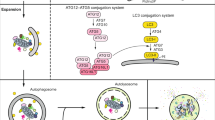
Abstract
Autophagy is an essential recycling and quality control pathway which preserves cellular and organismal homeostasis. As a catabolic process, autophagy degrades damaged and aged intracellular components in response to conditions of stress, including nutrient deprivation, oxidative and genotoxic stress. Autophagy is a highly adaptive and dynamic process which requires an intricately coordinated molecular control. Here we provide an overview of how autophagy is regulated post-transcriptionally, through RNA processing events, epitranscriptomic modifications and non-coding RNAs. We further discuss newly revealed RNA-binding properties of core autophagy machinery proteins and review recent indications of autophagy’s ability to impact cellular RNA homeostasis. From a physiological perspective, we examine the biological implications of these emerging regulatory layers of autophagy, particularly in the context of nutrient deprivation and tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42.
Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J. 2021;40:e108863.
Debnath J, Gammoh N, Ryan KM. Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. 2023;24:1–16.
Abildgaard MH, Brynjolfsdottir SH, Frankel LB. The autophagy-RNA interplay: degradation and beyond. Trends Biochem Sci. 2020;45:845–57.
Gonzalez-Rodriguez P, Klionsky DJ, Joseph B. Autophagy regulation by RNA alternative splicing and implications in human diseases. Nat Commun. 2022;13:2735.
Frankel LB, Lubas M, Lund AH. Emerging connections between RNA and autophagy. Autophagy. 2017;13:3–23.
Ghafouri-Fard S, Shoorei H, Mohaqiq M, Majidpoor J, Moosavi MA, Taheri M. Exploring the role of non-coding RNAs in autophagy. Autophagy. 2022;18:949–70.
Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22.
Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med. 2020;52:400–8.
Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428–41.
Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, et al. m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol. 2022;40:1210–9.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.
Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203.
Cho S, Lee G, Pickering BF, Jang C, Park JH, He L, et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Mol Cell. 2021;81:2064–75.e2068.
Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, et al. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell. 2017;171:1545–58.e1518.
Nandagopal N, Roux PP. Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation (Austin). 2015;3:e983402.
Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305.
Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14.
Tang HW, Weng JH, Lee WX, Hu Y, Gu L, Cho S, et al. mTORC1-chaperonin CCT signaling regulates m(6)A RNA methylation to suppress autophagy. Proc Natl Acad Sci USA. 2021;118:e2021945118.
Kim AR, Choi KW. TRiC/CCT chaperonins are essential for organ growth by interacting with insulin/TOR signaling in Drosophila. Oncogene. 2019;38:4739–54.
Hao W, Dian M, Zhou Y, Zhong Q, Pang W, Li Z, et al. Autophagy induction promoted by m(6)A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat Commun. 2022;13:5845.
Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129:2475–81.
Jin S, Zhang X, Miao Y, Liang P, Zhu K, She Y, et al. m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018;28:955–7.
Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–35.
Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419–37.
Gulati P, Cheung MK, Antrobus R, Church CD, Harding HP, Tung YC, et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc Natl Acad Sci USA. 2013;110:2557–62.
Chen Y, Wang J, Xu D, Xiang Z, Ding J, Yang X, et al. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy. 2021;17:457–75.
Wang K, Wang S, Zhang Y, Xie L, Song X, Song X. SNORD88C guided 2'-O-methylation of 28S rRNA regulates SCD1 translation to inhibit autophagy and promote growth and metastasis in non-small cell lung cancer. Cell Death Differ. 2023;30:341–55.
Hocine S, Singer RH, Grunwald D. RNA processing and export. Cold Spring Harb Perspect Biol. 2010;2:a000752.
Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–66.
Liu Y, Gonzalez-Porta M, Santos S, Brazma A, Marioni JC, Aebersold R, et al. Impact of alternative splicing on the human proteome. Cell Rep. 2017;20:1229–41.
Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–51.
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7.
Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–33.
Sakellariou D, Tiberti M, Kleiber TH, Blazquez L, Lopez AR, Abildgaard MH, et al. eIF4A3 regulates the TFEB-mediated transcriptional response via GSK3B to control autophagy. Cell Death Differ. 2021;28:3344–56.
Sakellariou D, Frankel LB. EIF4A3: a gatekeeper of autophagy. Autophagy. 2021;17:4504–5.
Park JY, Sohn HY, Koh YH, Jo C. A splicing variant of TFEB negatively regulates the TFEB-autophagy pathway. Sci Rep. 2021;11:21119.
B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–99.
Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, et al. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem. 2013;288:859–72.
Zhang Z, Qian Q, Li M, Shao F, Ding WX, Lira VA, et al. The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB. Autophagy. 2021;17:1841–55.
Wang CC, Peng H, Wang Z, Yang J, Hu RG, Li CY, et al. TRIM72-mediated degradation of the short form of p62/SQSTM1 rheostatically controls selective autophagy in human cells. Mil Med Res. 2022;9:35.
Moharir SC, Bansal M, Ramachandran G, Ramaswamy R, Rawat S, Raychaudhuri S, et al. Identification of a splice variant of optineurin which is defective in autophagy and phosphorylation. Biochim Biophys Acta Mol Cell Res. 2018;1865:1526–38.
Berkovits BD, Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–7.
Malka Y, Alkan F, Ju S, Korner PR, Pataskar A, Shulman E, et al. Alternative cleavage and polyadenylation generates downstream uncapped RNA isoforms with translation potential. Mol Cell. 2022;82:3840–55.e3848.
Tang HW, Hu Y, Chen CL, Xia B, Zirin J, Yuan M, et al. The TORC1-regulated CPA complex rewires an RNA processing network to drive autophagy and metabolic reprogramming. Cell Metab. 2018;27:1040–54.e1048.
Hwang HJ, Ha H, Lee BS, Kim BH, Song HK, Kim YK. LC3B is an RNA-binding protein to trigger rapid mRNA degradation during autophagy. Nat Commun. 2022;13:1436.
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8.
Mizushima N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020;63:1–10.
Vats S, Galli T. Role of SNAREs in unconventional secretion-focus on the VAMP7-dependent secretion. Front Cell Dev Biol. 2022;10:884020.
Kakanj P, Bhide S, Moussian B, Leptin M. Autophagy-mediated plasma membrane removal promotes the formation of epithelial syncytia. EMBO J. 2022;41:e109992.
Reid SE, Kolapalli SP, Nielsen TM, Frankel LB. Canonical and non-canonical roles for ATG8 proteins in autophagy and beyond. Front Mol Biosci. 2022;9:1074701.
Zhou J, Ma J, Yang C, Zhu X, Li J, Zheng X, et al. A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat Plants. 2023;9:749–65.
Rojas-Rios P, Chartier A, Pierson S, Severac D, Dantec C, Busseau I, et al. Translational control of autophagy by Orb in the Drosophila Germline. Dev Cell. 2015;35:622–31.
Yamaguchi T, Suzuki T, Sato T, Takahashi A, Watanabe H, Kadowaki A, et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal. 2018;11:eaan3638.
Yin Z, Zhang Z, Lei Y, Klionsky DJ. Bidirectional roles of the Ccr4-Not complex in regulating autophagy before and after nitrogen starvation. Autophagy. 2023;19:415–25.
Cheng DD, Li J, Li SJ, Yang QC, Fan CY. CNOT1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol Oncol. 2017;11:388–404.
De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–90.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57.
Seiden-Long IM, Brown KR, Shih W, Wigle DA, Radulovich N, Jurisica I, et al. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25:91–102.
Horos R, Buscher M, Kleinendorst R, Alleaume AM, Tarafder AK, Schwarzl T, et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019;176:1054–67.e1012.
Buscher M, Horos R, Hentze MW. 'High vault-age': non-coding RNA control of autophagy. Open Biol. 2020;10:190307.
Buscher M, Horos R, Huppertz I, Haubrich K, Dobrev N, Baudin F, et al. Vault RNA1-1 riboregulates the autophagic function of p62 by binding to lysine 7 and arginine 21, both of which are critical for p62 oligomerization. RNA. 2022;28:742–55.
Karras P, Riveiro-Falkenbach E, Canon E, Tejedo C, Calvo TG, Martinez-Herranz R, et al. p62/SQSTM1 fuels melanoma progression by opposing mRNA decay of a selective set of pro-metastatic factors. Cancer Cell. 2019;35:46–63.e10.
Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, et al. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 2016;15:1634–47.
Lin WC, Chen LH, Hsieh YC, Yang PW, Lai LC, Chuang EY, et al. miR-338-5p inhibits cell proliferation, colony formation, migration and cisplatin resistance in esophageal squamous cancer cells by targeting FERMT2. Carcinogenesis. 2019;40:883–92.
Zhang X, Dai M, Li S, Li M, Cheng B, Ma T, et al. The emerging potential role of p62 in cancer treatment by regulating metabolism. Trends Endocrinol Metab. 2023;34:474–88.
Wu X, Xu L. The RNA-binding protein HuR in human cancer: a friend or foe? Adv Drug Deliv Rev. 2022;184:114179.
Ji E, Kim C, Kang H, Ahn S, Jung M, Hong Y, et al. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol Cell Biol. 2019;39:e00508–18.
Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A, et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083–103.
Viiri J, Amadio M, Marchesi N, Hyttinen JM, Kivinen N, Sironen R, et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013;8:e69563.
Li XX, Xiao L, Chung HK, Ma XX, Liu X, Song JL, et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. 2020;40:e00492–19.
Shao Z, Ni L, Hu S, Xu T, Meftah Z, Yu Z, et al. RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration. Cell Prolif. 2021;54:e12975.
Bishayee K, Habib K, Nazim UM, Kang J, Szabo A, Huh SO, et al. RNA binding protein HuD promotes autophagy and tumor stress survival by suppressing mTORC1 activity and augmenting ARL6IP1 levels. J Exp Clin Cancer Res. 2022;41:18.
Liu M, Bai J, He S, Villarreal R, Hu D, Zhang C, et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell Metab. 2014;19:967–80.
Yang C, Wang Z, Kang Y, Yi Q, Wang T, Bai Y, et al. Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules. Autophagy. 2023;19:1934–51.
Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–74.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45.
An H, Harper JW. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2018;20:135–43.
Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, et al. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015;34:154–68.
Makino S, Kawamata T, Iwasaki S, Ohsumi Y. Selectivity of mRNA degradation by autophagy in yeast. Nat Commun. 2021;12:2316.
Hickl D, Drews F, Girke C, Zimmer D, Muhlhaus T, Hauth J, et al. Differential degradation of RNA species by autophagy-related pathways in Arabidopsis. J Exp Bot. 2021;72:6867–81.
Asadi MR, Rahmanpour D, Moslehian MS, Sabaie H, Hassani M, Ghafouri-Fard S, et al. Stress granules involved in formation, progression and metastasis of cancer: a scoping review. Front Cell Dev Biol. 2021;9:745394.
Kobayashi H, Shoji K, Kiyokawa K, Negishi L, Tomari Y. VCP machinery mediates autophagic degradation of empty argonaute. Cell Rep. 2019;28:1144–53.e1144.
Michaeli S, Clavel M, Lechner E, Viotti C, Wu J, Dubois M, et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci USA. 2019;116:22872–83.
Kobayashi H, Shoji K, Kiyokawa K, Negishi L, Tomari Y. Iruka eliminates dysfunctional argonaute by selective ubiquitination of its empty state. Mol Cell. 2019;73:119–29.e115.
Lai HH, Li JN, Wang MY, Huang HY, Croce CM, Sun HL, et al. HIF-1alpha promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J Clin Invest. 2018;128:625–43.
Tong X, Liu SY, Zou JZ, Zhao JJ, Zhu FF, Chai LX, et al. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021;40:e108050.
Santovito D, Egea V, Bidzhekov K, Natarelli L, Mourao A, Blanchet X, et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci Transl Med. 2020;12:eaaz2294.
Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24:430–47.
Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in Oncology. Cell. 2019;179:1033–55.
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41.
Lin Z, Tang X, Wan J, Zhang X, Liu C, Liu T. Functions and mechanisms of circular RNAs in regulating stem cell differentiation. RNA Biol. 2021;18:2136–49.
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149.
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.
Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36.
Karedath T, Ahmed I, Al Ameri W, Al-Dasim FM, Andrews SS, Samuel S, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19:565.
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan Q, et al. The influence of circular RNAs on autophagy and disease progression. Autophagy. 2022;18:240–53.
Ma L, Wang Z, Xie M, Quan Y, Zhu W, Yang F, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 2020;11:169.
Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int. 2020;44:1945–56.
He Z, Cai K, Zeng Z, Lei S, Cao W, Li X. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. 2022;13:233.
Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12:219.
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–42.
Xu J, Xu J, Liu X, Jiang J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell Death Discov. 2022;8:287.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61.
Tiessen I, Abildgaard MH, Lubas M, Gylling HM, Steinhauer C, Pietras EJ, et al. A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene. 2019;38:5127–41.
Sun Y, Ren D, Zhou Y, Shen J, Wu H, Jin X. Histone acetyltransferase 1 promotes gemcitabine resistance by regulating the PVT1/EZH2 complex in pancreatic cancer. Cell Death Dis. 2021;12:878.
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X, et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19:118.
Yang S, Yuan ZJ, Zhu YH, Chen X, Wang W. lncRNA PVT1 promotes cetuximab resistance of head and neck squamous cell carcinoma cells by inhibiting miR-124-3p. Head Neck. 2021;43:2712–23.
Wang J, Dong Z, Sheng Z, Cai Y. Hypoxia-induced PVT1 promotes lung cancer chemoresistance to cisplatin by autophagy via PVT1/miR-140-3p/ATG5 axis. Cell Death Discov. 2022;8:104.
Yi J, Wang L, Hu GS, Zhang YY, Du J, Ding JC, et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. EMBO J. 2023;42:e112408.
Shi J, Guo C, Li Y, Ma J. The long noncoding RNA TINCR promotes self-renewal of human liver cancer stem cells through autophagy activation. Cell Death Dis. 2022;13:961.
Zhou Y, Shao Y, Hu W, Zhang J, Shi Y, Kong X, et al. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ. 2023;30:111–24.
Coe EA, Tan JY, Shapiro M, Louphrasitthiphol P, Bassett AR, Marques AC, et al. The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor. PLoS Genet. 2019;15:e1008501.
Frixa T, Sacconi A, Cioce M, Roscilli G, Ferrara FF, Aurisicchio L, et al. MicroRNA-128-3p-mediated depletion of Drosha promotes lung cancer cell migration. Carcinogenesis. 2018;39:293–304.
Zhao C, Guo R, Guan F, Ma S, Li M, Wu J, et al. MicroRNA-128-3p enhances the chemosensitivity of temozolomide in glioblastoma by targeting c-Met and EMT. Sci Rep. 2020;10:9471.
Xu J, Song J, Xiao M, Wang C, Zhang Q, Yuan X, et al. RUNX1 (RUNX family transcription factor 1), a target of microRNA miR-128-3p, promotes temozolomide resistance in glioblastoma multiform by upregulating multidrug resistance-associated protein 1 (MRP1). Bioengineered. 2021;12:11768–81.
Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol. 2016;17:677–86.
Hu X, Li F, He J, Yang J, Jiang Y, Jiang M, et al. LncRNA NEAT1 recruits SFPQ to regulate MITF splicing and control RPE cell proliferation. Invest Ophthalmol Vis Sci. 2021;62:18.
Acknowledgements
Figures in this review were created using BioRender.com. We thank Steven E. Reid for critical reading and commenting of the manuscript. This work was supported by the Lundbeck Foundation (R272-2017-3872), the Novo Nordisk Foundation (NNF19OC0057772) and the Danish Cancer Society (R269-A15420 and R209-A13011).
Author information
Authors and Affiliations
Contributions
SPK, TMN, and LBF conceived the topic, discussed its contents, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kolapalli, S.P., Nielsen, T.M. & Frankel, L.B. Post-transcriptional dynamics and RNA homeostasis in autophagy and cancer. Cell Death Differ (2023). https://doi.org/10.1038/s41418-023-01201-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-023-01201-5