Abstract
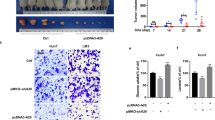
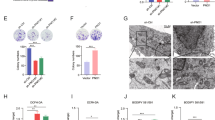
Oxoglutarate dehydrogenase-like (OGDHL) is considered to be the isoenzyme of oxyglutarate dehydrogenase (OGDH) in the OGDH complex, which degrades glucose and glutamate. OGDHL was reported to reprogram glutamine metabolism to suppress HCC progression in an enzyme-activity-dependent manner. However, the potential subcellular localization and non-canonical function of OGDHL is poorly understood. We investigated the expression of OGDHL and its effect on HCC progression. By employing a variety of molecular biology techniques, we revealed the underlying mechanism of OGDHL-induced DNA damage in HCC cells in vitro and in vivo. AAV loaded with OGDHL exerts therapeutic effect on mouse HCC and prolongs survival time. OGDHL induces DNA damage in HCC cells in vitro and in vivo. We also observed that OGDHL possesses nuclear localization in HCC cells and OGDHL-induced DNA damage was independent of its enzymatic activity. Mechanistically, it was demonstrated that OGDHL binds to CDK4 in the nucleus to inhibit the phosphorylation of CDK4 by CAK, which in turn attenuates E2F1 signaling. Inhibition of E2F1 signaling downregulates pyrimidine and purine synthesis, thereby inducing DNA damage through dNTP depletion. We clarified the nuclear localization of OGDHL and its non-canonical function to induce DNA damage, which demonstrated that OGDHL may serve as a select potential therapeutic target for HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Villanueva A. Hepatocellular Carcinoma. N. Engl J Med. 2019;380:1450–62.
Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–24.
Zou H, Li M, Lei Q, Luo Z, Xue Y, Yao D, et al. Economic Burden and Quality of Life of Hepatocellular Carcinoma in Greater China: A Systematic Review. Front Public Health. 2022;10:801981.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47.
WARBURG O. On the origin of cancer cells. Science. 1956;123:309–14.
Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr Metab (Lond). 2010;7:7.
Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8.
Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85.
Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–54.
Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61.
Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–34.
Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84.
Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4.
Currie E, Schulze A, Zechner R, Walther TC, Farese RJ. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61.
Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70.
Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, et al. Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin Cancer Res. 2009;15:1184–91.
Jiao Y, Li Y, Fu Z, Hou L, Chen Q, Cai Y, et al. OGDHL Expression as a Prognostic Biomarker for Liver Cancer Patients. Dis Markers. 2019;2019:9037131.
Sen T, Sen N, Noordhuis MG, Ravi R, Wu TC, Ha PK, et al. OGDHL is a modifier of AKT-dependent signaling and NF-kappaB function. Plos One. 2012;7:e48770.
Dai W, Xu L, Yu X, Zhang G, Guo H, Liu H, et al. OGDHL silencing promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J Hepatol. 2020;72:909–23.
Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The Evolving Landscape of Noncanonical Functions of Metabolic Enzymes in Cancer and Other Pathologies. Cell Metab. 2021;33:33–50.
Liu Y, Guo JZ, Liu Y, Wang K, Ding W, Wang H, et al. Nuclear lactate dehydrogenase A senses ROS to produce alpha-hydroxybutyrate for HPV-induced cervical tumor growth. Nat Commun. 2018;9:4429.
Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–66.
Huangyang P, Li F, Lee P, Nissim I, Weljie AM, Mancuso A, et al. Fructose-1,6-Bisphosphatase 2 Inhibits Sarcoma Progression by Restraining Mitochondrial Biogenesis. Cell Metab. 2020;31:174–88.
Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97.
DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–24.
Qin XQ, Livingston DM, Kaelin WJ, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci. 1994;91:10918–22.
Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19:326–38.
Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–8.
Nguyen BA, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. Bmc Bioinforma. 2009;10:202.
Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6:353–67.
Kato JY, Matsuoka M, Strom DK, Sherr CJ. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol. 1994;14:2713–21.
Matsuoka M, Kato JY, Fisher RP, Morgan DO, Sherr CJ. Activation of cyclin-dependent kinase 4 (cdk4) by mouse MO15-associated kinase. Mol Cell Biol. 1994;14:7265–75.
Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–5.
Bradley KJ, Bowl MR, Williams SE, Ahmad BN, Partridge CJ, Patmanidi AL, et al. Parafibromin is a nuclear protein with a functional monopartite nuclear localization signal. Oncogene 2007;26:1213–21.
Willis AN, Dean SE, Habbouche JA, Kempers BT, Ludwig ML, Sayfie AD, et al. Nuclear localization signal sequence is required for VACM-1/CUL5-dependent regulation of cellular growth. Cell Tissue Res. 2017;368:105–14.
Sharma M, Jamieson C, Johnson M, Molloy MP, Henderson BR. Specific armadillo repeat sequences facilitate beta-catenin nuclear transport in live cells via direct binding to nucleoporins Nup62, Nup153, and RanBP2/Nup358. J Biol Chem. 2012;287:819–31.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81871967, 82173129).
Author information
Authors and Affiliations
Contributions
Conceptualization: XJ, DY. Project administration: DY. Funding acquisition: DY; performed majority of the experiments and Writing original draft: XJ, YX, QL, YX and JH; Writing – review & editing: XJ, DY, SX; Investigation: CC, YW, LZ; Data curation: HL, YL, BL, JP; Supervision: DY.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics aprroval
The authors declare no competing interests. The research was approved by Experimental Animal Ethics Committee of Nanjing Hospital Affiliated to Nanjing Medical University (DWSY-2105599).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, X., Peng, J., Xie, Y. et al. Oxoglutarate dehydrogenase-like inhibits the progression of hepatocellular carcinoma by inducing DNA damage through non-canonical function. Cell Death Differ 30, 1931–1942 (2023). https://doi.org/10.1038/s41418-023-01186-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01186-1