Abstract
Exercise in later life is important for bone health and delays the progression of osteoporotic bone loss. Osteocytes are the major bone cells responsible for transforming mechanical stimuli into cellular signals through their highly specialized lacunocanalicular networks (LCN). Osteocyte activity and LCN degenerate with aging, thus might impair the effectiveness of exercise on bone health; however, the underlying mechanism and clinical implications remain elusive. Herein, we showed that deletion of Sirt3 in osteocytes could impair the formation of osteocyte dendritic processes and inhibit bone gain in response to exercise in vivo. Mechanistic studies revealed that Sirt3 regulates E11/gp38 through the protein kinase A (PKA)/cAMP response element-binding protein (CREB) signaling pathway. Additionally, the Sirt3 activator honokiol enhanced the sensitivity of osteocytes to fluid shear stress in vitro, and intraperitoneal injection of honokiol reduced bone loss in aged mice in a dose-dependent manner. Collectively, Sirt3 in osteocytes regulates bone mass and mechanical responses through the regulation of E11/gp38. Therefore, targeting Sirt3 could be a novel therapeutic strategy to prevent age-related bone loss and augment the benefits of exercise on the senescent skeleton.
Similar content being viewed by others
Introduction
Osteocytes constitute 90–95% of the total bone cells. The total number of osteocytes in the human adult skeleton is ~42 billion [1], making them the major source of circulating bone factors. For instance, osteocytes secrete sclerostin and receptor activator of nuclear factor-κB ligand (RANKL), which regulate bone homeostasis, and fibroblast growth factor 23, an important endocrine factor that regulates phosphate metabolism via action on the kidney [2]. Furthermore, they are embedded inside the bone matrix and are surrounded by fluid-filled spaces known as lacunocanalicular networks (LCN) [3], formed by interconnected dendritic processes connecting neighboring osteocytes and cells on the bone surface, such as osteoblasts and osteoclasts [4]. The structural integrity of the LCN is believed to influence bone quality and mechanosensitivity to mechanical loading [5]. The shear stress generated by fluid flow induced by mechanical loading, such as physical exercise, is regarded as a major source of biochemical signaling pathway within osteocytes [6, 7], thus conveying these signals to osteoblasts and osteoclasts.
Imbalanced bone formation and resorption with aging lead to reduced bone mass and strength, resulting in a higher risk of fracture. Exercise is an effective approach to counteract skeletal fragility; however, its effectiveness is diminished in older adults [8,9,10]. Reduced osteocyte density and decreased ability of osteocytes to transduce mechanical stimuli into biochemical signals have been suggested to contribute to the diminished response of bone to mechanical loading [11, 12]. Recently, findings on altered LCN structure and reduced dendrite number per osteocyte in human and mice aged bone have raised questions about the underlying causes and clinical implications of these morphological changes in the aged bone [5, 13]. Understanding the drivers of the diminished bone response to exercise during aging could provide new insights into novel treatment approaches to prevent age-related bone loss and synergize the benefits of exercise on the senescent skeleton.
Sirt3, a longevity gene, is predominantly expressed in mitochondria and is involved in various mitochondrial functions, including energy metabolism, reactive oxygen species (ROS) homeostasis, and mitochondrial biogenesis [14,15,16]. Decreased Sirt3 expression has been implicated in age-related disorders such as sarcopenia and metabolic syndromes [17, 18]. Current evidence shows that the number of mitochondria within osteocyte dendritic processes significantly decreases with aging [19], and Sirt3 deficiency inhibits osteoblast differentiation [20]. In addition, Sirt3 inhibition reduces the beneficial effects of exercise on skeletal muscle and neurons [21, 22]. These findings prompted us to test the hypothesis that declined Sirt3 expression in osteocytes is responsible for the altered LCN structure and the diminished response of the aging bone to exercise.
Results
Concomitant age-related degenerative change of bone quality, osteocyte dendritic processes, and Sirt3 expression in osteocyte
Micro-CT analysis showed that the cortical bone area (BArea) and thickness and finite element analysis predicted stiffness peaked at 6 months, then declined progressively with aging, and reached the lowest level at 24 months in the study period (Fig. 1A). Age-related changes in the trabecular bone followed a similar pattern, with a peak in the trabecular bone mass occurring earlier than that of the cortical bone (Supplementary Fig. 1A, B). Confocal imaging of fluorescein isothiocyanate (FITC)-stained cortical bone showed significant changes in osteocyte cell morphology and number and total length of dendrites per osteocyte with aging, which follows a pattern similar to the cortical bone mass and strength. Generally, most of the osteocytes in the young mice (aged 3 and 6 months) were spindle shaped. When mice aged, osteocytes changed to an elliptical shape at 12 months and then to a more roundish shape at 24 months. Spindle-shaped osteocytes tend to have more and longer dendritic processes per osteocyte (Fig. 1B). Quantitative analysis using Imaris software showed similar findings. The osteocyte density, number, and total length of dendrites per osteocyte reached a peak at 6 months and then decreased gradually with aging (Fig. 1C). Notably, the number and total length of dendrites per osteocyte reduced by 46% and 65%, respectively, at 24 months than at 3 months, indicating a dynamic morphological change of osteocyte dendritic processes with aging. In addition, cell body area and volume decreased with aging, whereas sphericity increased with aging (Supplementary Fig. 1C).
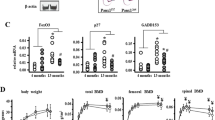
A Age-related changes in cortical bone area (BArea), cortical thickness (Ct.Th), and finite element analysis (FEA) predicted-stiffness in the midshaft femur of mice at different ages (n = 8–12). B Representative images of confocal imaging of fluorescein isothiocyanate (FITC)-stained (green) midshaft femur sections showing osteocyte connectivity in mice of different ages. Scale bar = 20 μm (upper panel) and 5 μm (lower panel). C Quantification of osteocyte density, dendrite number per osteocyte, and total dendrite length per osteocyte in mice at different ages (n = 4). D qPCR analysis of mRNA expression of Sirt3 and osteocyte markers, including E11/gp38, dentin matrix acidic phosphoprotein 1 (Dmp1), Sost, receptor activator of nuclear factor kappa-B ligand (Rankl) and osteoprotegerin (Opg), in osteocyte-enriched bone in mice at different ages (n = 4). E qPCR analysis of mRNA expressions of Sirt1-7 in osteocyte-enriched bone tissue in mice at different ages (n = 4). F A representative immunoblot of Sirt3, E11/gp38, Dmp1, and β-actin in osteocyte-enriched bone in mice at different ages (n = 4). G Semiquantification of protein levels of Sirt3, E11/gp38, and Dmp1 in F (n = 4). H qPCR analysis of Sirt3 mRNA expression in different tissues in mice at different ages (n = 4). I Mitochondrial DNA content (mt-DNA) and cellular adenosine triphosphate (ATP) levels in ostoecyte-enriched bone in mice at different ages (n = 3). All data were presented in mean ± SEM and analyzed with one-way ANOVA. *p < 0.05, **p < 0.01, and ***p < 0.001, ns not significant.
In osteocyte-enriched bone, Sirt3 mRNA expression was consistent with the expression of E11/gp38, dentin matrix protein 1 (Dmp1), Sost, Rankl and osteoprotegerin (Opg), and the mRNA levels of these genes increased significantly when mice grew from 3 to 6 months and then decreased and reached the lowest level at 24 months (Fig. 1D). We observed that the change in Sirt3 mRNA was among the most predominant during aging (Fig. 1E). The Sirt3 protein level followed the similar pattern (Fig. 1F, G). Notably, the E11/gp38 protein level at 24 months was even lower than that of 3 months by ~60% (Fig. 1F, G). Unlike Sirt3, Dmp1 protein levels peaked at 12 months. Furthermore, prominent age-related changes in Sirt3 mRNA expression were observed only in cortical bone tissues compared with other vital organs (Fig. 1H). For mitochondrial biogenesis, there was a significant drop in both mitochondrial DNA (mt-DNA) content and adenosine triphosphate (ATP) levels when mice aged from 3 to 24 months (Fig. 1I).
Blunted bone remodeling in response to exercise in the aged mice
To determine whether reduced Sirt3 expression can affect bone response to mechanical stimulation, young (4 months) and aged (20 months) mice were subjected to running exercise or control conditions for 6 consecutive weeks (Supplementary Fig. 1D). Six weeks of running exercise caused a significant increase in trabecular bone mineral density (BMD), the ratio of bone volume to tissue volume (BV/TV), and trabecular thickness (Tb.Th) as well as cortical BArea of the young mice (Supplementary Fig. 1E–G). In contrast, the benefits of running exercise on aged bones were inconspicuous, as indicated by undetectable changes in trabecular and cortical bone mass (Supplementary Fig. 1E–G). Consistently, bone quality deteriorates with aging regardless of running conditions (Supplementary Fig. 1E–G).
Confocal imaging of FITC-stained cortical bone showed that running exercise increased the number of dendrites per osteocyte by 9% and the length of dendrites by 12% in 4 months mice, whereas the percentage changes were subtle and insignificant in 20 months mice (Supplementary Fig. 2A, B). Notably, the osteocyte density was not affected by running exercise in both groups (Supplementary Fig. 2B). Moreover, the protective effect of running exercise on osteocytes appears to be associated with the simultaneous upregulation of Sirt3 and mitochondrial transcription factor A (Tfam) mRNA expression (Supplementary Fig. 2C) and higher mitochondrial biogenesis, as indicated by a significant increase in mt-DNA content (Supplementary Fig. 2D).
Bone histomorphometry of H&E staining revealed a significant increase in the number and surface area of bone-lining cells on the trabecular bone surface of young mice after the running exercise (Supplementary Fig. 2E, F). In addition, running exercise resulted in a significantly larger surface area of tartrate resistant acid phosphatase (TRAP)-positive osteoclasts on the trabecular bone surface in young mice (Supplementary Fig. 2G, H). The observed increased bone turnover in young mice after running exercise was further supported by the significantly elevated serum levels of bone formation marker [Procollagen I N-Terminal Propeptide (PINP)] and bone resorption marker [C-telopeptide of type I collagen (CTX-1)] (Supplementary Fig. 2I, J). However, such changes were not detectable in the aged mice after running exercise. At the transcription level in the femoral bone, osteoblast markers, including osteocalcin (Ocn), axis inhibition protein 2 (Axin2), and T-cell factor 1 (Tcf1), were significantly upregulated in young mice, but not in aged mice (Supplementary Fig. 2K). Notably, bone formation-associated markers were significantly lower in aged mice than in young mice (Supplementary Fig. 2K). Although serum CTX-1 levels remain relatively stable during aging (Supplementary Fig. 2J), N.Oc/B.Pm and Oc.S/B.S levels were significantly higher in aged mice than in young mice (Supplementary Fig. 2H). Taken together, running exercise increased bone turnover and mass in young mice. However, the aged mice failed to exhibit similar responses to mechanical loading of the same magnitude and duration.
Sirt3 mediated the formation of dendritic processes and the response of osteocyte-like cells to fluid shear stress in vitro
MLO-Y4 cells embedded in a collagen type I gel were used to examine the effect of Sirt3 on osteocyte activity in vitro. We labeled the cytoskeleton with phalloidin and mitochondria with MitoTracker Red and then examined their changes at different timepoints. At 12 h, the cells started to form dendrite-like structures, which progressively extended further in length and number per cell as the cells differentiated (Fig. 2A). The presence of mitochondria in the osteocyte dendritic processes suggests increased energy demand supported by increased cellular ATP content (Fig. 2B). Notably, Sirt3 mRNA and protein expressions increased simultaneously (Fig. 2C). To confirm whether Sirt3 regulates osteocyte dendritic formation, MLO-Y4 cells with Sirt3 knockdown (KD-Sirt3) were used to examine whether dendritic processes were impaired. The knockdown efficiency was shown in Supplementary Fig. 3A, B. Confocal imaging and quantitative analysis showed a remarkable reduction in the number and length of dendritic processes per cell in KD-Sirt3 cells than in control cells (Fig. 2D–F). These changes were accompanied by decreased mitochondrial function (Supplementary Fig. 3C–E).
A Representative images of confocal imaging of phalloidin-stained (green) (upper panel), Mitotracker Red-stained (red) (middle panel), and merged (lower panel) MLO-Y4 cells at different timepoints cultured in a 3D gel model. Scale bars = 10 μm. B Cellular adenosine triphosphate (ATP) level (relative to protein level) of 3D cultured MLO-Y4 cells at different timepoints (n = 4). C qPCR and Western blot analysis of Sirt3 and β-actin expression in 3D cultured MLO-Y4 cells at different timepoints (n = 3). D Representative images of confocal imaging of phalloidin-stained (green) and DAPI-stained (blue) KD-Ctrl and KD-Sirt3 MLO-Y4 cells. Scale bar = 20 μm. E Semiquantification of dendrite number per osteocyte and F dendrite length per osteocyte in KD-Ctrl and KD-Sirt3 cells (n = 4). G qPCR analysis of prostaglandin-endoperoxide synthase (Ptgs2), receptor activator of nuclear factor kappa-B ligand (Rankl), and matrix metallopeptidase 13 (Mmp13) mRNA expressions in KD-Ctrl and KD-Sirt3 cells in static or fluid shear stress condition (n = 4). All data were presented in mean ± SEM and analyzed with one-way ANOVA in B, C, Student’s t test in E, F, and two-way ANOVA in G. *p < 0.05, **p < 0.01, and ***p < 0.001, ns not significant.
Considering the essential role of osteocyte dendritic processes in mechanotransduction, MLO-Y4 cells were subjected to fluid shear stress to examine whether Sirt3 knockdown affects cellular response to fluid shear stress in vitro. Similar to previous studies, MLO-Y4 cells responded to fluid shear stress, as indicated by the significant upregulation of prostaglandin-endoperoxide synthase 2 (Ptgs2) and downregulation of Rankl and matrix metallopeptidase 13 (Mmp13) (Fig. 2G). However, Sirt3 knockdown significantly impaired the sensitivity of MLO-Y4 cells to fluid shear stress stimulation; for instance, Ptgs2 upregulation reduced from 4.8-fold increase in control cells to 1.2-fold increase in KD-Sirt3 cells, and Rankl and Mmp13 downregulation decreased from 25.1% to 10.4% and 33.3% to 17.4%, respectively (Fig. 2G). Interestingly, the expression of Rankl and Mmp13 increased with Sirt3 deficiency (Fig. 2G). However, co-culture of KD-Sirt3 MLO-Y4 cells with RAW264.7 cells did not increase the expression of osteoclastic markers (Supplementary Fig. 3F).
Sirt3 regulated the formation of osteocyte dendritic processes and the anabolic response of bone to exercise in vivo
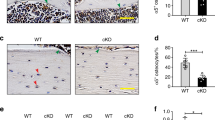
To validate the biological function of Sirt3 in osteocyte dendritic processes and mechanotransduction in vivo, osteocyte-specific Sirt3 knockout mice (cKO) were generated by crossing Dmp1-Cre with Sirt3fl/fl mice (Fig. 3A), causing a specific reduction in Sirt3 expression in osteocyte-enriched bone tissue (Fig. 3B), which was further confirmed by immunostaining Sirt3 in the cortical bone tissue (Fig. 3C). Sirt3 cKO mice were viable and indistinguishable from control littermates at birth and showed normal growth (Supplementary Fig. 4A, B). In the cortical bone of 3 months cKO mice, osteocyte dendritic processes were abrogated and visibly blunted (Fig. 3D). Compared with the control mice, the osteocyte density, number, and total length of dendrites per osteocyte in the cKO mice were reduced by 26%, 69% and 75%, respectively (Fig. 3E).
A Breeding strategy for generation of the osteocyte-specific Sirt3 knockout (cKO) mice and control (Ctrl) mice. B qPCR analysis of Sirt3 expression in different tissues of 3 months male Ctrl and cKO mice (n = 4). C Immunohistochemistry (IHC) of Sirt3 in the cortical bone of 3 months Ctrl and cKO mice. Scale bars = 50 μm. D Representative images of confocal imaging of fluorescein isothiocyanate (FITC)-stained (green) osteocyte in the midshaft femur sections showing osteocyte connectivity in 3 months male Ctrl and cKO mice. Scale bars = 20 μm (upper panel) and 5 μm (lower panel). E Quantification of osteocyte density, dendrite number per osteocyte, and the total dendrite length per osteocyte of 3 months male Ctrl and cKO mice (n = 4). All data were presented in mean ± SEM and analyzed with Student’s t test. *p < 0.05, **p < 0.01 and ***p < 0.001, ns not significant.
We then investigated whether Sirt3 deficiency in osteocytes affects the anabolic response of bone to running exercise in vivo (Fig. 4A). Running exercise promoted trabecular bone mass in control mice as indicated by a significant increase in trabecular BMD, BV/TV, and Tb.Th in both femur and tibia (Fig. 4B, C and Supplementary Fig. 5A, B). In addition, there was a significant increase in cortical BArea in the control mice after running exercise (Fig. 4D). In contrast, the anabolic effect of running exercise on bone remodeling was remarkably diminished in the cKO mice, as indicated by the unchanged cortical and trabecular bone parameters (Fig. 4C, D and Supplementary Fig. 5B). Notably, there was no significant difference in cortical and trabecular bone quality between the Ctrl and cKO mice at the basal level (Fig. 4B–D). Confocal imaging of FITC-stained cortical bone showed that running exercise increased the number of dendrites per osteocyte by 13% and the length of dendrites by 9% in the control mice; however, the percentage changes were subtle and insignificant in the cKO mice (Fig. 4E, F). Moreover, the osteocyte density was not affected by running exercise in both groups (Fig. 4F). Consistent with the results in Fig. 3D, osteocyte dendritic processes were significantly impaired by Sirt3 deletion. It is interesting to observe that Tfam transcript level and mt-DNA content did not change after running exercise in the absence of Sirt3 (Fig. 4G, H), suggesting that Sirt3 downregulation might be the major contributor of the blunted response of the bone to exercise in the aged mice. In addition, the cKO mice had a significantly lower mt-DNA content than the control mice (Fig. 4H), which was consistent with the in vitro results shown in Supplementary Fig. 3C.
A A schematic diagram depicted the procedural steps. B Representative micro-CT reconstruction images of trabecular bone in the distal femur (upper panels) and cortical bone in the midshaft femur (lower panels) of Ctrl and cKO mice with or without treadmill running exercise. Scale bars = 100 μm. C Quantification of trabecular bone parameters including bone mineral density (BMD), ratio of bone volume to tissue volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) of Ctrl and cKO mice with or without treadmill running (n = 8). D Quantification of cortical parameters including BMD, bone area (BArea), tissue area (TArea), and cortical thickness (Ct.Th) of Ctrl and cKO mice with or without treadmill running exercise (n = 8). E Representative images of confocal imaging of fluorescein isothiocyanate (FITC)-stained (green) midshaft femur sections showing osteocyte connectivity in Ctrl and Sirt3 cKO with or without treadmill running exercise. Scale bar = 20 μm (upper panel) and 5 μm (lower panel). F Quantification of osteocyte density, dendrite number per osteocyte, and total dendrite length per osteocyte in Ctrl and Sirt3 cKO mice with or without treadmill running (n = 4). G qPCR analysis of Sirt3 and mitochondrial transcription factor A (Tfam) expression in the bone collected from Ctrl and cKO mice with or without treadmill running (n = 4). H Mitochondrial DNA (mt-DNA) content was measured in the bone collected from Ctrl and cKO mice with or without treadmill running (n = 3). All data were presented in mean ± SEM and analyzed with two-way ANOVA. *p < 0.05, **p < 0.01 and ***p < 0.001, ns not significant.
H&E staining revealed a significant increase in the number and surface area of lining cells by 22.6% and 10.4%, respectively, in the trabecular bone of the control mice after running exercise (Supplementary Fig. 5C, D), whereas no significant change was detected in the cKO mice. Consistently, serum PINP and femoral transcript levels of osteoblast markers including Runx2, Axin2, and Tcf1 were significantly increased upon running in the control mice but not in the cKO mice (Supplementary Fig. 5G, I). In addition, serum CTX-1 levels were significantly increased with a numerical increase in the number and surface area of TRAP-positive osteoclasts on the trabecular bone surface in the control mice (Supplementary Fig. 5E, F, H). Taken together, we observed that the Sirt3 cKO mice failed to exhibit similar responses to mechanical stimulation of the same magnitude and duration.
Sirt3 regulated E11/gp38 expression through the protein kinase A (PKA)/cAMP response element-binding protein (CREB) pathway
To determine how Sirt3 mediates the response of osteocytes to running exercise, we speculated that E11/gp38 might be a potential target of Sirt3-mediated response. E11/gp38 protein expression was significantly reduced in the cortical bone of the cKO mice compared to the control mice (Fig. 5A and Supplementary Fig. 6A). In addition, the Western blot analysis showed persistently lower protein levels of E11/gp38 and connexin 43 (Cx43) in the cKO mice at the age of 5 weeks and 3 months (Fig. 5B and Supplementary Fig. 6B). Sirt3 knockdown significantly suppressed E11/gp38 transcript level in MLO-Y4 cells (Supplementary Fig. 6C). Then bioinformatics was used to identify the potential transcription factors bridging Sirt3 and E11/gp38. CREB was selected for the subsequent experiments due to its high binding probability to the E11/gp38 core promoter region. We first tested whether Sirt3 regulates CREB phosphorylation in vitro. E11/gp38 expression was significantly reduced after Sirt3 knockdown in MLO-Y4 cells and this was accompanied by inhibited phosphorylation of CREB (Supplementary Fig. 6D, E). Reportedly, CREB is phosphorylated by PKA, mitogen-activated protein kinases, and several other kinases [23]. Hence, our results showed that PKA phosphorylation was significantly decreased compared to total PKA after Sirt3 knockdown (Supplementary Fig. 6D, E), suggesting that the PKA-CREB signaling pathway could mediate the regulatory effect of Sirt3 on E11/gp38 expression. We then treated Sirt3 knockdown MLO-Y4 cells with forskolin, a potent activator of the PKA-CREB signaling pathway [24]. The treatment had no effect on Sirt3 levels but it caused a marked increase in E11/gp38 expression and enhanced the phosphorylation of CREB and PKA in Sirt3 KD cells (Fig. 5C, D), implying that activation of PKA-CREB signaling rescued the decreased E11/gp38 after Sirt3 knockdown.
A Immunohistochemistry of E11/gp38 in the cortical bone of 3 months male Ctrl and cKO mice. Scale bars = 50 μm. B Western blot analysis of osteocyte markers E11/gp38, connexin 43 (Cx43), dentin matrix protein 1 (Dmp1), and β-actin in osteocyte-enriched bone tissue of 5-week-old (left) and 3-month-old (right) male Ctrl and cKO mice (n = 4). C A representative immunoblot of Sirt3, phosphorlyated protein kinase A (p-PKA), protein kinase A (PKA), phosphorylated cAMP-response element-binding protein (p-CREB), cAMP-response element-binding protein (CREB), E11/gp38, and β-actin in KD-Ctrl and KD-Sirt3 MLO-Y4 cells with or without the treatment of forskolin (FSK) (n = 4). D Semiquantification of protein levels of Sirt3, E11/gp38, p-CREB/CREB, and p-PKA/PKA in C (n = 4). E Representative images of confocal imaging of phalloidin-stained (green) and DAPI-stained (blue) KD-Ctrl, KD-Sirt3, and KD-Sirt3 MLO-Y4 cells treated with FSK (n = 4). Scale bar = 50 μm. F Semiquantification of dendrite number per osteocyte and dendrite length per osteocyte in E (n = 4). G qPCR analysis of prostaglandin-endoperoxide synthase (Ptgs2), receptor activator of nuclear factor kappa-B ligand (Rankl), and matrix metallopeptidase 13 (Mmp13) mRNA expression in KD-Ctrl and KD-Sirt3 MLO-Y4 cells with or without the treatment of FSK in static or fluid shear stress condition (n = 4). All data were presented in mean ± SEM and analyzed with one-way ANOVA test in D, F and Student’s t test in G. *p < 0.05, **p < 0.01 and ***p < 0.001, ns not significant.
We then tested whether PKA-CREB activation could rescue the phenotypes of Sirt3-deficient osteocytes. Confocal imaging of phalloidin-stained osteocytes showed that PKA-CREB activation alleviated the inhibition of dendritic processes after Sirt3 knockdown (Fig. 5E, F). Consistently, the sensitivity of Sirt3-deficient osteocytes to fluid shear stress was also enhanced by forskolin treatment, and this was proved by the higher fold/percentage change of mechanically responsive markers in forskolin treatment group upon fluid shear stress (e.g. the fold increase in Ptgs2 increased from 0.7 to 2.1; the percentage change of Rankl and Mmp13 increased from 10.9% to 23.3% and 10.6% to 22.0%, respectively, after the treatment) (Fig. 5G). Interestingly, overexpression of E11/gp38 in Sirt3-deficiency cells also rescued the phenotypic changes after Sirt3 knockdown in osteocytes (Supplementary Fig. 6F–H), suggesting that PKA-CREB-E11/gp38 are downstream factors mediating the regulatory effect of Sirt3 on osteocyte dendritic processes.
Phosphorylated CREB (p-CREB) directly bound to the E11/gp38 promoter and facilitated its transcriptional activity
We tested whether CREB directly regulates E11/gp38 expression in vitro. qPCR results showed that CREB inhibitor, KG-501, significantly decreased E11/gp38 expression, whereas CREB activator, forskolin, increased E11/gp38 expression (Supplementary Fig. 7A). Furthermore, E11/gp38 protein expression was also significantly reduced in KG-501-treated cells (Fig. 6A, B). Concomitantly, p-CREB/CREB levels decreased after the treatment with KG-501 (Fig. 6A, B). These results showed that CREB can positively regulate E11/gp38 expression in osteocytes.
A A representative immunoblot of phosphorylated cAMP-response element-binding protein (p-CREB), cAMP-response element-binding protein (CREB), E11/gp38, and β-actin in MLO-Y4 cells treated with 10 μM KG-501 or vehicle control (Ctrl) (n = 4). B Semiquantification of p-CREB/CREB and E11/gp38 levels in A (n = 4). C Chromatin immunoprecipitation (ChIP)-PCR analysis of the engagement of phosphorylated CREB and CREB in E11/gp38 promoter in MLO-Y4 cells. Results are representatives of three independent experiments. D p-CREB promoter fold enrichment from immunoprecipitated DNA with antibody against p-CREB was subjected to qPCR (n = 3). E Putative p-CREB-binding motifs (labeled with green) were predicted by the online informatics software JASPAR, and the E11/gp38 transcription start site (TSS) is underlined in the genomic sequence. F Promoter-luciferase reporter assay was performed in MLO-Y4 cells by transfecting the cells with blank, wildtype (WT), and deletion mutant plasmids (n = 4). All data were presented in mean ± SEM and analyzed with Student’s t test in B, D and one-way ANOVA in F. *p < 0.05, **p < 0.01 and ***p < 0.001, ns not significant.
We further investigated whether CREB binds directly to the promoter region of E11/gp38. ChIP-PCR and ChIP-qPCR analysis were performed in MLO-Y4 cells and revealed a phosphorylated CREB-binding site in the E11/gp38 promoter (Fig. 6C, D). A schematic representation of the p-CREB-binding sites with respect to the E11/gp38 transcription start site was shown in Fig. 6E. As shown in Fig. 6F, the luciferase reporter assay confirmed that plasmids containing phosphorylated CREB-binding sites showed high promoter activity, and deletion mutation strongly inhibited E11/gp38 promoter activation. Therefore, phosphorylated CREB regulates the activation of the E11/gp38 promoter by directly engaging its binding motif.
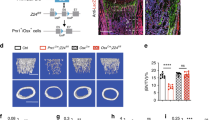
Sirt3 activator, honokiol, prevented age-related bone loss and increased osteocyte response to fluid shear stress in vitro
To determine whether Sirt3 activation could prevent age-related bone loss, aged mice were treated with a low or high dose of honokiol or vehicle control for 2 months (Fig. 7A). Honokiol treatment did not affect the body weight of mice (Supplementary Fig. 8A). A high-dose of honokiol significantly increased trabecular BMD, BV/TV, and Tb.N, while such beneficial effects were not observed with the low-dose honokiol treatment (Fig. 7B, C), suggesting a dose-dependent effect of honokiol on aged mice bone, There was no significant change in the cortical parameters between the low- and high-dose honokiol groups and the control groups (Supplementary Fig. 8B, C). Notably, high-dose honokiol treatment enhanced the dendritic processes of osteocytes, as evidenced by confocal imaging and quantitative Imaris analysis showing that the osteocyte density, number, and length of dendrites were significantly higher in the high-dose honokiol treatment group (Fig. 7D, E). As expected, the protective effect of honokiol on bone was associated with significantly increased Sirt3 expression (Fig. 7F and Supplementary Fig. 8E). Concomitantly, E11/gp38, p-CREB/CREB, and mt-DNA content increased following honokiol treatment (Fig. 7F and Supplementary Fig. 8D). For the other osteocyte markers, Cx43, Ocn, and Opg/Rankl mRNA expressions increased after honokiol treatment, while Dmp1 and Sost expression remained unchanged (Supplementary Fig. 8E). Consistently, honokiol treatment increased the protein expression of Sirt3, p-CREB/CREB, and E11/gp38 in MLO-Y4 cells in vitro (Fig. 7G and Supplementary Fig. 8F). Also, honokiol treatment increased the response of osteocytes to fluid shear stress. The fold increase in Ptgs2 expression was higher in the honokiol-treated cells (4.5 fold) than in control cells (3.2 fold), and the percentage change in Rankl expression was also higher in the honokiol-treated cells (43.6%) than in the control cells (27.0%) upon fluid shear stress (Fig. 7H).
A A schematic diagram depicts the procedural steps. B Representative micro-CT reconstruction images of trabecular bone in the distal femur of mice treated with vehicle control and high-dose HKL. Scale bars = 100 μm. C Quantification of trabecular bone parameters of mice treated with low- and high-dose HKL, including bone mineral density (BMD), ratio of bone volume to tissue volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) (n = 5 for low-dose-group and n = 7 for high-dose group). D Representative images of confocal imaging of FITC-stained (green) midshaft femur sections showing osteocyte connectivity in aged mice with or without the treatment of high-dose HKL. Scale bar = 20 μm (upper panel) and 5 μm (lower panel). E Quantification of osteocyte density, dendrite number per osteocyte, and total dendrite length per osteocyte in aged mice with or without the treatment of HKL (n = 4). F A representative immunoblot and semiquantification analysis of the protein level of Sirt3, E11/gp38, phosphorylated cAMP-response element-binding protein (p-CREB), and cAMP-response element-binding protein (CREB) in bone tissue of aged mice with or without the treatment of high-dose HKL (n = 4). G A representative immunoblot and semiquantification of the protein levels of Sirt3, E11/gp38, p-CREB, and CREB in MLO-Y4 cells treated with 5 and 10 μM HKL after 24 h culture (n = 4). H qPCR analysis of prostaglandin-endoperoxide synthase (Ptgs2) and receptor activator of nuclear factor kappa-B ligand (Rankl) in MLO-Y4 cells treated with Ctrl or 5 μM HKL in static or fluid shear stress condition (n = 4). All data were presented in mean ± SEM and analyzed with Student’s t test in C, E, F, and two-way ANOVA in H. *p < 0.05, **p < 0.01, and ***p < 0.001, ns not significant.
Discussion
Our findings with a wide age spectrum showed degenerative changes in mice bones, evidenced by the natural aging mouse and exercise model, which is consistent with previous findings [25]. In addition, observation of reduced osteocyte density, dendrite number and length with aging are comparable to previous studies on mice [13], rats [26], and humans [5]. E11/gp38 is a critical regulator of dendritic elongation and is selectively expressed during osteocyte differentiation [27, 28]. The essential function of E11/gp38 in bone homeostasis and loading response was evidenced by the bone specific E11/gp38 KO mice, which showed significant impairment in the osteocyte dendritic network and a declined response to mechanical loading [29]. Generally, the expression of E11/gp38 is regulated by mechanical strain. Unlike E11/gp38, Dmp1 protein level peaked at 12 months, which could be due to its accumulation in bone matrix because the measured Dmp1 protein level in our study included Dmp1 both in the bone matrix and inside the osteocytes. In cellular studies, Rankl and Mmp13 expression increased following Sirt3 deficiency, which requires further investigation to clarify their potential roles in bone and perilacunar remodeling.
Our results showed that the other two mitochondrial sirtuins (Sirt4 and Sirt5) remain relatively steady throughout life, implying the unique function of Sirt3 in the aging bone. One possible explanation for the decline in Sirt3 levels could be the decrease in cellular NAD+ levels with aging [30]. NAD+ is an endogenous enzyme cofactor and a substrate for several families of enzymes, including Sirt3 [31]. Therefore, the expression pattern of Sirt3 in the bone, which appears similar to bone mass increase, might be influenced by NAD+ level or other potential upstream factors. Our study together with previous studies show the transition of the bone phenotype after Sirt3 deletion from young to adulthood [32]. Further investigations with wider age spectra and both sexes are warranted to investigate the biological function of Sirt3 in bone metabolism and aging.
Our results showed a reduced anabolic response of bone to running exercise in aged mice, which is consistent with clinical studies [8]. This might partly be attributed to the defects in osteocyte dendritic network during aging. In addition, age-related alterations in cellular function can impair the ability of the bone to respond to mechanical stimulation. A recent study reported that the application of exercise intervention in mtDNA mutant mice with impaired mitochondrial function had no effect on the bone mass increase [33], suggesting the important role of the mitochondria in bone response to exercise. Furthermore, it was demonstrated that mitochondrial transfer from healthy osteocytes could restore the metabolic function of metabolically stressed osteocytes [19]. This suggests a new biological function for LCN and mitochondria in osteocytes and bone health. Although the involvement of muscle-bone crosstalk is beyond the scope of this study, it should be noted that exercises can also lead to higher levels of mitochondrial enzymes and oxidative capacity in the muscle [34], suggesting muscle involvement in the observed changes after exercise. Further investigation using tibial loading, which is lacking in this study, is warranted to address the confounding effect of the muscle.
Previous studies have demonstrated the important roles of CREB in bone remodeling [35,36,37]. In addition to its function as a transcription factor, CREB is important for mitochondrial biogenesis by increasing the expression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) [38]. Since dendritic formation is accompanied by increased energy supply, we hypothesized that Sirt3 regulates osteocyte dendritic processes by direct regulation of E11/gp38 expression and modulation of mitochondrial biogenesis. However, how Sirt3 regulates gene transcription may be a more complex molecular activity. Previous studies have indicated that Sirt3 exists in a full-length form, which is further cleaved to a short form that is localized specifically to the mitochondria due to its mitochondrial localization sequences [39]. However, reports show that the full-length Sirt3 also exists in the nucleus and exhibits histone deacetylase activity [31], which is further supported by the finding that Sirt3 regulates the expression of nuclear genes such as PGC-1α, MnSOD and Forkhead box O3a (Foxo3a) [40]. Further understanding of how Sirt3 regulates osteocyte gene expression, protein modification, and signal transduction may help unveil critical mechanisms that regulate osteocyte function.
Preclinical studies have shown that Sirt3 activation using the small molecule honokiol is beneficial for several age-related diseases [41,42,43]. Reportedly, honokiol can diffuse across the membrane into the mitochondria and physically bind to Sirt3 [44]. The physical interaction of honokiol with Sirt3 may enhance Sirt3’s deacetylase activity and thus the increased mitochondrial biogenesis and activation of MnSOD, which is responsible for maintaining the balance between ROS generation and clearance. The decline in Sirt3 expression during aging may contribute to increased ROS production in aged bones [45, 46], further inducing the apoptosis of osteocytes [47]. Hence, honokiol could protect osteocytes from apoptosis through alleviating oxidative stress and maintaining mitochondrial biogenesis. Future studies are needed to determine the exact effector cells of honokiol and explore the potential side/adverse effects of honokiol treatment.
In summary, our findings demonstrate that Sirt3, a gene linked to human longevity, is critical for osteocyte dendritic processes and the response of bone to exercise. The Sirt3 activator, honokiol, may be used to improve bone health and amplify the effect of exercise on the bone especially in the aged population.
Materials and methods
Animals
The experimental procedures were conducted in accordance with the guidelines of the National Institutes of Health and were approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong (ethical approval number 19-159-MIS). Mice were housed and bred in conventional conditions in our animal facility.
To assess the impact of aging on bone qualities, osteocyte dendritic processes, and mRNA/protein expression, male C57BL/6 mice (age: 3-, 6-, 12-, and 24-month-old) were used. At the time of sacrifice, mice were euthanized, femora and tibiae were dissected for micro-CT analysis, histomorphometry analysis, and RNA/protein isolation.
Osteocyte-specific deletion of Sirt3 was obtained by crossing Sirt3fl/fl mice with transgenic mice expressing Cre recombinase under the control of the Dmp1 gene promoter [48], which were imported from the Jackson Laboratory (Stock No: 031201; Bar Harbor, ME, USA). Dmp1-Cre+ Sirt3fl/fl mice (referred to cKO mice) and Dmp1-Cre- Sirt3fl/fl (referred to control mice) were used in the experiments. All mice were on a 100% C57BL/6 background. Analysis of bone quality and osteocyte dendritic processes was performed on 3 months male mice. The primers for genotyping were listed in Supplementary Table 1.
Treadmill running was performed with 4 months (young), 20 months (aged) male C57BL/6 mice, 4 months control mice, and 4 months cKO mice on a treadmill machine (76–0896, Panlab, Harvard Apparatus). Briefly, weight-matched mice were randomly assigned to control vs. treadmill running group. Treadmill running regimen consisted of 3 days’ training and 6 weeks’ treadmill running. In the training program, mice run on the treadmill (8 meters/min at a 5° incline) for 30 min/day. Then, mice were subjected to treadmill running program (15 meters/min at a 5° incline) for 60 min/day, 7 days/week for 6 consecutive weeks. The control group remained caged with free movement in the same room at the same time. At the end of running program, mice were euthanized and the whole-body weight was measured. Blood was collected and both femora and tibiae were collected for micro-CT analysis, histomorphometry analysis, and RNA/protein isolation.
To study the effect of honokiol treatment (S2310, Selleck) on bone mass in aged mice (20 months male C57BL/6), honokiol at 0.2 mg/kg/day (low-dose) or 2 mg/kg/day (high dose) was injected intraperitoneally for 2 months. Body weight was recorded before and after the treatment. At the end of treatment, mice were euthanized, blood was collected and both femora and tibiae were collected for micro-CT analysis, histomorphometry analysis, and RNA/protein isolation.
Micro-CT analysis
Mice femora and tibiae were collected for ex vivo micro-CT analysis (Scanco Medical, Brüttisellen, Switzerland). The resolution was set at 8 μm per voxel and 1024 × 1024 pixels. Scanner parameters were set as follows: 114 μA current, 70 kV voltage and 0.5-mm-thick aluminum filter. The cortical bone analysis of the midshaft femora was performed in the diaphyseal region of the femora at the inferior border of the great trochanter and extended 50 slices (400 μm) longitudinally. The parameter sigma and support were set 0.8 and 1, and a global threshold was set to 260. Ct.Th., BArea, TArea, and BMD were measured for comparison. The trabecular bone, which is proximal to the distal growth plate for the femora and distal to the proximal growth plate for the tibiae, was measured starting at the inferior border of the growth plate and extended a further longitudinal distance of 100 slices (800 μm) with the parameter sigma and support equal to 0.8 and 1, using a global threshold set to equal 220. BMD, BV/TV, Tb.N, Tb.Th, Tb.Sp, Conn.D and SMI were measured and evaluated. All measurements were performed according to the guidelines of the American Society for Bone and Mineral Research [49]. Mechanical properties of the cortical bone in the midshaft femur were calculated through using finite element analysis software provided by manufacturer (μFE Element Analysis Solver v.1.15; Scanco Medical AG). In this linear finite model, it is assumed that bone tissue is an isotropic and linear material with Young’s modulus as 10 GPa and a Poisson’s ratio as 0.3 [50]. Uniaxial compression test with 1% strain along the axial direction was performed using the software. Stiffness was calculated in simulation. The analyzers were blinded to the group allocation.
Histology and immunostaining
For H&E, TRAP and immunohistochemical staining, femora were fixed overnight at 4 °C in 4% paraformaldehyde, decalcified for 2 weeks in 0.5 M EDTA (pH 7.4), and embedded in paraffin. Embedded femora were cut into 5 μm-thick sections with a rotary microtome (RM2255, Leica Microsystems, Buffalo Grove, IL, USA).
Osteoblast parameters were measured on H&E-stained sections. Briefly, paraffin sections were stained with haematoxylin for 5 min, followed by staining with eosin for 3 min. Five fields of view in the distal femora were randomly selected for each bone slice for further analysis. Bone histomorphometry parameters, including the number of osteoblasts per bone surface (N.Ob/B.S, /mm2) and osteoblast surface per bone surface (Ob.S/B.S, %), were analyzed using an OsteoMeasure Image Analysis System (Osteometrics, Decatur, GA).
Osteoclasts with TRAP activity and counterstained with Fast Green (C.I. 42053, Merck) were counted on paraffin sections. Five fields of view in the distal femora were randomly selected for each bone slice for further analysis. Bone histomorphometry parameters, including the number of osteoclast per bone perimeter (N.Oc/B.Pm, /mm) and osteoclast surface per bone surface (Oc.S/B.S, %), were analyzed using an OsteoMeasure Image Analysis System (Osteometrics, Decatur, GA).
For immunohistochemical staining, paraffin-embedded sections were de-waxed, rehydrated, incubated with 0.1% trypsin (25200056, Thermo Fisher Scientific) for 30 min at 37 °C and washed in TBS. Endogenous peroxidase activity was blocked by immersing the sections in 3% (v/v) H2O2 in dark for 10 min at room temperature, followed by blocking with 2% of bovine serum albumin (BSA) in Tris-buffered saline with 0.1% tween 20 (TBST) for 1 h at room temperature. Then, sections were incubated overnight with anti-Sirt3 antibody (1:100, ab86671, Abcam) and anti-Podoplanin antibody (1:50, AF3244, R&D systems). Then the sections were washed with TBST 3 times and incubated with HRP-coupled secondary antibodies. HRP detection was performed using 3,3’-diaminobenzidine (DAB; TA-060-QHDX, Thermo Fisher Scientific) for 5–10 min. Afterwards, slides were counterstained with haematoxylin and imaged under DM550 microscope (Leica Microsystems). Semi-quantitative analysis of IHC staining was conducted by ImageJ according to a previously published protocol [51].
3D confocal imaging and quantitative analysis of LCN properties
Osteocytes were visualized on FITC-stained confocal imaging and Imaris analysis as we previously described [52]. In brief, bone samples were stained in 1% (w/v) FITC isomer I (F7250, MilliporeSigma) solution in absolute ethanol for 8 h at room temperature with gentle mixing before embedding in Technovit® 9100 methyl methacrylate. The embedded bone tissues were then trimmed and cut into ~200‐μm‐thick sections with a diamond inner‐hole saw (Leica SP1600, Leica, Wetzlar, Germany). Finally, the 200‐μm‐thick sections were sanded down on a polishing machine to achieve ~70–100‐μm‐thick bone sections for confocal microscope imaging. Confocal stacking images were obtained in a single‐blinded manner with SP8 confocal microscope (Leica). FITC was excited by a 488‐nm argon laser, and emission at 519 ± 5 nm was acquired by a hybrid detector. The z-stack images with a thickness of around 40 μm were obtained with a pixel size of 0.180 × 0.180 × 0.208 μm at a resolution of 1024 × 1024 with linear z compensation. The confocal images were constructed into 3D images with Imaris software (v.8.0; Bitplane, Zürich, Switzerland) at the same dimension with a pixel size of 0.180 × 0.180 × 0.208 μm. The 3D reconstructions were used to count osteocyte number using ImageJ software with the cell counting plugin (Author: Kurt De Vos, University of Sheffield Academic Neurology). Only osteocytes with complete cell bodies and dendrites were selected for analysis. The mean number of osteocytes per mm3 was determined for each animal in the midshaft regions. To measure the number of dendrite per osteocyte and total dendrite length per osteocyte, 3D reconstructions of 63× stacks of FITC labeled sections were generated in Imaris software for filament tracing and statistical quantification. Dendrites were mapped with the same threshold (0.5–8 μm for filament tracing). The dendrite number, dendrite length, cell body surface area, cell body volume, and sphericity of cell body were analyzed as previously described in Ren et al. [53].
Whole blood analysis and serum biochemistry
Blood was collected with EDTA tube and separated by centrifugation at 3200 × g for 10 min, then the serum was stored at −80 °C until use. Serum level of PINP (AC-33F1, Immunodiagnostic Systems Limited, Gaithersburg, MD, USA) and CTX-1 (AC-02F1, Immunodiagnostic Systems Limited, Gaithersburg, MD, USA) were tested according to the manufacturer’s instructions.
MLO-Y4 cell culture
MLO-Y4 cells were kindly provided by Prof. Lynda F. Bonewald (Indiana University, USA) and cultured in α-MEM supplemented with 2.5% fetal bovine serum (FBS), 2.5% calf serum (CS), and 1% penicillin/streptomycin/glutamine on rat-tail collagen type I-coated plates (0.15 mg/ml collagen in 0.02 M acetic acid). The type I collagen was from Corning (354236).
Sirt3 knockdown (KD-Sirt3) was achieved by transfecting cells with lentivirus carrying shRNA against Sirt3. The target sequences for Sirt3 were 5’-GCCCAATGTCACTCACTACTT-3’ and 5’-AGACAGCTCCAACACGTTTAC-3’. A lentivirus carrying a nonsense scrambled shRNA sequence (TTCTCCGAACGTGTCACGT) was used as control. E11/gp38 overexpression (OE-E11/gp38) was achieved by transfecting cells with lentiviral vector encoding the mouse podoplanin (pdpn) gene, transcript variant 1 (NCBI reference sequence ID, NM_010329.3). The lentiviruses were provided by Shanghai GeneChem (Shanghai, China; http://www.genechem.com.cn/). For viral infection, cells were incubated in medium containing virus at MOI 25 and 8 μg/ml polybrene for 16 h and then virus-containing medium was changed to growth medium for further experiments. Sirt3 knockdown and overexpression efficiency was determined by qRT-PCR and Western blot.
For the observation of dendrite formation of osteocyte, 3D culture model was used [54]. Briefly, 1 ml of cell suspension/gel solution containing 8 × 106 cells was prepared by mixing 670 μl of 3 mg/ml type I collagen solution (354236, Corning), 100 μl of 10× α-MEM, 17 μl of 1 N NaOH (to adjust the pH to 7.4), 13 μl of PBS and 200 μl of cells in PBS. Then, 200 μl cell suspension/gel mixture per well was immediately transferred into 96-well-plate and allowed to polymerize at 37 °C for 5 min. The cells embedded in collagen gel were cultured with α-MEM supplemented with 5% FBS and 5% CS and 1% antibiotics (penicillin/streptomycin/glutamine). The cells were collected for molecular tests, including qPCR, Western blot, and ATP level examination after 1 h, 12 h, 1 day and 3 days of culture. For confocal imaging of the osteocyte dendrite, MLO-Y4 cells were plated in the confocal dish directly and collected for confocal imaging after 1 h, 12 h, 1 day and 3 days of culture. To assess the effect of Sirt3 deficiency on osteocyte dendrite formation, MLO-Y4 cells transfected with KD-Ctrl and KD-Sirt3 lentiviruses were cultured in the collagen type I gel. After 3 days of culture, cells were washed with cold PBS, fixed with 4% paraformaldehyde for 10 min, and then carefully permeabilized with 0.2% (v/v) Triton X-100 for 10 min. The cells were then incubated with FITC-conjugated Phalloidin (A34055, Invitrogen) at room temperature for 30 min. Mitochondria were labeled with mito-tracker red (M7512, Thermo Fisher Scientific) at room temperature for 10 min. The stained cells were then mounted for confocal imaging. The analysis of cell number, dendrite number per cell, and dendrite length per cell was conducted using ImageJ. Data from five randomly selected fields of view per sample were averaged for statistical analysis.
To assess the impact of Sirt3 deficiency and activation on osteocyte response to fluid shear stress, KD-Ctrl and KD-Sirt3 cells, honokiol (5 uM; S2310, Selleck) and DMSO (1/1000 dilution, Sigma-Aldrich) treated MLO-Y4 cells (cell number 1 × 105) were seeded in a µ-Slide I Luer (0.4 mm) fluid chamber slide (80176, IBIDI, Germany) overnight and then exposed to 15 dynes/cm2 fluid shear stress at 1 Hz for 1 h using a Manostat Carter Cassette Pump System (740-000-12131, Barnant Company, USA). Then the cells were collected for qPCR analysis.
To assess the effect of CREB inhibition and activation on E11/gp38 expression, MLO-Y4 cells in 6-well and 12-well plates were treated with KG-501 (10 μM; 8409, Selleck), forskolin (5 μM, S2449, Selleck), or DMSO as vehicle control (Sigma-Aldrich). After 24 h of treatment, cells were collected for qPCR and Western blot analysis.
To assess the effect of activation of PKA-CREB signaling or overexpression of E11/gp38 on dendritic formation, MLO-Y4 cells transfected with KD-Ctrl and KD-Sirt3 lentiviruses were cultured overnight and then the cells were treated with forskolin (5 μM, S2449, Selleck) and DMSO as vehicle control (Sigma-Aldrich), or OE-Sirt3 and control lentiviruses. After 2 days (forskolin) or 3 days (OE-Sirt3) of culture, cells were stained for FITC-conjugated Phalloidin (A34055, Invitrogen) and mounted for confocal imaging and subsequent analysis.
To assess the effect of activation of PKA-CREB or overexpression of E11/gp38 on osteocyte response to fluid shear stress, MLO-Y4 cells transfected with KD-Ctrl and KD-Sirt3 lentiviruses were cultured in a µ-Slide I Luer (0.4 mm) fluid chamber slide (80176, ibidi, Germany) overnight and then the cells were treated with forskolin (5 μM, S2449, Selleck) and DMSO as vehicle control (Sigma-Aldrich), or OE-Sirt3 and control lentiviruses. After 2 days (forskolin) or 3 days (OE-Sirt3) culture, the cells were exposed to 15 dynes/cm2 fluid shear stress at 1 Hz for 1 h using a Manostat Carter Cassette Pump System (740-000-12131, Barnant Company, USA). Then the cells were collected for qPCR analysis.
Quantitative polymerase chain reaction (qPCR)
To get the osteocyte-enriched bone tissue, freshly collected bone tissues with muscle and soft tissues removed by scalpel and distal and proximal epiphyses excised were subjected to centrifugation (15,000 × g for 1 min at room temperature) to move bone marrow as previously described [55]. The resultant hollow bone shafts undergo a series of digestion in collagenase solution (300 active U/ml collagenase type-I dissolved in α-minimal essential medium) and EDTA solution (5 mM, pH = 7.4) for three times separately [56]. The remaining bone shafts were snap frozen in liquid nitrogen. Samples were stored at −80 °C until RNA extraction. Cells cultured in 3D collagen gel were washed with cold PBS and snap frozen in liquid nitrogen and stored in −80 °C until RNA extraction. RNA isolation and qPCR analysis were performed as described before [57]. Briefly, total RNA was collected and purified with the Trizol (15596026, Thermo Fisher Scientific), according to the manufacturer’s protocol. After the isolation of the RNA, the concentration of RNA was determined using Nanodrop 2000. To perform real-time PCR, cDNA was synthesized from 1 µg RNA with reverse transcriptase (RR036A, Takara Bio). Gene expression was analyzed by qPCR with Power SYBR Green (4367659, Thermo Fisher Scientific) in a 10-μl reaction system (for 384-well plate) using the specific primers (Supplementary Table 2). The relative expression level of target genes was normalized to that of β-actin and was analyzed by the 2−ΔΔCT method. The final results were presented as a percentage of the control.
Protein analysis by Western blot
The total soluble protein of the osteocyte-enriched bone tissues and cells was extracted with RIPA lysis buffer (9806S, Cell Signaling Technology). Then the samples were centrifuged at 12,000 × g at 4 °C for 15 min, and the supernatants were collected and stored below −80 °C. After measuring the concentration by the BCA assay (23225, Thermo Fisher Scientific), the proteins were denatured by boiling for 5 min. In total, 30 μg proteins and protein ladder ranging from 10–180 kDa (26616, Thermo Fisher Scientific) were separated on SDS-PAGE (the concentration used was dependent on molecular weight of the targets) and transferred onto PVDF membrane (1620177, Bio-Rad). The membrane was blocked with 5% (w/v) BSA (A1933, Sigma-Aldrich) in Tris-buffered saline (20 mM Tris, pH 7.4, 135 mM NaCl) containing 0.1% Tween 20 (TBS-T) for 1 h at room temperature. Membranes were probed with indicated primary antibodies overnight at 4 °C (Supplementary Table 3). Then the membranes were washed three times with TBS-T. The membranes were incubated with the appropriate secondary antibodies for 1 h according to the vendor’s recommendation, followed with washing with TBS-T. Protein bands were developed with ECL (WBKLS0500, Millipore). β-actin was used as the internal housekeeping gene for the Western blot assay. Densitometric quantification was performed using ImageJ. Uncropped Western blots are provided in Supplementary Material.
Mitochondrial DNA (mt-DNA) content
Total DNA was extracted from cells or osteocyte-enriched bone tissues using DNAzol (10503027, Thermo Fisher Scientific) according to the manufacturer’s instructions. Concentrations of total DNA were measured using a Nanodrop 2000 spectrophotometer (Thermo Scientific). Total DNA stocks were subsequently diluted in water to a final concentration of 40 ng/ml. mt-DNA content was estimated by calculating mtDNA/nDNA ratio through qRT-PCR. The cytochrome c oxidase subunit I (COX1) gene of the mtDNA and the 18S nDNA gene were amplified by qPCR as described before [58]. The final results are presented as a percentage of the control.
Mitochondrial membrane potential (MMP)
MLO-Y4 cells were plated on confocal dish (100350, SPL) and transfected with KD-Ctrl and KD-Sirt3 lentivirus. Then the cells were stained with 200 nM MitoTracker Red CMXRos (M7512, Thermo Fisher Scientific) diluted in serum-free culture medium for 30 min at 37 °C. Then the cells were washed with cold PBS, fixed with 4% paraformaldehyde for 10 min, and then permeabilized with 0.2% Triton X-100 for 10 min and stained with DAPI (62448, Thermo Fisher Scientific). The fluorescence images of mitochondria were visualized with confocal microscope. Five fields of view were randomly selected for each well for analysis with ImageJ.
Mitochondrial ROS level
MLO-Y4 cells were plated on confocal dish (100350, SPL) and transfected with KD-Ctrl and KD-Sirt3 lentivirus. Then the cells were stained with 200 nM MitoTracker Red CM-H2Xros (M7513, Thermo Fisher Scientific) diluted in serum-free culture medium for 20 min at 37 °C. Then the cells were stained with DAPI and visualized with confocal microscope. Five fields of view were randomly selected for each well for analysis with ImageJ.
Quantification of ATP level
ATP level was measured with an ATP Fluorometric Assay Kit according to the manufacturer’s instruction (K354-100, BioVision). Prior to the assay, cells were deproteinized immediately with a PCA deproteinization kit (K808-200, BioVision). Fluorescence intensity (Ex/Em = 535/587 nm) was measured in a VICTOR X Series Multilabel Plate Readers (PerkinElmer, Waltham, USA). ATP content was finally normalized to the protein level and expressed as percentage of control.
Chromatin immunoprecipitation PCR (ChIP-PCR) and qPCR (ChIP-qPCR)
MLO-Y4 cells were first expanded and upon 95–100% confluency, cells were fixed using 1% formaldehyde, washed, and collected by centrifugation (1000 × g for 5 min at 4 °C). The pellet was resuspended in RIPA buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 1% SDS, 1% protease inhibitors), and then vortexed and incubated on ice for 10 min and sonicated by Bioruptor Sonification System (UCD-200). The product of shear cross-linked DNA was determined through electrophoresis. The samples were then centrifuged (16,000 × g for 10 min at 4 °C) and from the supernatant, sheared chromatin (1%) was used as input and immunoprecipitation was performed with anti-CREB antibody (9197, Cell Signaling Technology) and anti-Phospho-CREB antibody (9198S, Cell Signaling Technology). Normal rabbit IgG was used as a negative control (Sc-2027, Santa Cruz). The positive control is a mouse monoclonal antibody with specificity for the CTD region of RNA polymerase II (05-623B, Merck). After precipitation using Pierce Protein A/G Magnetic Beads (17-10085, Merck), followed by RNA and protein digestion, DNA was purified using ChIP kit (17-10085, Merck) according to the manufacturer’s instructions.
PCR was performed using a PCR kit (GoTAq Hot Start Green Master Mix, M7422, Promega). Electrophoresis was performed in a 2% agarose gel under 100 V voltage for 40 min. The primers used for detection of the promoter region of E11/gp38 were shown in Supplementary Table 4. The electrophoretic bands were observed and photographed under UV light. ChIP-qPCR was performed using SYBR® GreenER™ qPCR SuperMix Universal (Thermo Fisher Scientific) and primers for specific E11/gp38 promoter regions.
Plasmids, transfection, and dual-luciferase reporter assays
Truncated promoters of E11/gp38 were cloned into the pGL3 basic plasmid (Promega) by using primers listed in Supplementary Table 5. All plasmids were sequenced to confirm truncations of sequences. The E11/gp38 promoter activity assay was carried out in MLO-Y4 cells by using LipofectamineTM3000 transfection reagent (L3000-001, Invitrogen) according to the manufacturer’s instructions. Cells in 96-well plates were co-transfected with 100 ng of each of the luciferase containing plasmids together with 4 ng of a control pRL-TK plasmid (Renilla luciferase) as an internal control. Blank pGL3 basic plasmids served as negative controls. After 8 h of transfection, cells were administered of indicated drugs and incubated for another 24 h. Cells were harvested and luciferase assay was performed by using the Dual Reporter assay system (E1910, Promega) according to the manufacturer’s instruction. Briefly, cells were lysed with 20 μl of Passive Lysis Buffer (E1910, Promega), and luciferase activities were measured on a VICTOR X Series Multilabel Plate Readers (PerkinElmer, Waltham, USA).
Statistical analysis
The investigators were blinded to genotypes for micro-CT analysis and confocal imaging and related analysis. Data are presented as mean ± SEM, with n as the number of tissue preparations, cells, or animals. No animals or samples were excluded from analysis. The experiments were randomized and mice/cells were randomly assigned to treatment groups. For each independent in vitro experiment, at least three technical replicates were analyzed. Data distribution was previously tested with the Shapiro–Wilk test. Unpaired two-tailed Student’s t test was used for comparison between two groups when data are normally distributed, and Mann–Whitney U test was used when data are not normally distributed. Variance between groups was similar as assessed by the Bartlett’s test. One-way or two-way ANOVA with relevant post hoc tests were used for multiple-group comparisons. A p < 0.05 was considered to be statistically significant. SPSS software (Version 19.0) was used for above statistical analysis.
Data availability
All data relevant to the study are included in the article or uploaded as Supplementary Information.
References
Buenzli PR, Sims NA. Quantifying the osteocyte network in the human skeleton. Bone 2015;75:144–50.
Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26:229–38.
Schneider P, Meier M, Wepf R, Müller R. Towards quantitative 3D imaging of the osteocyte lacuno-canalicular network. Bone 2010;47:848–58.
Bonewald L. Generation and function of osteocyte dendritic processes. J Musculoskelet Neuronal Interact. 2005;5:321.
Milovanovic P, Zimmermann EA, Hahn M, Djonic D, Püschel K, Djuric M, et al. Osteocytic canalicular networks: morphological implications for altered mechanosensitivity. ACS Nano. 2013;7:7542–51.
You L, Cowin SC, Schaffler MB, Weinbaum S. A model for strain amplification in the actin cytoskeleton of osteocytes due to fluid drag on pericellular matrix. J Biomech. 2001;34:1375–86.
Nicolella DP, Moravits DE, Gale AM, Bonewald LF, Lankford J. Osteocyte lacunae tissue strain in cortical bone. J Biomech. 2006;39:1735–43.
Srinivasan S, Gross TS, Bain SD. Bone mechanotransduction may require augmentation in order to strengthen the senescent skeleton. Ageing Res Rev. 2012;11:353–60.
Howe TE, Shea B, Dawson LJ, Downie F, Murray A, Ross C, et al. Exercise for preventing and treating osteoporosis in postmenopausal women. Cochrane Database Syst Rev. 2011;7:CD000333.
Korpelainen R, Keinänen-Kiukaanniemi S, Heikkinen J, Väänänen K, Korpelainen J. Effect of impact exercise on bone mineral density in elderly women with low BMD: a population-based randomized controlled 30-month intervention. Osteoporos Int. 2006;17:109–18.
Busse B, Djonic D, Milovanovic P, Hahn M, Püschel K, Ritchie RO, et al. Decrease in the osteocyte lacunar density accompanied by hypermineralized lacunar occlusion reveals failure and delay of remodeling in aged human bone. Aging Cell. 2010;9:1065–75.
Klein-Nulend J, Sterck J, Semeins C, Lips P, Joldersma M, Baart J, et al. Donor age and mechanosensitivity of human bone cells. Osteoporos Int. 2002;13:137–46.
Tiede-Lewis LM, Xie Y, Hulbert MA, Campos R, Dallas MR, Dusevich V, et al. Degeneration of the osteocyte network in the C57BL/6 mouse model of aging. Aging 2017;9:2190.
Masgras I, Cannino G, Ciscato F, Sanchez-Martin C, Darvishi FB, Scantamburlo F, et al. Tumor growth of neurofibromin-deficient cells is driven by decreased respiration and hampered by NAD+ and SIRT3. Cell Death Differ. 2022;29:1–13.
Kincaid B, Bossy-Wetzel E. Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci. 2013;5:48.
Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010;464:121–5.
Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA. 2011;108:14608–13.
Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–90.
Gao J, Qin A, Liu D, Ruan R, Wang Q, Yuan J, et al. Endoplasmic reticulum mediates mitochondrial transfer within the osteocyte dendritic network. Sci Adv. 2019;5:eaaw7215.
Gao J, Feng Z, Wang X, Zeng M, Liu J, Han S, et al. SIRT3/SOD2 maintains osteoblast differentiation and bone formation by regulating mitochondrial stress. Cell Death Differ. 2018;25:229–40.
Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL III, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging 2009;1:771.
Cheng A, Yang Y, Zhou Y, Maharana C, Lu D, Peng W, et al. Mitochondrial SIRT3 mediates adaptive responses of neurons to exercise and metabolic and excitatory challenges. Cell Metab. 2016;23:128–42.
Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiological Rev. 2009;89:121–45.
Delghandi MP, Johannessen M, Moens U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal. 2005;17:1343–51.
Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age‐related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007;22:1197–207.
Okada S, Yoshida S, Ashrafi SH, Schraufnagel DE. The canalicular structure of compact bone in the rat at different ages. Microsc Microanalysis. 2002;8:104.
Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, et al. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol. 2006;26:4539–52.
Prideaux M, Loveridge N, Pitsillides AA, Farquharson C. Extracellular matrix mineralization promotes E11/gp38 glycoprotein expression and drives osteocytic differentiation. PLoS ONE. 2012;7:e36786.
Staines KA, Javaheri B, Hohenstein P, Fleming R, Ikpegbu E, Unger E, et al. Hypomorphic conditional deletion of E11/Podoplanin reveals a role in osteocyte dendrite elongation. J Cell Physiol. 2017;232:3006–19.
Covarrubias AJ, Perrone R, Grozio A, Verdin E. NAD+ metabolism and its roles in cellular processes during ageing. Nat Rev Mol Cell Biol. 2021;22:119–41.
Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8.
Li Q, Cheng JCY, Jiang Q, Lee WYW. Role of sirtuins in bone biology: Potential implications for novel therapeutic strategies for osteoporosis. Aging Cell. 2021;20:e13301.
Dobson PF, Dennis EP, Hipps D, Reeve A, Laude A, Bradshaw C, et al. Mitochondrial dysfunction impairs osteogenesis, increases osteoclast activity, and accelerates age related bone loss. Sci Rep. 2020;10:1–14.
Figueiredo PA, Powers SK, Ferreira RM, Amado F, Appell HJ, Duarte JA. Impact of lifelong sedentary behavior on mitochondrial function of mice skeletal muscle. J Gerontol Ser A Biomed Sci Med Sci. 2009;64:927–39.
Kim JM, Choi JS, Kim YH, Jin SH, Lim S, Jang HJ, et al. An activator of the cAMP/PKA/CREB pathway promotes osteogenesis from human mesenchymal stem cells. J Cell Physiol. 2013;228:617–26.
Long F, Schipani E, Asahara H, Kronenberg H, Montminy M. The CREB family of activators is required for endochondral bone development. Development 2001;128:541–50.
Sato K, Suematsu A, Nakashima T, Takemoto-Kimura S, Aoki K, Morishita Y, et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med. 2006;12:1410–6.
Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1α transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci USA. 2006;103:14379–84.
Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir) 2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide–dependent deacetylase. J Cell Biol. 2002;158:647–57.
Kim H-S, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52.
Matsui N, Takahashi K, Takeichi M, Kuroshita T, Noguchi K, Yamazaki K, et al. Magnolol and honokiol prevent learning and memory impairment and cholinergic deficit in SAMP8 mice. Brain Res. 2009;1305:108–17.
Liou K-T, Shen Y-C, Chen C-F, Tsao C-M, Tsai S-K. Honokiol protects rat brain from focal cerebral ischemia–reperfusion injury by inhibiting neutrophil infiltration and reactive oxygen species production. Brain Res. 2003;992:159–66.
Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, et al. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun. 2015;6:1–16.
Zhang L, Wang X. Hydrophobic ionic liquid‐based ultrasound‐assisted extraction of magnolol and honokiol from cortex Magnoliae officinalis. J Sep Sci. 2010;33:2035–8.
Bause AS, Haigis MC. SIRT3 regulation of mitochondrial oxidative stress. Exp Gerontol. 2013;48:634–9.
Almeida M, Han L, Martin-Millan M, Plotkin LI, Stewart SA, Roberson PK, et al. Skeletal involution by age-associated oxidative stress and its acceleration by loss of sex steroids. J Biol Chem. 2007;282:27285–97.
Galliera E, Massaccesi L, Banfi G, De Vecchi E, Ragone V, Corsi Romanelli MM, et al. Effect of oxidative stress on bone remodeling in periprosthetic osteolysis. Clin Rev Bone Miner Metab. 2021;19:14–23.
Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald L, Feng J. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320–5.
Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro–computed tomography. J Bone Min Res. 2010;25:1468–86.
Cheuk KY, Wang XF, Wang J, Zhang Z, Yu FWP, Tam EMS, et al. Sexual dimorphism in cortical and trabecular bone microstructure appears during puberty in Chinese children. J Bone Miner Res. 2018;33:1948–55.
Crowe AR, Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc. 2019;9:e3465.
Chen H, Zhang J, Wang Y, Cheuk KY, Hung AL, Lam TP, et al. Abnormal lacuno‐canalicular network and negative correlation between serum osteocalcin and Cobb angle indicate abnormal osteocyte function in adolescent idiopathic scoliosis. FASEB J. 2019;33:13882–92.
Ren Y, Lin S, Jing Y, Dechow P, Feng JQ. A novel way to statistically analyze morphologic changes in Dmp1-null osteocytes. Connect Tissue Res. 2014;55:129–33.
Zhang J, Chen H, Leung RK, Choy KW, Lam TP, Ng BK, et al. Aberrant miR‐145–5p/β‐catenin signal impairs osteocyte function in adolescent idiopathic scoliosis. FASEB J. 2018;32:6537–49.
Kelly NH, Schimenti JC, Ross FP, van der Meulen MC. A method for isolating high quality RNA from mouse cortical and cancellous bone. Bone 2014;68:1–5.
Stern AR, Stern MM, Van Dyke ME, Jähn K, Prideaux M, Bonewald LF. Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques 2012;52:361–73.
Stegen S, van Gastel N, Eelen G, Ghesquière B, D’Anna F, Thienpont B, et al. HIF-1α promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab. 2016;23:265–79.
Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu‐Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator–activated receptor γ coactivator 1α. Arthritis Rheumatol. 2015;67:2141–53.
Acknowledgements
We thank Johan Auwerx for providing the Sirt3 floxed mice and Lynda Bonewald for providing the MLO-Y4 cell lines. We thank Dr. Cao Qian (visiting scholar from Nanjing Medical University) for helping with the animal breeding.
Funding
This work was substantially supported by Start-up grant from Chinese University of Hong Kong (Ref Nos. 4930991 and 4930992), and partly supported by the General Research Fund (Ref Nos. 14163517, 14120818 and 14104620) and Research Matching Grant Scheme, University Grants Committee, HKSAR; Health and Medical Research Fund, The Food and Health Bureau, HKSAR (Ref No. 06170546); and Area of Excellence, University Grants Committee, HKSAR (AoE/M-402/20), 2020 The American Society for Bone and Mineral Research (ASBMR) Rising Star Award, Major Project of Natural Science Foundation of China (Ref No. 81991514), Young Scientists Fund of the Natural Science Foundation of China (Ref No. 82202755), and Young Scientists Fund of the Natural Science Foundation of Jiangsu Province, China (Ref No. BK20220183).
Author information
Authors and Affiliations
Contributions
WYWL and QQL conceived the project. WYWL and QQL designed the study. QQL conducted most assays and acquired and analyzed data. RLW, ZZ, HXW, XML, and JJZ participated in some experiments. APKK, XYT, HFC, ACKC, and QJ provided technical and material support. QQL drafted the manuscript. QJ, JCYC and WYWL revised the manuscript. All authors approved the final version of the manuscript. QQL and WYWL take responsibility for the integrity of the data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Our studies did not include human participants or human tissue. Animal studies were approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong (ethical approval number 19-159-MIS).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by: M. Piacentini
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Q., Wang, R., Zhang, Z. et al. Sirt3 mediates the benefits of exercise on bone in aged mice. Cell Death Differ 30, 152–167 (2023). https://doi.org/10.1038/s41418-022-01053-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01053-5
This article is cited by
-
Bone-targeting engineered small extracellular vesicles carrying anti-miR-6359-CGGGAGC prevent valproic acid-induced bone loss
Signal Transduction and Targeted Therapy (2024)