
References
Marszalek, R.J., John Solaro, R. & Wolska, B.M. Coronary arterial vasculature in the pathophysiology of hypertrophic cardiomyopathy. Pflugers Archiv : Eur. J. physiol. 471, 769–780 (2019).
Ping, S. et al. Simultaneous increases in proliferation and apoptosis of vascular smooth muscle cells accelerate diabetic mouse venous atherosclerosis. PloS ONE 10, e0141375 (2015).
Te Riet, L., van Esch, J. H., Roks, A. J., van den Meiracker, A. H. & Danser, A. H. Hypertension: renin-angiotensin-aldosterone system alterations. Circ. Res. 116, 960–975 (2015).
Intengan, H. D. & Schiffrin, E. L. Vascular remodeling in hypertension: roles of apoptosis, inflammation, and fibrosis. Hypertension 38, 581–587 (2001).
van Thiel, B. S., van der Pluijm, I., te Riet, L., Essers, J. & Danser, A. H. The renin-angiotensin system and its involvement in vascular disease. Eur. J. Pharmacol. 763, 3–14 (2015).
Zhang, M., Xu, Y., Qiu, Z. & Jiang, L. Sulforaphane attenuates angiotensin II-Induced vascular smooth muscle cell migration via suppression of NOX4/ROS/Nrf2 signaling. Int. J. Biol. Sci. 15, 148–157 (2019).
Xu, M. M., Deng, H. Y. & Li, H. H. MicroRNA-27a regulates angiotensin II-induced vascular smooth muscle cell proliferation and migration by targeting alpha-smooth muscle-actin in vitro. Biochem. Biophys. Res. Commun. 509, 973–977 (2019).
Pollman, M. J., Yamada, T., Horiuchi, M. & Gibbons, G. H. Vasoactive substances regulate vascular smooth muscle cell apoptosis. Countervailing influences of nitric oxide and angiotensin II. Circ. Res. 79, 748–756 (1996).
Dimmeler, S., Rippmann, V., Weiland, U., Haendeler, J. & Zeiher, A. M. Angiotensin II induces apoptosis of human endothelial cells. Protective effect of nitric oxide. Circ. Res. 81, 970–976 (1997).
Antoniades, C., Antonopoulos, A. S., Bendall, J. K. & Channon, K. M. Targeting redox signaling in the vascular wall: from basic science to clinical practice. Curr. Pharm. Des. 15, 329–342 (2009).
Lemmens, K., Doggen, K. & De Keulenaer, G. W. Role of neuregulin-1/ErbB signaling in cardiovascular physiology and disease: implications for therapy of heart failure. Circulation 116, 954–960 (2007).
Murphy, S., Krainock, R. & Tham, M. Neuregulin signaling via erbB receptor assemblies in the nervous system. Mol. Neurobiol. 25, 67–77 (2002).
Parodi, E. M. & Kuhn, B. Signalling between microvascular endothelium and cardiomyocytes through neuregulin. Cardiovasc. Res. 102, 194–204 (2014).
Bao, J., Wolpowitz, D., Role, L. W. & Talmage, D. A. Back signaling by the Nrg-1 intracellular domain. J. Cell Biol. 161, 1133–1141 (2003).
Sun, Y. et al. A novel regulatory mechanism of smooth muscle alpha-actin expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. 121, 628–635 (2017).
Odiete, O., Hill, M. F. & Sawyer, D. B. Neuregulin in cardiovascular development and disease. Circ. Res. 111, 1376–1385 (2012).
Kyotani, Y. et al. Intermittent hypoxia induces the proliferation of rat vascular smooth muscle cell with the increases in epidermal growth factor family and erbB2 receptor. Exp. Cell Res. 319, 3042–3050 (2013).
Clement, C. M. et al. Neuregulin-1 attenuates neointimal formation following vascular injury and inhibits the proliferation of vascular smooth muscle cells. J. Vasc. Res. 44, 303–312 (2007).
Wu, C. et al. Expression and secretion of neuregulin-1 in cardiac microvascular endothelial cells treated with angiogenic factors. Exp. Ther. Med. 15, 3577–3581 (2018).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Yang, W., Du, W. W., Li, X., Yee, A. J. & Yang, B. B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 35, 3919–3931 (2016).
Zang, J., Lu, D. & Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. Preprint at https://doi.org/10.1002/jnr.24356 (2018).
Zhang, Y. et al. Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806 (2013).
Zheng, Q. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 (2016).
Li, Z. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015).
Wang, C., Han, M., Zhao, X. M. & Wen, J. K. Kruppel-like factor 4 is required for the expression of vascular smooth muscle cell differentiation marker genes induced by all-trans retinoic acid. J. Biochem. 144, 313–321 (2008).
Li, H. X. et al. Kruppel-like factor 4 promotes differentiation by transforming growth factor-beta receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells. J. Biol. Chem. 285, 17846–17856 (2010).
Hedhli, N. et al. Endothelium-derived neuregulin protects the heart against ischemic injury. Circulation 123, 2254–2262 (2011).
Xu, G. et al. Preventive effects of heregulin-beta1 on macrophage foam cell formation and atherosclerosis. Circ. Res. 105, 500–510 (2009).
Hedhli, N., Kalinowski, A. & K, S. R. Cardiovascular effects of neuregulin-1/ErbB signaling: role in vascular signaling and angiogenesis. Curr. Pharm. Des. 20, 4899–4905 (2014).
Sen, R., Ghosal, S., Das, S., Balti, S. & Chakrabarti, J. Competing endogenous RNA: the key to posttranscriptional regulation. Sci. World J. 2014, 896206 (2014).
Cai, W. et al. Angiotensin II confers resistance to apoptosis in cardiac myofibroblasts through the AT1/ERK1/2/RSK1 pathway. IUBMB life 71, 261–276 (2019).
Filippatos, G., Tilak, M., Pinillos, H. & Uhal, B. D. Regulation of apoptosis by angiotensin II in the heart and lungs (review). Int. J. Mol. Med. 7, 273–280 (2001).
Li, Y., Song, Y. H., Mohler, J. & Delafontaine, P. ANG II induces apoptosis of human vascular smooth muscle via extrinsic pathway involving inhibition of Akt phosphorylation and increased FasL expression. Am. J. Physiol. Heart Circ. Physiol. 290, H2116–2123 (2006).
Pei, N. et al. Angiotensin II type 2 receptor promotes apoptosis and inhibits angiogenesis in bladder cancer. J. Exp. Clin. Cancer Res. 36, 77 (2017).
Liu, T. J., Shi, Y. Y., Wang, E. B., Zhu, T. & Zhao, Q. AT1R blocker losartan attenuates intestinal epithelial cell apoptosis in a mouse model of Crohn’s disease. Mol. Med. Rep. 13, 1156–1162 (2016).
Tan, N. Y., Li, J. M., Stocker, R. & Khachigian, L. M. Angiotensin II-inducible smooth muscle cell apoptosis involves the angiotensin II type 2 receptor, GATA-6 activation, and FasL-Fas engagement. Circ. Res. 105, 422–430 (2009).
Acknowledgements
This work is supported by the Major Program of the National Natural Science Foundation of China (No. 91539205); the National Natural Science Foundation of China (No. 81800425); the Postdoctoral Science Foundation of China (No. 2018M630178); and Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (No. KZ201810025039).
Author information
Authors and Affiliations
Contributions
H.L. and Y.S. contributed to the study concept and design. Y.S., M.Y., Y.L. and J.B. carried out the experiments and analyzed the data. Y.S. and S.Z. were involved in drafting the manuscript and writing the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Stephanou
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Y., Zhang, S., Yue, M. et al. Angiotensin II inhibits apoptosis of mouse aortic smooth muscle cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell Death Dis 10, 362 (2019). https://doi.org/10.1038/s41419-019-1590-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1590-5
This article is cited by
-
G6PD maintains the VSMC synthetic phenotype and accelerates vascular neointimal hyperplasia by inhibiting the VDAC1–Bax-mediated mitochondrial apoptosis pathway
Cellular & Molecular Biology Letters (2024)
-
Circular RNAs: regulators of vascular smooth muscle cells in cardiovascular diseases
Journal of Molecular Medicine (2022)
-
CircRNA–miRNA interactions in atherogenesis
Molecular and Cellular Biochemistry (2022)
-
Increased AT2R expression is induced by AT1R autoantibody via two axes, Klf-5/IRF-1 and circErbB4/miR-29a-5p, to promote VSMC migration
Cell Death & Disease (2020)