Abstract
Hepatic stellate cells activate upon liver injury and help at restoring damaged tissue by producing extracellular matrix proteins. A drastic increase in matrix proteins results in liver fibrosis and we hypothesize that this sudden increase leads to accumulation of proteins in the endoplasmic reticulum and its compensatory mechanism, the unfolded protein response. We indeed observe a very early, but transient induction of unfolded protein response genes during activation of primary mouse hepatic stellate cells in vitro and in vivo, prior to induction of classical stellate cell activation genes. This unfolded protein response does not seem sufficient to drive stellate cell activation on its own, as chemical induction of endoplasmic reticulum stress with tunicamycin in 3D cultured, quiescent stellate cells is not able to induce stellate cell activation. Inhibition of Jnk is important for the transduction of the unfolded protein response. Stellate cells isolated from Jnk knockout mice do not activate as much as their wild-type counterparts and do not have an induced expression of unfolded protein response genes. A timely termination of the unfolded protein response is essential to prevent endoplasmic reticulum stress-related apoptosis. A pathway known to be involved in this termination is the non-sense-mediated decay pathway. Non-sense-mediated decay inhibitors influence the unfolded protein response at early time points during stellate cell activation. Our data suggest that UPR in HSCs is differentially regulated between acute and chronic stages of the activation process. In conclusion, our data demonstrates that the unfolded protein response is a JNK1-dependent early event during hepatic stellate cell activation, which is counteracted by non-sense-mediated decay and is not sufficient to drive the stellate cell activation process. Therapeutic strategies based on UPR or NMD modulation might interfere with fibrosis, but will remain challenging because of the feedback mechanisms between the stress pathways.
Similar content being viewed by others
Introduction
Sustained chronic liver injury leading to fibrosis, cirrhosis and finally organ failure causes significant morbidity and mortality world-wide1. Liver fibrosis is defined by hardening and scarring of the liver due to an excessive deposition of extracellular matrix (ECM) components. The major cellular source for the ECM production are the hepatic stellate cells (HSCs). During chronic liver injury, HSCs undergo a transdifferentiation from quiescent, lipid droplet containing cells towards activated myofibroblast-like HSCs with an increased proliferation rate and high production of ECM2. HSC activation is a critical step in the fibrotic response to liver injury3. Novel insights into mechanisms regulating HSC activation are considered key in developing new treatments for hepatic fibrosis.
Sensing and responding to stress is essential for maintaining cellular homeostasis. There are many triggers that can cause stress in a cell, e.g., viral infections, hypoxia, chemical insults and alterations in substrates and energy. The process of protein folding is particularly sensitive to such insults4. An unfolded protein response (UPR) is initiated to restore cellular homeostasis upon acute stress exposure, while chronic activation of the UPR leads to endoplasmic reticulum (ER) stress and ultimately to apoptosis. UPR transmits survival signals through three sensory systems, the PERK (protein kinase R (PKR)-like endoplasmic reticulum kinase), IRE1α (inositol-requiring enzyme-1α) and ATF6 (activating transcription factor 6) cascades, which aim at reducing ER stress by increasing the folding and export capacity and by lowering general translation5.
The three UPR arms have been associated with chronic liver disease6,7,8,9. Numerous studies report on UPR induction in hepatocytes in, for example, non-alcoholic fatty liver disease, but more recent studies have also attributed a role for the UPR to HSC activation and fibrotic wound healing. In general, it is found that chronic injury or HSC activation correlates with high levels of ER stress related genes and that chemical induction of ER stress further increases HSC activation10,11,12,13,14. Non-sense-mediated mRNA decay (NMD) is a mechanism to remove aberrant messenger RNA (mRNA) transcripts, but also to finetune the expression of certain normal mRNAs. As unfolded protein response will block translation, mRNA accumulation is expected, and this can be controlled by NMD. It was shown that NMD can buffer cells from an overactive UPR. In addition, there is evidence that NMD directly targets the mRNAs encoding several UPR components15,16,17.
In this study, we confirm that there is an UPR in chronically in vivo activated HSCs by showing increased expression of BIP, Chop and XBP1 spliced and that these high UPR levels are paralleled by low NMD marker expression. However, more interestingly, we also observe a transient, endogenous induction of these genes very early during in vitro and in vivo HSC activation. We propose that this early UPR is a compensatory mechanism to cope with the increased needs for protein production and secretion of, for example, collagens, which is regulated by NMD to prevent disastrous ER stress levels which could lead to cell death.
Materials and methods
Isolation, culturing and treatment of mouse HSCs
The animal experiments were approved by the animal care and use committee of Vrije Universiteit Brussel (Permits 13-212-1 and 14-212-4) and the institution’s guidelines for the care and use of laboratory animals in research were strictly followed. The HSC isolation method for male BALB/c mice (aged 15–20 weeks) was performed as previously described18. Total c-Jun N-terminal kinase (JNK) mice were provided by Professor Christian Trautwein and were originally described in Zhao et al.19 and Cubero et al.20. After isolation, a fraction of the cells was collected for RNA or cytospinned and the rest was cultured in Dulbecco’s modified Eagle’s medium (Lonza, Braine-l’Alleud, Belgium) supplemented with 10% fetal bovine serum (Lonza) at 37 °C in a humidified atmosphere with 5% CO2. Treatment with 5 and 10 µM JNK V inhibitor (Calbiochem, Merck, Overijse, Belgium) was started from the moment of seeding and refreshed every day. VG-1 (Gift from Prof. Coldham) and NMDi-14 (R&D systems) treatments were initiated from the moment of seeding. Solvent controls were treated with 0.1% dimethyl sulfoxide. Bright-field images were taken with an Axioskop light microscope (Carl Zeiss, Zaventem, Belgium).
CCl4-induced liver fibrosis
An acute liver injury in BALB/c mice was induced by a single intraperitoneal injection of carbon tetrachloride (CCl4, Sigma (Diegem, Belgium) 50 µl CCl4/100 g body weight in mineral oil). At different time points after the injection, i.e., 2, 6, 10 and 24 h, three livers were pooled for each time point followed by HSC isolation using nycodenz. Chronic liver injury was induced by two intraperitoneal injections of CCl4 per week for 2 up to 8 weeks. HSC isolation after chronic injury in BALB/c mice and from the JNK1 and JNK2 knockout (KO) mice was done based on ultraviolet positivity using fluorescently activated cell sorting (FACS Aria BD (Erembodegem, Belgium)21. After isolation, HSCs were not cultured, but immediately collected for mRNA analysis or cytospinned and formalin-fixed.
Three-dimensional (3D)-spheroid cultivation
Primary mHSCs were seeded in U-bottom 96-well plates at a density of 4000 HSCs/cm2 and cultured under gentle rotation on a shaker at a speed of 80 rpm. As a result, 3D spheroids are formed with a diameter of 200 µm. At least 6 spheroids per time point were pooled for mRNA analysis. For transforming growth factor-β (TGF-β; R&D Systems, Oxon, UK) or Tunicamycin (Calbiochem) treatment, spheroids were first serum starved overnight followed by treatment with 10 ng/ml TGF-β or 0.5 µg/ml tunicamycin for 10 h.
Messenger RNA analysis
Total RNA from cultured cells was extracted using ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI, USA). RNA was reverse-transcribed into complementary DNA using the Revert Aid kit (Thermofisher) and performed at 25 °C for 10 min, 30 min at 50 °C. For quantitative real-time polymerase chain reaction (qPCR), GoTaq qPCR Master Mix with BRYTE green (Promega) was used and the samples were subsequently subjected to qPCR in an ABI 7500 real-time PCR system and analyzed using System SDS software (Applied Biosystems). For analysis according to the comparative Ct (δδCt) method, each Ct value was first normalized to the house-keeping gene Gapdh. Gene-specific primers were produced by Integrated DNA Technologies (Suppl. table 1).
Immunocytochemistry
Cells were formalin-fixed followed by washing, permeabilization and overnight incubation at 4 °C with the following primary antibodies: rabbit monoclonal anti-BIP (1:200, Cell Signaling Technology, Leiden, The Netherlands) and mouse monoclonal anti-CHOP (1:1000, Cell Signaling technology). Incubation with Alexa-coupled secondary antibody (1:200) for 1 h at room temperature was used to visualize the antibody binding. Following mounting with ProLong® Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI), images were taken using confocal microscopy (Zeiss LSM 710 NLO) and quantification of the mean intensity was carried out using Adobe Photoshop CS6 software. In each condition, the mean intensity of at least 25 cells was analyzed and normalized to the control. For each experimental repeat, at least 25 cells were manually selected from the pictures. The mean intensity for the green channel was extracted from the histogram window in Adobe Photoshop. The mean intensity value for each experimental condition was corrected for the average mean intensity of all 0 h control cells (Freshly isolated, without seeding) and plotted as a percentage.
To visualize α-smooth muscle actin (α-SMA), cells were formalin-fixed, washed, permeabilized and incubated with a Cy-3 coupled mouse monoclonal anti-α-SMA (1:400, Sigma-Aldrich) for 90 min at room temperature. Finally, cells were mounted with the DAPI containing medium and pictures were taken using confocal microscopy.
Statistical analysis
Results are presented as the mean ± SEM. Comparisons between more than two groups were analyzed for statistical significance by one-way analysis of variance or Kruskal–Wallis followed by Dunnett’s multiple comparison test. Statistical analysis of values for comparison between two groups was performed using a single-sided t-test or Mann–Whitney test depending on sample distribution. Results were considered significant when p < 0.05. For in vitro experiments with treatment over time, statistical significance was determined using the multiple t-test, with Sidak–Bonferroni correction for multiple comparisons.
Results
Unfolded protein response is an immediate early response during stellate cell activation
The process of HSC activation, as a result of liver injury, can partially be mimicked in vitro by plating primary mouse HSCs (mHSCs) on plastic tissue culture dishes. HSC activation markers like α-SMA (encoded by Acta2) and Lysyl oxidase (Lox) are significantly up-regulated from 4 days onwards showing that HSCs undergo activation in vitro (Fig. 1a). Earlier studies have shown that stress pathways such as the Hippo pathway are dysregulated very early during HSC activation, even prior to the appearance of typical activation markers22. We observe that in primary mouse HSCs the UPR genes are strongly induced as soon as 2 h after seeding of the cells (Fig. 1a). Genes from each of the UPR transduction cascades were evaluated by qPCR and we observed an up-regulation of IRE1α, specific spliced XBP1 (XBP1Spl) and PERK downstream genes Atf4 and Herpud1 as a marker for ATF6 activation. In addition to those branch-specific markers, we evaluated mRNA and protein levels of two proteins that are regulated by the different UPR tracks. Bip and Chop RNA levels peaked at 10 h after seeding, after which levels returned to below starting levels after 10 days in culture (Fig. 1a). Immunofluorescent staining and western blot analysis showed that BIP protein levels however stay high in fully activated cultured cells (Fig. 1b). Importantly, CHOP is known as a pro-apoptotic protein when present in the nucleus. We did not observe any nuclear CHOP throughout the culture of primary mouse HSCs (Suppl. Fig. 1), suggesting that the UPR levels remain subtoxic.
a Bright-field images of freshly isolated mouse HSCs (mHSCs), plated on plastic culture dishes, were taken after 2, 10 and 24 h and 4, 7 and 10 days in culture (upper right panel). At the same time points, expression of HSC activation markers Acta2 and Lox was determined at the mRNA level by quantitative real-time polymerase chain reaction (qPCR; upper left panel). The lower panel shows mRNA levels of UPR genes Bip, Chop, Xbp1s, Atf4 and Herpud1 at the indicated time points during HSC culture; n = 3. b Immunofluorescent staining for Bip (green) of HSCs fixed at different time points during cultivation. 4′,6-Diamidino-2-phenylindole (DAPI; blue) was used as a nuclear staining. Graph representing the quantification of % mean intensity in at least 25 cells for each time point. c Western blot analysis of HSC activation markers PDGFRβ and α-smooth muscle actin (α-SMA) and of UPR marker BIP. GAPDH was used as a loading control. Cells were collected at the indicated time points, and samples for the 0 h time point were not seeded, but lysed immediately after cell isolation. Images and graphics are representatives of at least two different HSC isolations. *P < 0.05, **P < 0.01 and ***P < 0.001 versus 0 h
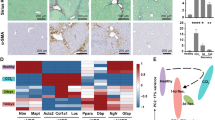
One might argue that the procedure of stellate cell isolation by organ perfusion, followed by in situ digestion and several washing steps causes stress and that the observations are isolation-induced artefacts. In addition, culture-induced HSC activation only partly reflects the in vivo activation of HSCs22,23. These important issues were addressed by isolating primary mHSCs from CCl4-treated BalbC mice, a well-established mouse model of liver fibrosis induction. Mice were given a single dose of the hepatotoxin, and stellate cells were isolated from these mice at different time points after CCl4 for immediate RNA and protein analysis. While stellate cell activation markers Acta2 and Lox are not up-regulated until 24 h after the exposure (Fig. 2a), UPR response markers Bip and Chop show the same kinetics as during in vitro mHSC activation, with peak mRNA and protein levels 2–6 h after CCl4 administration (Fig. 2b). Together, these data show that during culture-induced HSC activation and CCl4-induced HSC activation in vivo, an UPR is rapidly and transiently induced.
a BALB/c mice received a single CCl4 injection. After the indicated time points, i.e., 2, 6, 10 and 24 h, HSCs were isolated and immediately collected for mRNA analysis of activation and endoplasmic reticulum (ER) stress markers. The graphics show the results of two different HSC isolations (each time point = 3 mice). b Immunofluorescent staining for Bip (green) of HSCs which were isolated, cytospinned and fixed at different time points after a single CCl4 injection. 4′,6-Diamidino-2-phenylindole (DAPI; blue) was used as a nuclear staining. Graph representing the quantification of % mean intensity in at least 25 cells for each time point, from 3 independent isolations. c mRNA analysis of ER stress markers in HSCs isolated after multiple CCl4 injections (harvested 24 h after the last injection); n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 versus control
A more vigorous mHSC activation and subsequent liver fibrosis can be obtained by repeated injections of CCl4. When mice were exposed to a chronic liver injury model, induced by two injections of CCl4 per week for 2 to 8 weeks, the UPR was sustained in isolated HSCs from mice that received up to 8 CCl4 injections, followed by a limited decrease after 12 and 16 injections (6–8 weeks respectively, Fig. 2c). This suggests that only repeated liver injury increases and maintains high levels of the UPR in HSCs.
ER stress induction is not sufficient to drive HSC activation in a model for quiescence
Since in vitro HSC activation is known to be influenced by matrix stiffness24,25, we hypothesized that preventing adherence to the hard tissue culture plastic could avoid UPR and subsequent HSC activation. Spheroid cultures are well accepted in cancer studies, gain interest in liver toxicity studies and could be useful for fibrosis studies as well. Here, we grew freshly isolated mHSCs in 3D spheroids which maintains HSCs quiescent for at least 10 days22. Primary mHSCs grown in 3D spheroids showed a strong inhibition of activation marker expression compared to HSCs seeded on plastic (two dimensional (2D)) (Fig. 3a). Concomitantly, the early UPR observed in 2D was clearly inhibited when mHSCs were cultured directly in 3D (Fig. 3b). Thus, these 3D cultures allow to study HSC activation in vitro without the influence of the “stiff” environment of tissue culture plates. Indeed, incubating 3D spheroids for 10 h withTGF-β, an imperative fibrogenic growth factor26, induces the expression of HSC activation markers Acta2 and Lox. Analysis of UPR genes only shows a modest increase after TGF-β treatment (Fig. 3c). The use of an ER stress inducer, Tunicamycin (Tm)27, on the other hand, significantly induced ER stress in 3D spheroids, but did not induce HSC activation (Fig. 3d). This demonstrates that in the absence of other stimuli, the UPR is not sufficient to drive the HSC activation process.
a Freshly isolated mouse HSCs (mHSCs) were cultured in 96-well plates under gentle rotation to prevent attachment and induce the formation of 3D spheroids. In the upper panel, bright-field images of the 3D spheroids are shown at several time points in culture. At the same time points, 3D spheroids (3D) and plastic controls (2D) were collected for mRNA analysis of HSC activation markers (lower panel). b Graphics show mRNA expression of UPR genes during cultivation of 3D and 2D plated cells. Primary mHSCs were seeded as 3D spheroids followed by transforming growth factor-β (TGF-β) (c) or 0.5 µg/ml tunicamycin (Tm) (d) treatment. After 10 h, cells were collected and expression of activation and UPR genes was determined and compared with control-treated cells at the mRNA level by quantitative real-time polymerase chain reaction (qPCR). *P < 0.05, **P < 0.01 and ***P < 0.001 versus 0 h. N = 3–4 biological repeats
The early UPR during in vitro HSC activation is JNK1 dependent
As a downstream integrin target, JNK is involved in cell-substrate adhesion and influences adherent junction formation in a substrate-stiffness-dependent manner. In addition, clear links between stress responses and JNK28 and between JNK and HSC activation and liver fibrogenesis19 have been described. To investigate a potential link between JNK and ER stress during the in vitro HSC activation process, we treated primary mHSCs with non-toxic (Suppl. Fig. 2) concentrations of an adenosine triphosphate (ATP)-competitive inhibitor of JNK (JNK V inhibitor) from the moment of seeding. After 7 days in culture, JNK inhibitor-treated HSCs maintain a more quiescent phenotype compared to control cells (Fig. 4a) and show a dose-dependent reduction in mRNA and protein expression of α-SMA, which was not related to toxicity (Fig. 4b, d). In addition, JNK inhibition reduced the early UPR as demonstrated by reduced mRNA and protein levels of UPR markers Bip and Chop (Fig. 4b, c). Next, we isolated HSCs from JNK1 and JNK2 KO mice and cultured them on regular plastic culture dishes to investigate the importance of JNK in the early UPR during culture-induced HSC activation independent of inhibitors. Only HSCs from JNK1 KO mice showed a decreased mRNA expression level of Acta2, confirming earlier work by Zhao et al.19. Furthermore, the early peak in Bip, Chop and Atf4 expression was significantly down-regulated in JNK1 KO HSCs, while no effect could be seen in Xbp1s expression and Herpud1 was only slightly up-regulated. No reduction in ER stress and activation was seen in HSCs from JNK2 KO mice (Fig. 4e). Altogether, these results suggest that the early UPR observed during culture-induced HSC activation is JNK1 dependent.
a Freshly isolated mouse HSCs (mHSCs) were treated with the JNK V inhibitor starting at the moment of seeding. Bright-field images of control (days 1 and 7) and JNK inhibitor-treated HSCs (day 7) were taken. b At regular intervals during culture, expression of activation and endoplasmic reticulum (ER) stress markers was determined and compared with untreated cells at the mRNA level by quantitative real-time polymerase chain reaction (qPCR). c Immunofluorescent images for Bip (green, upper left panel) of JNK inhibitor-treated and control cells after 10 h in culture were taken with a confocal microscope. 4′,6-Diamidino-2-phenylindole (DAPI; blue) was used as a nuclear staining. Graph representing the quantification of % mean intensity in at least 25 cells for each time point (right panel). d Control cells (days 1 and 7) and 7-day treated HSCs with JNK inhibitor were fixed and stained for α-smooth muscle actin (α-SMA; red) and DAPI (blue). Images were taken with a confocal microscope. We had to slightly overexpose all groups to be able to observe α-SMA staining in the 10 µM JNK inhibitor-treated group and to keep the same exposure time between the different conditions. Images and graphics are representatives of at least two different HSC isolations. *P < 0.05 and ***P < 0.001 versus control. e HSCs were isolated from JNK1 knockout (KO) mice, JNK2 KO mice and their littermate controls followed by plating on plastic culture dishes. After 0, 10 and 24 h in culture, cells were collected and analyzed for mRNA expression levels of activation and UPR genes by qPCR. *P < 0.05 and **P < 0.01 versus control. N = 3 biological repeats
Fine tuning of UPR by non-sense-mediated decay
We hypothesized that the transient character of the UPR gene expression peaks reflects a tightly regulated process, possibly to prevent the cell from devastating UPR levels that would lead to premature HSC death. One mechanism that can regulate the kinetics of UPR is the NMD pathway29. We first evaluated the expression of several NMD components during HSC activation. UPF3 is a peripheral NMD factor and is only marginally up-regulated early during in vitro activation. The recruitment of UPF1 is considered an essential step in the NMD complex assembly and this gene shows a moderate up-regulation in the first hours after HSC seeding. UPF1 on its turn can attract SMG5 which will recruit ribonucleases for the actual RNA decay, and the Smg5 mRNA level is transiently up-regulated during HSC activation similar to the induction of UPR genes. Finally, we investigated the expression of a known NMD-downstream gene with a role in HSC activation, Gadd45α, which followed the same trend (Fig. 5a). Like the culture-induced HSC activation, NMD-related genes are only affected at the early stages of in vivo HSC activation upon a single dose of CCl4 (Fig. 5b), while no changes are observed during fibrogenesis upon chronic injury (Fig. 5c). These observations support the hypothesis of interaction between UPR and NMD in activating HSCs, and next we confirmed this interplay between the stress responses.
The expression of NMD core components Upf1 and Upf3a and of NMD-regulated genes Gadd45α and Smg5 was determined by quantitative real-time polymerase chain reaction (qPCR) on primary mouse HSCs. a HSCs from healthy mouse livers were collected at the moment of seeding and after 4, 10 and 24 h in culture to allow HSC activation. *P < 0.05 versus 0 h, n = 4. b HSCs were isolated after a single administration of CCl4 and analyzed immediately after the isolation. *P < 0.05, **P < 0.01 and ****P < 0.0001 versus Ctr (N = 3 biological repeats and for each repeat cells from 3 mice were pooled). c mRNA changes were examined on HSCs sorted from fibrotic mouse liver. *P < 0.05 versus Ctr. N = 3 biological repeats
In Fig. 3d we have shown that tunicamycin treatment induces UPR but not HSC activation in quiescent 3D spheroids. In these cultures, we do observe a slight elevation of NMD genes, indicative of a compensatory response (Fig. 6a). Several compounds that interfere with NMD activity such as wortmannin, NMDi-14 and VG-1 have been reported. Here, we treated primary mouse HSCs with VG-1 as it is described as a very selective NMD inhibitor30. VG-1 treatment from the moment of seeding of the HSCs resulted in moderately increased UPR at the 10 h time point (Fig. 6b) and significantly increased the expression of two NMD-associated genes Gadd45α and Smg5, of which the latter itself is part of the NMD machinery, after 4–10 h of treatment (Fig. 6b). Comparable effects were obtained with the commercial inhibitor NMDi-14 (Suppl Fig. 3). These experiments suggest that UPR during early stellate cell activation is controlled by NMD.
a In three-dimensional (3D) spheroids formed with quiescent, freshly isolated HSCs, UPR was induced by 10 h exposure to tunicamycin (Tm). The effect on NMD markers was evaluated by quantitative real-time polymerase chain reaction (qPCR). *P < 0.05 versus Ctr; n = 4. b Freshly isolated mouse HSCs were treated with the NMD inhibitor VG-1 at the moment of seeding. At regular intervals during culture, expression of endoplasmic reticulum (ER) stress markers and NMD marker genes was determined and compared with solvent cells at the mRNA level by qPCR. *P < 0.05 versus control. N = 6 biological repeats. c Schematic overview of potential interaction between UPR and NMD pathways during acute and chronic HSC activation
Discussion
HSC activation is a key process in the onset of liver fibrosis. The mechanisms underlying HSC activation are not completely understood, but it is clear that several stress pathways are involved11,21,31. We report four major findings with respect to the UPR and HSC activation: (1) the UPR is a very early event during in vitro and in vivo HSC activation, (2) culture of HSCs in 3D spheroids can prevent the UPR and a forced UPR alone cannot drive HSC activation, (3) the early induction of the UPR during HSC activation is JNK1 dependent and (4) the NMD pathway controls the UPR at the initiation of HSC activation (Fig. 6c).
Our analysis on freshly isolated HSCs from chronically treated CCl4 mice is in line with several reports that show an increased UPR in livers (or hepatocytes) in response to both acute and chronic liver injury induced by N-acetyl-p-aminophenol, CCl4 or thioacetamide32,33. In addition, several groups described a role for ER stress during HSC apoptosis; in most cases, a high dose of a chemical ER stress inducer was used to achieve an extremely strong UPR which then resulted in apoptosis27,34,35. In our study we focused on the endogenous UPR in HSCs and demonstrate that UPR genes, related to all three major UPR sensory pathways, are up-regulated very early during HSC activation. This temporarily increased UPR occurred in HSCs after an acute liver injury in vivo and during culture-induced activation in vitro. Such an early UPR has been reported in neurodegenerative diseases during the first stages of accumulation and aggregation of toxic proteins involved in Alzheimer and Parkinson disease36. In addition, a previous study by Kim et al.14 also observed an activation of the XBP1 arm in HSCs 2 days after seeding.
Activation of JNK by IRE1α has been extensively described in different cell culture models37,38. Our data confirm that JNK can directly affect the UPR, since we observe a complete inhibition of ER stress by chemical abrogation of JNK in primary mouse HSCs. Furthermore, JNK1-deficient HSCs do not activate in culture and the more general UPR genes Bip and Chop are not induced (Fig. 4). In these JNK1 KO HSCs, only Atf4 mRNA levels are not induced when compared to JNK2 KO mice or control mice, suggesting that JNK1 is involved in the PERK pathway of the UPR in these cells. Taken together, these results indicate that the early UPR that occurs during HSC activation is JNK1 dependent.
By using 3D cultures of primary HSCs we clearly demonstrated that ER stress alone is not sufficient to drive HSC activation, while others have shown that when seeded on a rigid surface, ER stress inducers can increase HSC activation markers11,12,27. The existence and cooperation of multiple stress pathways to induce HSC activation is most likely the reason why we do not observe HSC activation in 3D spheroids upon tunicamycin-induced ER stress. It is tempting to speculate that simultaneous specific induction of other stress pathways, which on their own would also not induce HSC activation in 3D culture conditions, could lead to HSC activation in combination with ER stress stimulation. In regular 2D cultures of HSC cell lines, ectopic XBP1 overexpression can promote fibrogenic gene expression without inducing the TGF-β pathway, indicating that under these culture conditions ER stress is sufficient to activate HSCs14.
While several studies showed that UPR plays a role during HSC activation, experimental identification and validation of the different UPR branches remains challenging. Different papers describe conflicting data and either PERK13, IRE1α10,11,12,14,39 or even ATF612 have been declared the most important UPR tract during HSC activation. These discrepancies are most likely due to differences in the used cell types (primary versus cell lines, human versus rodent) and to the sampling at different culture time points. This is of utmost importance as the UPR seems to be tightly controlled in time and extent by different counteracting mechanisms to avoid cellular death. Examples of these pathways that could relief UPR stress are autophagy11 and non-sense-mediated decay29.
Long-term treatment of rats with ethanol increased both ER stress and autophagy and blocking of the IRE1α pathway in JS1 cells by using a dominant-negative mutant for IRE1α revealed an inhibitory effect on autophagy in this HSC cell line. This suggested a direct effect of ER stress induction on autophagy in HSCs11. Previously, we showed that autophagy is only significantly up-regulated during in vitro HSC activation from day 4 onwards21. Since we observe in this study an UPR already after 10 h which rapidly decreases after 24 h, we believe that it is unlikely that this relatively low UPR induces the autophagy observed in these cultures 3 days later. Autophagy inhibition in HSP47-KO HSCs does cause ER stress-mediated apoptosis, indicating that in such extreme circumstances these processes are clearly linked in HSCs40. One of the characteristics of an active UPR is a general inhibition of mRNA translation into protein through the PERK branch and eIF2alpha phosphorylation. This would result in an increase in cellular mRNA transcripts, which can be removed by NMD. NMD, which was originally described as a mechanism to remove aberrant RNAs, can also finetune the amounts of normal mRNA transcripts in a cell. Karam et al.29 showed that IRE1α can be an NMD target. In our study, we observe a modest effect of NMD inhibition on UPR genes, and a significant effect on NMD-sensitive RNA Gadd45α at the evaluated early time points. Gadd45α is one of the few conserved NMD targets and was shown to play a role in reactive oxygen species-mediated TGF-β signaling in HSCs41. Our data support the hypothesis that NMD, at the initiation of HSC activation, might counteract the UPR on the one hand, but on the other hand also cause decay of Gadd45α which on its turn is involved in reactive oxygen species-mediated HSC activation. Finally, we observe increased UPR levels and low NMD gene expression in fibrotic mice following chronic CCl4 treatment. NMD activator therapy could thus potentially be used to treat patients with advanced liver fibrosis and prolonged HSC activation, but will require more in-depth in vivo studies when NMD activating molecules become available.
Taken together, our data show that there is an early JNK1-dependent UPR during HSC activation, but that this UPR is not enough to drive the activation process and is at least partly countered at the early activation stages by NMD. Hence, we can speculate that targeting the UPR as potential anti-fibrotic therapy will be challenging because of the close interplay and feedback mechanisms that exist and it might not suffice to observe relevant changes in scar deposition.
References
Friedman S., Sheppard D., Duffield, J. S., Violette, S. Therapy for fibrotic diseases: nearing the starting line. Sci. Transl. Med. 5, 167sr1 (2013).
Friedman S., Sheppard D., Duffield, J. S., Violette, S. Therapy for fibrotic diseases: nearing the starting line. Sci. Transl. Med. Physiol. Rev. 88, 125–172 (2008).
Mederacke, I. et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 4, 2823 (2013).
Senft, D. & Ronai, Z. A. UPR, autophagy, and mitochondria crosstalk underlies the er stress response. Trends Biochem. Sci. 40, 141–148 (2015).
Rutkowski, D. T. & Kaufman, R. J. A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28 (2004).
Wu, F. L. et al. Targeting endoplasmic reticulum stress in liver disease. Expert Rev. Gastroenterol. Hepatol. 10, 1041–1052 (2016).
Chusri, P. et al. HCv induces transforming growth factor beta1 through activation of endoplasmic reticulum stress and the unfolded protein response. Sci. Rep. 6, 22487 (2016).
Li, X. et al. Emodin ameliorates hepatic steatosis through endoplasmic reticulum-stress sterol regulatory element-binding protein 1c pathway in liquid fructose-feeding rats. Hepatol. Res. 46, E105–E117 (2016).
Kwanten, W. J. et al. Hepatocellular autophagy modulates the unfolded protein response and fasting-induced steatosis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G599–G609 (2016).
Borkham-Kamphorst, E. et al. CCN1/CYR61 overexpression in hepatic stellate cells induces er stress-related apoptosis. Cell. Signal. 28, 34–42 (2016).
Hernandez-Gea, V. et al. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J. Hepatol. 59, 98–104 (2013).
de Galarreta, M. R. et al. Unfolded protein response induced by brefeldin A increases collagen type I levels in hepatic stellate cells through an IRE1alpha, p38 MAPK and SMAD-dependent pathway. Biochim. Biophys. Acta 1863, 2115–2123 (2016).
Koo, J. H., Lee, H. J., Kim, W. & Kim, S. G. Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of HNRNPA1 and up-regulation of SMAD2. Gastroenterology 150, 181–193 e188 (2016).
Kim, R. S. et al. The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci. Rep. 6, 39342 (2016).
Li, Z., Vuong, J. K., Zhang, M., Stork, C. & Zheng, S. Inhibition of nonsense-mediated RNA decay by ER stress. RNA 23, 378–394 (2017).
Carreras-Sureda, A. & Hetz, C. RNA metabolism: putting the brake on the UPR. EMBO Rep. 16, 545–546 (2015).
Bhuvanagiri, M. et al. 5-azacytidine inhibits nonsense-mediated decay in a myc-dependent fashion. EMBO Mol. Med. 6, 1593–1609 (2014).
Mannaerts, I. et al. Chronic administration of valproic acid inhibits activation of mouse hepatic stellate cells in vitro and in vivo. Hepatology 51, 603–614 (2010).
Zhao, G. et al. Jnk1 in murine hepatic stellate cells is a crucial mediator of liver fibrogenesis. Gut 63, 1159–1172 (2014).
Cubero, F. J. et al. Combined activities of JNK1 and JNK2 in hepatocytes protect against toxic liver injury. Gastroenterology 150, 968–981 (2016).
Thoen, L. F. R. et al. A role for autophagy during hepatic stellate cell activation. J. Hepatol. 55, 1353–1360 (2011).
Mannaerts, I. et al. The Hippo pathway effector YAP controls mouse hepatic stellate cell activation. J. Hepatol. 63, 679–688 (2015).
De Minicis, S. et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 132, 1937–1946 (2007).
Olsen, A. L. et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G110–G118 (2011).
Guvendiren, M., Perepelyuk, M., Wells, R. G. & Burdick, J. A. Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J. Mech. Behav. Biomed. Mater. 38, 198–208 (2014).
Pinzani, M. & Marra, F. Cytokine receptors and signaling in hepatic stellate cells. Semin. Liver Dis. 21, 397–416 (2001).
De Minicis, S. et al. Endoplasmic reticulum stress induces hepatic stellate cell apoptosis and contributes to fibrosis resolution. Liver Int. 32, 1574–1584 (2012).
Urano, F. et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 (2000).
Karam, R. et al. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 16, 599–609 (2015).
Gotham, V. J. et al. Synthesis and activity of a novel inhibitor of nonsense-mediated mRNA decay. Org. Biomol. Chem. 14, 1559–1563 (2016).
Hernández–Gea, V. et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 142, 938–946 (2012).
Campos, G. et al. The transcription factor CHOP, a central component of the transcriptional regulatory network induced upon CCL4 intoxication in mouse liver, is not a critical mediator of hepatotoxicity. Arch. Toxicol. 88, 1267–1280 (2014).
Jin, C. F. et al. Mechanism of the inhibitory effects of Eucommia ulmoides Oliv. Cortex Extracts (EUCE) in the CCL4-induced acute liver lipid accumulation in rats. Int. J. Endocrinol. 2013, 751854 (2013).
Lim, M. P., Devi, L. A. & Rozenfeld, R. Cannabidiol causes activated hepatic stellate cell death through a mechanism of endoplasmic reticulum stress-induced apoptosis. Cell Death Dis. 2, e170 (2011).
Huang, Y. et al. Endoplasmic reticulum stress-induced hepatic stellate cell apoptosis through calcium-mediated JNK/p38 MAPK and calpain/caspase-12 pathways. Mol. Cell Biochem. 394, 1–12 (2014).
Hoozemans, J. J., van Haastert, E. S., Nijholt, D. A., Rozemuller, A. J. & Scheper, W. Activation of the unfolded protein response is an early event in Alzheimer's and Parkinson's disease. Neurodegener. Dis. 10, 212–215 (2012).
Ozcan, U. et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306, 457–461 (2004).
Sun, R. Q. et al. IRE1 impairs insulin signaling transduction of fructose-fed mice via JNK independent of excess lipid. Biochim Biophys. Acta 1852, 156–165 (2015).
Heindryckx, F. et al. Endoplasmic reticulum stress enhances fibrosis through IRE1alpha-mediated degradation of miR-150 and XBP-1 splicing. EMBO Mol. Med. 8, 729–744 (2016).
Kawasaki, K. et al. Deletion of the collagen-specific molecular chaperone HSP47 causes endoplasmic reticulum stress-mediated apoptosis of hepatic stellate cells. J. Biol. Chem. 290, 3639–3646 (2015).
Hong, L. et al. New role and molecular mechanism of Gadd45a in hepatic fibrosis. World J. Gastroenterol. 22, 2779–2788 (2016).
Acknowledgements
We would like to acknowledge Jean Marc Lazou, Danielle Blyweert and Eddy Himpe for technical support. L.F.R.T. was funded by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen; SB/101577). I.M. is supported by a Fund of Scientific Research Flanders FWO-V post-doctoral fellowship (117300/12N5415N LV). L.A.v.G and C.T. were supported by an Interuniversity Attraction Poles (IAP)–phase VII–project P7/47 (Federal Science Policy–BELSPO) and L.A.v.G. and I.C. by an FWO grant (G.0751.12N).
Authors' contributions
I.M.: study concept and design; acquisition of data; analysis and interpretation of data; writing of the manuscript. L.F.R.T.: study concept and design; acquisition of data; analysis and interpretation of data; statistical analysis; drafting of the manuscript. N.E.: acquisition and analysis of data. F.J.C., C.T. and I. Coldham: provision of materials; critical revision of the manuscript. S.B.L.: acquisition of data; data analysis and interpretation of data; critical revision of the manuscript. I. Colle: funding; critical revision of the manuscript. L.A.v.G.: study concept and design, funding; interpretation of data, critical revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Daugaard
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mannaerts, I., Thoen, L.F.R., Eysackers, N. et al. Unfolded protein response is an early, non-critical event during hepatic stellate cell activation. Cell Death Dis 10, 98 (2019). https://doi.org/10.1038/s41419-019-1327-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1327-5
This article is cited by
-
Cellular stress in the pathogenesis of nonalcoholic steatohepatitis and liver fibrosis
Nature Reviews Gastroenterology & Hepatology (2023)
-
Initiation of hepatic stellate cell activation extends into chronic liver disease
Cell Death & Disease (2021)
-
Cytoplasmic vacuolation with endoplasmic reticulum stress directs sorafenib induced non-apoptotic cell death in hepatic stellate cells
Scientific Reports (2021)