Abstract
NFATc3 is the predominant member of the NFAT family of transcription factors in neurons, where it plays a pro-apoptotic role. Mechanisms controlling NFAT protein stability are poorly understood. Here we identify Trim39 as an E3 ubiquitin-ligase of NFATc3. Indeed, Trim39 binds and ubiquitinates NFATc3 in vitro and in cells where it reduces NFATc3 protein level and transcriptional activity. In contrast, silencing of endogenous Trim39 decreases NFATc3 ubiquitination and increases its activity, thereby resulting in enhanced neuronal apoptosis. We also show that Trim17 inhibits Trim39-mediated ubiquitination of NFATc3 by reducing both the E3 ubiquitin-ligase activity of Trim39 and the NFATc3/Trim39 interaction. Moreover, we identify Trim39 as a new SUMO-targeted E3 ubiquitin-ligase (STUbL). Indeed, mutation of SUMOylation sites in NFATc3 or SUMO-interacting motifs in Trim39 reduces NFATc3/Trim39 interaction and Trim39-induced ubiquitination of NFATc3. In addition, Trim39 preferentially ubiquitinates SUMOylated forms of NFATc3 in vitro. As a consequence, a SUMOylation-deficient mutant of NFATc3 exhibits increased stability and pro-apoptotic activity in neurons. Taken together, these data indicate that Trim39 modulates neuronal apoptosis by acting as a STUbL for NFATc3.
Similar content being viewed by others
Introduction
The NFAT (Nuclear Factor of Activated T cells) family of transcription factors is a key player in a wide range of physiological and pathological processes. Initially discovered in activated T cells, the different members of the NFAT family have been identified in most tissues where they play both redundant and specific roles [1,2,3,4]. They are implicated in the development and the function of the immune system, brain and other organs by regulating the expression of target genes involved in cytokine production but also in cell proliferation, differentiation and apoptosis. NFAT deregulation is involved in many pathologies including auto-immune diseases, cancer and neurodegenerative diseases [2, 5]. A better understanding of NFAT regulation is therefore of crucial importance.
The cytoplasmic-nuclear shuttling of NFATc1, NFATc2, NFATc3 and NFATc4 has been extensively studied. These NFAT members are normally found in the cytoplasm in a hyperphosphorylated and inactive state. When intracellular calcium increases, they are dephosphorylated by the calcium-dependent phosphatase calcineurin, which triggers their nuclear import and activation. Once in the nucleus, NFATs regulates the transcription of target genes, in cooperation with partner transcription factors such as AP-1 [3, 5]. In contrast, the regulation of NFAT stability remains elusive. However, different studies have shown that interfering with the regulation of NFAT levels by the ubiquitin-proteasome system can have a marked impact on the physiology of various cell types [6,7,8,9,10,11]. In addition to phosphorylation and ubiquitination, NFAT proteins can be regulated by SUMOylation. Indeed, covalent conjugation of SUMO to NFATs has been shown to impact their cytoplasmic-nuclear shuttling and activity [12,13,14,15]. SUMOylation can have many consequences on its substrate proteins, including interaction properties and subcellular localization [16, 17] but also stability [18]. Indeed, a few SUMO-targeted E3 ubiquitin-ligases (STUbLs) that specifically recognize and ubiquitinate SUMOylated proteins have been described [19,20,21], raising the possibility that SUMOylation might also modulate NFAT ubiquitination and degradation.
NFATc3 is the predominant NFAT family member expressed in neurons [22,23,24]. We have previously shown that NFATc3 is involved in neuronal apoptosis [24]. Two independent studies have also implicated NFATc3 in α-synuclein-induced degeneration of dopaminergic neurons in Parkinson’s disease [23, 25]. Activation of protein kinases such as GSK3β does not seem to be sufficient to induce NFATc3 nuclear exclusion in neurons [22, 24]. Proteasomal degradation could therefore be an alternative to reduce its activity. However, only one study on the ubiquitination and degradation of NFATc3 has been reported so far, in cardiomyoblasts [11]. In previous work, we have shown that NFATc3 can be SUMOylated and bind to the E3 ubiquitin-ligase Trim17 [24]. However, Trim17 does not induce but rather reduces the ubiquitination of NFATc3 and increases its protein level [24]. Since TRIM17 can prevent ubiquitination of some of its binding partners by inhibiting other E3 ubiquitin-ligases from the TRIM family [26, 27], we hypothesized that the stability of NFATc3 might be regulated by a TRIM protein interacting with Trim17, such as Trim39.
In the present study, we demonstrate that Trim39 is a genuine E3 ubiquitin-ligase for NFATc3 and that Trim39-mediated ubiquitination of NFATc3 is inhibited by Trim17. Moreover, we show that SUMOylation of NFATc3 favors its ubiquitination and degradation, as well as its binding to Trim39. Further data confirm that Trim39 acts as a STUbL for NFATc3. As a result, SUMOylation and Trim39 modulate the transcription factor activity of NFATc3 and its pro-apoptotic effect in neurons. Therefore, our study provides the identification of a new STUbL and a first insight into complex mechanisms regulating the stability of NFATc3 in neurons.
Results
Trim39 is an E3 ubiquitin-ligase for NFATc3
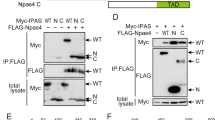
Human TRIM39 and TRIM17 proteins have been found to bind to each other in three independent proteome-scale yeast two-hybrid screens [28,29,30]. We confirmed that mouse Trim39 and Trim17 proteins also interact, using co-immunoprecipitation. Indeed, in cells co-transfected with Trim17-GFP and Flag-Trim39, the two proteins were reciprocally co-immunoprecipitated using either an anti-Flag antibody or GFP-Trap beads (Fig. 1A). Using in situ proximity ligation assay (PLA) in mouse Neuro2A cells, we also found that Trim17 and Trim39 are in close proximity, primarily in the cytoplasm, suggesting that the two endogenous proteins do interact (Supplementary Fig. S1A). Overexpression of Trim17-GFP increased the PLA signal, confirming the specificity of the assay (Supplementary Fig. S1A). Importantly, Trim39 can also bind to NFATc3. Indeed, overexpressed HA-NFATc3 and Flag-Trim39 were reciprocally co-immunoprecipitated (Fig. 1B) and endogenous Trim39 was co-immunoprecipitated with endogenous NFATc3 (Fig. 1C). Moreover, a PLA signal was detected between endogenous NFATc3 and endogenous Trim39, predominantly in the cytoplasm (Fig. 1D). To increase protein amounts and the resulting signal in this assay, cells were treated with the proteasome inhibitor MG-132 for 4 h before fixation. Overexpression of Trim39 increased the PLA signal (Fig. 1D), whereas reducing the level of endogenous Trim39 protein by using a specific shRNA (Supplementary Fig. S1B) strongly decreased the PLA signal (Fig. 1E), therefore confirming the specificity of the assay. Taken together, these data indicate that Trim39 interacts with both Trim17 and NFATc3.
A BHK cells were transfected with Trim17-GFP together with Flag-Trim39 or empty plasmid for 24 h. Cells were then treated with 20 μM MG-132 for 5 h. The cells were subsequently harvested and lysates were subjected to immunoprecipitation using anti-Flag agarose beads (left panel) or GFP-Trap beads (right panel). Immunoprecipitates and total lysates were analyzed by western blot using anti-GFP and anti-Flag antibodies. B Neuro2A cells were transfected with HA-NFATc3 together with Flag-Trim39 or empty plasmid for 24 h. Cells were then treated as in A and lysates were subjected to immunoprecipitation using anti-Flag (left panel) or anti-HA (right panel) antibodies. Immunoprecipitates and total lysates were analyzed by western blot using anti-HA and anti-Flag antibodies. C Total lysate from Neuro2A cells was subjected to immunoprecipitation using normal rabbit IgG (as a negative control) or anti-NFATc3 antibody. Immunoprecipitates and input total lysates were analyzed by western blot using anti-NFATc3 and anti-Trim39 antibodies. D Neuro2A cells were treated with 10 μM MG-132 for 4 h and then fixed and subjected to in situ PLA using rabbit anti-NFATc3 and mouse anti-Trim39 antibodies. Each bright red spot indicates that the two proteins are in close proximity. Negative control was obtained by omitting anti-Trim39 antibody. When indicated, cells were previously transfected with Trim39 for 24 h (overexpressed). Images were analyzed by confocal microscopy. To better visualize the differences in PLA intensity, maximum intensity projection was applied to the z-stacks of images on the left panel. To better determine the subcellular location of the NFATc3/Trim39 interaction, a single slice of the z-stack is presented on the right panel (endo 1 slice). Nuclear staining was performed using DAPI and visualized in merged pictures with PLA signal (merge). The number of dots per cell was determined in one slice of each image using Fiji. The graph shows the mean ± SEM of more than 12 images per condition, including a total of more than 140 cells, from one experiment representative of two independent experiments. **P < 0.01, ****P < 0.0001 significantly different (one way ANOVA followed by Tukey’s multiple comparison test). E Neuro2A cells were transduced with lentiviral particles expressing a control shRNA (sheGFP) or a specific shRNA against Trim39 (shTrim39#1) for 24 h. Transduced cells were selected using puromycin for two additional days and plated onto coverslips. The day after plating, cells were treated and analyzed by PLA as in D. The graph shows the mean ± SEM of 6 images per condition, including more than 75 cells, from one experiment representative of two independent experiments. ***P < 0.001 significantly different (one way ANOVA followed by Tukey’s multiple comparison test).
We next examined whether Trim39 could mediate the ubiquitination of NFATc3. In Neuro2A cells co-transfected with His-tagged ubiquitin, the ubiquitination level of NFATc3 was significantly increased by Trim39 but not by an inactive mutant deleted of its RING domain (Trim39-ΔRING; Fig. 2A). In contrast, silencing of Trim39 using three different specific shRNAs, strongly decreased NFATc3 ubiquitination (Fig. 2B). Then, in vitro ubiquitination assay was carried out using in vitro translated/immuno-purified NFATc3 and purified recombinant proteins. GST-Trim39 ubiquitinated NFATc3, whereas an inactive mutant in which two Cys residues of the RING domain were mutated (GST-Trim39-C49S/C52S) did not (Fig. 2C). To determine the type of ubiquitin chains promoted by Trim39, in vitro ubiquitination of NFATc3 was performed with ubiquitin mutants in which all but one of the Lys residues were replaced by Arg, so that ubiquitin chains can be formed with only one type of linkage. Data suggest that Trim39 mediates the formation of K48 and, to a lesser extent, K63 ubiquitin chains on NFATc3, but not K6, K27, K29 or K33 ubiquitin chains (Fig. 2D). As no modification of NFATc3 was visible when the reaction was performed in the absence of ubiquitin or with inactive Trim39, these data strongly suggest that NFATc3 is a direct substrate for the E3 ubiquitin-ligase activity of Trim39.
A Neuro2A cells were transfected with HA-NFATc3 together with empty plasmid or His-tagged ubiquitin, in the presence or absence of Flag-Trim39 or the inactive mutant Flag-Trim39-ΔRING for 24 h. Cells were then incubated with 20 μM MG-132 for 6 h before harvesting. The ubiquitinated proteins were purified using nickel beads and analyzed by western blotting using anti-HA antibody to detect ubiquitin-conjugated HA-NFATc3. In a separate SDS-PAGE, samples of the input lysates used for the purification were analyzed with antibodies against HA and Flag. B Neuro2A cells were transduced with lentiviruses expressing a control shRNA (sheGFP) or three different shRNAs targeting Trim39. Following 48 h transduction and 48 h selection of transduced cells using puromycin, cells were kept in culture for one week and plated in new dishes. Then, cells were transfected with HA-NFATc3 or His-tagged ubiquitin or both for 24 h, and treated as in (A). C In vitro translated HA-NFATc3 was first immunopurified from wheat germ extract using anti-HA antibody. Then, beads used for immunopurification of NFATc3 were incubated for 1 h at 37 °C in the in vitro ubiquitination reaction mix (containing E1 and E2 enzymes) with purified recombinant GST-Trim39 (WT) or its inactive mutant GST-Trim39-C49S/C52S (mut) in the presence or the absence of ubiquitin as indicated. Poly-ubiquitinated forms of NFATc3 were detected by immunoblotting using an anti-NFATc3 antibody. The same membrane was immunoblotted with an anti-TRIM39 antibody. D In vitro translated HA-NFATc3 was purified as in (C). and incubated for 2 h at 37 °C in the ubiquitination reaction mix with purified recombinant His-TRIM39 (WT), in the presence of WT ubiquitin or different ubiquitin mutants, as indicated. Poly-ubiquitinated NFATc3 and His-TRIM39 were detected as in (C).
Trim39 induces the degradation of NFATc3 and decreases its transcriptional activity
Ubiquitination often targets proteins for proteasomal degradation, in particular when K48 ubiquitin chains are formed. Indeed, the protein level of NFATc3 progressively decreased when co-transfected with increasing amounts of Trim39 (Fig. 3A). Interestingly, inactive Trim39-ΔRING did not decrease but rather increased NFATc3 protein, as did proteasome inhibition (Fig. 3A). As mutation of the RING of E3 ubiquitin-ligases generally induces a dominant-negative effect [31, 32], this increase may be due to the inhibition of endogenous Trim39 by Trim39-ΔRING. Consistently, silencing of endogenous Trim39 also significantly increased the protein level of endogenous NFATc3, without significantly modifying its mRNA level (Fig. 3B, Supplementary Fig. S2A). Therefore, these data strongly suggest that Trim39-mediated ubiquitination leads to the proteasomal degradation of NFATc3.
A BHK cells were transfected with a fixed amount of a HA-NFATc3 plasmid (1 μg) together with empty plasmid (-) or increasing amounts of Flag-Trim39 plasmid (0.1, 0.2, 0.5 and 1 μg) or 0.2 μg of Flag-Trim39-ΔRING plasmid. When indicated, the cells were treated with 10 μM MG-132 for 6 h before harvesting. Total lysates were analyzed by western blot using antibodies against HA, Flag and actin. B Neuro2A cells were left untreated (NT), or transfected twice with an siRNA against Trim39 (siTrim39#1) or with a negative control siRNA (siCtrl) for 48 h. Total lysates were analyzed by western blot using antibodies against NFATc3 and actin. The intensity of the NFATc3 bands was quantified, normalized by the intensity of the actin bands and expressed relative to the values obtained with the control siRNA. The graph shows mean and individual points from three independent experiments. *P < 0.05 significantly different from siCtrl (one-way ANOVA followed by Dunnett’s multiple comparisons test). C Neuro2A cells were co-transfected with empty plasmid or HA-NFATc3, together with empty plasmid, Flag-Trim39 or Flag-Trim39-ΔRING for 24 h. Then, total RNA was extracted and the mRNA level of Trim17 was estimated by quantitative PCR. Data are the means ± SEM of four independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 significantly different (two-way ANOVA followed by Tukey’s multiple comparisons test). D Neuro2A cells were transfected with empty plasmid, Flag-Trim39 or Flag-Trim39-ΔRING for 24 h. Then, cells were left untreated (control) or were deprived of serum for 3 h and subsequently treated with 1 μM A23187 and 100 nM PMA in serum-free medium for 1 h (A23 + PMA). Total RNA was extracted and the mRNA level of Trim17 and NFATc3 was estimated by quantitative PCR. Data are the means ± SEM of three independent experiments. *P < 0.05; **P < 0.01 significantly different (two-way ANOVA followed by Tukey’s multiple comparisons test). #P < 0.05 significantly different with A23 + PMA compared to the corresponding ctrl (two-way ANOVA followed by Sidak’s multiple comparisons test). E Neuro2A cells were transfected twice with two different siRNAs against Trim39 or with a control siRNA for 48 h. Then, cells were left untreated (control) or were treated with A23187 and PMA as in D. Total RNA was extracted and the mRNA level of Trim39, Trim17 and NFATc3 was estimated by quantitative PCR (NT = non transfected). Data are the means ± SEM of six independent experiments. *P < 0.05; **P < 0,005; ***P < 0.001; ****P < 0.0001 significantly different from cells transfected with control siRNA or non-transfected cells in the same condition (two-way ANOVA followed by Dunnet’s or Sidak’s multiple comparisons tests). #P < 0.05 significantly difference between A23 + PMA and control for the indicated condition (two-way ANOVA followed by Sidak’s multiple comparisons test). For unknown reason the control siRNA strongly decreased the mRNA levels of NFATc3. Note that this did not have a significant impact on the expression of Trim17 taken as a target gene of NFATc3, probably because the protein level of NFATc3 was not significantly reduced over the time course of the experiment despite the decrease in its mRNA levels.
To examine whether Trim39 have an impact on the transcription factor activity of NFATc3, we measured the expression of one of its target genes: Trim17 [24]. The increase in Trim17 mRNA levels induced by NFATc3 overexpression was completely abrogated by co-expression of wild type (WT) but not inactive Trim39-ΔRING (Fig. 3C). Even when NFATc3 was not transfected, Trim39-ΔRING significantly increased Trim17 expression (Fig. 3C), suggesting that the inhibition of endogenous Trim39 by the dominant-negative mutant Trim39-ΔRING is sufficient to increase the activity of endogenous NFATc3. To confirm this, Neuro2A cells were treated with the calcium ionophore A23187 and phorbol 12-myristate 13-acetate (PMA) to activate both endogenous NFATc3 and its transcriptional partner AP-1. As previously reported [24], Trim17 mRNA level was increased by this treatment (Fig. 3D), probably due to the activation of endogenous NFATc3, the NFAT transcription factor predominantly expressed in Neuro2A cells [24]. Again, this Trim17 induction was completely abrogated by overexpression of WT but not inactive Trim39 (Fig. 3D). In addition, silencing of Trim39 using two different specific siRNAs (Fig. 3E, left panel) induced Trim17 expression, in particular following treatment with A23187 and PMA (Fig. 3E). As we have previously shown that Trim17 can bind and inhibit NFATc3 by preventing its nuclear translocation [24], we examined whether Trim39 has the same effect. Indeed, under conditions where Trim17 decreased the nuclear translocation of NFATc3 by more than twofold, Trim39 had no impact on NFATc3 subcellular localization (Supplementary Fig. S2B,C). These data therefore suggest that Trim39 inhibits the transcription factor activity of NFATc3 by ubiquitinating it and by inducing its proteasomal degradation, but not by preventing its nuclear translocation.
Trim17 inhibits the ubiquitination of NFATc3 mediated by Trim39
As we initially observed that Trim17 decreases basal NFATc3 ubiquitination [24], we tested whether Trim17 could affect Trim39-mediated ubiquitination of NFATc3. Indeed, the increase in NFATc3 ubiquitination induced by Trim39 was abolished by co-transfection of Trim17 in cells (Fig. 4A). Moreover, in vitro ubiquitination of NFATc3 by His-TRIM39 was completely prevented by MBP-TRIM17 (Fig. 4B). As only purified proteins were used in this assay, this indicates that TRIM17 directly inhibits TRIM39-mediated ubiquitination of NFATc3. Interestingly, the ubiquitination level of Trim39 was strongly decreased in the presence of Trim17 both in cells (Fig. 4A) and in vitro (Fig. 4B). As in vitro auto-ubiquitination gives a measure of E3 ubiquitin-ligase activity [32], this suggests that TRIM17 directly inhibits the E3 activity of TRIM39. Trim17 ubiquitination was also reduced by Trim39 in cells (Supplementary Fig. S3), suggesting a reciprocal inhibition of the two TRIM proteins. Intriguingly, a RING-deletion mutant of TRIM17 was unable to inhibit TRIM39-mediated ubiquitination of NFATc3 and TRIM39 auto-ubiquitination (Fig. 4C). Moreover, Trim17-GFP almost completely prevented the reciprocal co-immunoprecipitation of Flag-Trim39 and HA-NFATc3, when co-transfected with the two partners (Fig. 5A, B). Consistently, the PLA signal between endogenous NFATc3 and Trim39 proteins was significantly reduced by transfection of Trim17-GFP compared to GFP (Fig. 5C, D). Taken together, these data strongly suggest that Trim17 inhibits Trim39-mediated ubiquitination of NFATc3 by inhibiting both the E3 ubiquitin-ligase activity of Trim39 and the interaction between NFATc3 and Trim39.
A BHK cells were transfected with HA-NFATc3 together with His-tagged ubiquitin, in the presence or the absence of Flag-Trim39, Trim17-GFP or both for 24 h. Then, cells were incubated with 20 μM MG-132 for 6 h before harvesting. The ubiquitinated proteins were purified using nickel beads and analyzed by western blotting using anti-HA and anti-Flag antibodies. In a separate SDS-PAGE, samples of the input lysates were analyzed with antibodies against HA, Flag and GFP. B In vitro translated HA-NFATc3 was first immunopurified from wheat germ extract using anti-HA antibody. Then, beads used for immunopurification of NFATc3 were incubated for 1 h at 37 °C in the in vitro ubiquitination reaction mix with purified recombinant His-TRIM39 or MBP-TRIM17 or both. Poly-ubiquitinated forms of NFATc3, TRIM39 and TRIM17 were detected by immunoblotting using anti-NFATc3, anti-TRIM39 and anti-TRIM17 antibodies revealed using high exposure times. Low exposure times were used to compare the level of TRIM39 and TRIM17 in the different conditions. C In vitro translated HA-NFATc3 was incubated for 1 h at 37 °C in the in vitro ubiquitination reaction mix with purified recombinant His-TRIM39 or MBP-TRIM17 (either WT or deleted from RING domain). Poly-ubiquitinated forms of NFATc3, TRIM39 and TRIM17 were detected by immunoblotting using anti-NFATc3, anti-TRIM39 and anti-TRIM17 antibodies.
A, B Neuro2A cells were transfected with HA-NFATc3 in the presence or the absence of Flag-Trim39, Trim17-GFP or both, for 24 h. Cells were then treated with 20 μM MG-132 for 7 h. The cells were subsequently harvested and lysates were subjected to immunoprecipitation using anti-HA (A) or anti-Flag (B) antibodies. Immunoprecipitates and total lysates were analyzed by western blot using anti-HA, anti-GFP and anti-Flag antibodies. The intensity of the bands containing Flag-Trim39 co-immunoprecipitated with HA-NFATc3 was normalized by the intensity of the bands corresponding to immunoprecipitated HA-NFATc3 (A). The intensity of the bands containing HA-NFATc3 co-immunoprecipitated with Flag-Trim39 was normalized by the intensity of the bands corresponding to immunoprecipitated Flag-Trim39 (B). Relative values, indicated in red, correspond to the experiment shown in the figure, which is representative of three independent experiments. C Neuro2A cells were transfected with GFP or Trim17-GFP for 24 h. Then cells were treated with 10 μM MG-132 for 4 h, fixed and subjected to in situ PLA using rabbit anti-NFATc3 and mouse anti-Trim39 antibodies. Images were analyzed by confocal microscopy and a single slice of the z-stacks is presented for each condition. Nuclear staining was performed using DAPI and merged with the PLA signal. Note that, in the Trim17-GFP condition, transfected cells (delineated by a yellow or red line) show less dots than neighboring non transfected cells, which is not the case in the GFP condition. D The number of dots was determined on one slice for individual cells transfected with either GFP or Trim17-GFP using Fiji. Data represent one experiment, including 68 transfected cells for each condition, representative of two independent experiments. ****p < 0.0001, significantly different from GFP transfected cells (unpaired t test).
SUMOylation of NFATc3 modulates its ubiquitination and stability
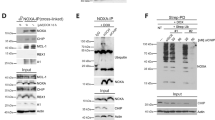
In a previous study, we have identified three consensus SUMOylation sites in NFATc3 [24]. We next tested whether alteration of NFATc3 SUMOylation has an impact on its ubiquitination and half-life. We had previously used NFATc3 K/R mutants in which the acceptor Lys residues of the SUMOylation consensus motifs (ψKXE [33]) were replaced by Arg [24]. However, large-scale mass spectrometry studies have shown that a quarter of SUMO acceptor Lys are also used for ubiquitination [18]. Therefore, additional NFATc3 E/A mutants were generated by substituting the Glu residues of the SUMOylation motifs for Ala, in order to prevent SUMOylation without affecting a possible ubiquitination at these sites. As expected, in vitro SUMOylation of NFATc3-EallA and NFATc3-KallR mutants (in which the Glu and Lys residues of the three consensus motifs were replaced by Ala and Arg respectively) was almost completely abrogated (Fig. 6A). The slow-migrating bands detected in this assay were SUMOylated forms of NFATc3 as they were not produced when the reaction mix did not contain SUMO-1 (Supplementary Fig. S4A). Interestingly, the ubiquitination level of NFATc3 in cells was not significantly decreased by single mutations or the double E437A/E706A mutation whereas it was more decreased by the E706A/E1015A mutation and strongly decreased by the triple mutation (Fig. 6B, C). Therefore, SUMOylation of at least one of the three consensus motifs seems to be necessary for NFATc3 ubiquitination. To further confirm the SUMOylation-dependent ubiquitination of NFATc3, cells were treated with the SUMO-activating enzyme inhibitor ML-792 [34]. This abrogated global SUMOylation (Supplementary Fig. S4B) and deeply decreased NFATc3 ubiquitination (Fig. 6D). Consistently, the half-life of NFATc3-EallA was significantly increased compared to WT NFATc3 (Fig. 6E, F). Taken as a whole, these data indicate that SUMOylation of NFATc3 favors its ubiquitination and subsequent degradation.
A In vitro translated HA-NFATc3 was incubated with in vitro SUMOylation reaction mix (containing SUMO-1, E1, E2 and E3 enzymes) for 1 h 30 or directly added to sample loading buffer together with reaction mix (time 0). Multi-SUMOylated forms of NFATc3 were detected by immunoblotting using anti-NFATc3 antibody. B Neuro2A cells were transfected with His-tagged ubiquitin or empty plasmid, together with WT HA-NFATc3 or the different HA-NFATc3 E/A mutants for 24 h. Then, cells were incubated with 20 μM MG-132 for 7 h before harvesting. The ubiquitinated proteins were purified using nickel beads and analyzed by western blot using anti-HA antibody to detect ubiquitin-conjugated HA-NFATc3. In a separate SDS-PAGE, input samples were analyzed with antibodies against HA and tubulin. C The intensity of the NFATc3 ladders from the nickel bead purification was quantified in different experiments performed as in (B). Data are the mean and individual points of four independent experiments. *p < 0.05, significantly different from WT NFATc3 (one-way ANOVA followed by Dunnett’s multiple comparisons test). The apparent discrepancy with the immunoblot shown in (B) is due to the fact that NFATc3 mutations have more or less impact from one experiment to another, because reducing the basal level of ubiquitination generates very weak signals that are more of less well detected. D Neuro2A cells were transfected with His-tagged ubiquitin or empty plasmid, together with HA-NFATc3 for 24 h. Then, cells were incubated with 20 μM MG-132 in the presence or the absence of 0.3 μM ML-792 for 8 h. The ubiquitination level of NFATc3 was assessed as in (B). E Neuro2A cells were transfected with WT HA-NFATc3 or NFATc3-EallA for 48 h. Then, cells were incubated with 20 μg/ml cycloheximide (CHX) for increasing times before harvesting. Proteins were analyzed by western blot using antibodies against HA and tubulin. F The intensity of the bands on the western blots of different experiments performed as in (C) was quantified. For each experiment, the amount of NFATc3 was normalized by the level of tubulin in each condition and plotted against CHX incubation time. Data are the mean ± SEM of three independent experiments. ***p < 0.0001, **p < 0.005 significantly different from WT NFATc3 at the same incubation time (two-way ANOVA followed by Sidak’s multiple comparisons test).
Trim39 acts as a SUMO-targeted E3 ubiquitin-ligase for NFATc3
We further examined whether SUMOylation of NFATc3 modulates its ubiquitination by Trim39. Indeed, the ubiquitination level of the NFATc3-EallA mutant was decreased compared to WT NFATc3 when co-expressed with Trim39 in cells (Fig. 7A). Inhibition of global SUMOylation by ML-792 (Supplementary Fig. S4B) also strongly reduced Trim39-mediated ubiquitination of NFATc3 (Fig. 7B). Moreover, mutation of the three SUMOylation sites of NFATc3 reduced its interaction with Trim39, as assessed by reciprocal co-immunoprecipitation experiments (Fig. 7C). Therefore, these data indicate that Trim39 binds and ubiquitinates preferentially SUMOylated forms of NFATc3.
A Neuro2A cells were transfected with His-tagged ubiquitin together with WT HA-NFATc3 or HA-NFATc3 EallA, in the presence or the absence of Flag-Trim39, for 24 h. Then, cells were incubated with 20 μM MG-132 for 6 h before harvesting. The ubiquitinated proteins were purified using nickel beads and analyzed by western blotting using anti-HA antibody. In a separate SDS-PAGE, samples of the input lysates were analyzed with antibodies against HA and Flag. B Neuro2A cells were transfected with HA-NFATc3, in the presence or the absence of His-tagged ubiquitin and Flag-Trim39, as indicated, for 24 h. Then, cells were incubated with 20 μM MG-132 in the presence or the absence of 0.3 μM ML-792 for 8 h before harvesting. The ubiquitination level of NFATc3 was assessed as in (A). C Neuro2A cells were transfected with Flag-Trim39 together with WT HA-NFATc3, HA-NFATc3-EallA or empty plasmid for 24 h. Cells were then treated with 10 μM MG-132 for 8 h and lysates were subjected to immunoprecipitation using anti-HA antibody (left panel) or anti-Flag beads (right panel). Immunoprecipitates and total lysates were analyzed by western blot using anti-HA and anti-Flag antibodies. Band intensities of co-immunoprecipitated Flag-Trim39 were quantified and normalized by band intensities of immunoprecipitated HA-NFATc3. Band intensities of co-immunoprecipitated HA-NFATc3 were normalized by band intensities of immunoprecipitated Flag-Trim39. Relative values, indicated in red, correspond to the experiment shown in the figure, which is representative of three independent experiments. D Schematic representation of mouse Trim39 depicting its different domains and its three putative SIMs. E Recombinant GST, GST-Trim39 and its different SIM mutants were purified using glutathione beads and subsequently incubated with purified recombinant SUMO-2 and SUMO-2 chains. Material bound to the beads was eluted and analyzed by western blot using anti-SUMO and anti-GST antibodies. A small fraction of the SUMO-2 chains was also loaded on the gel (input) for comparison. Note that SUMO bands are multiple of ≈15 kDa corresponding to mono-, di-, tri-, tetra-SUMO etc… (F) Neuro2A cells were transfected with HA-NFATc3 or empty plasmid together with WT Flag-Trim39 or its SIM3 mutant for 24 h. Then, cells were treated as in (C) and lysates were subjected to immunoprecipitation using anti-HA antibody. Immunoprecipitates and total lysates were analyzed as in (C). G Neuro2A cells were transfected with His-tagged ubiquitin (or empty plasmid) together with HA-NFATc3 in the presence or the absence of Flag-Trim39 or its SIM3 mutant, for 24 h. Then, cells were treated and ubiquitinated proteins and input lysates were analyzed as in (A). H In vitro translated HA-NFATc3 was first incubated (+) in SUMOylation reaction mix or not (−) for 1 h at 37 °C. Then, NFATc3 was immunopurified from the reaction mix using anti-HA antibody. Beads used for immunopurification of NFATc3 were then incubated for 1 h at 37 °C in the in vitro ubiquitination reaction mix with purified recombinant His-Trim39 (WT) or its SIM3 mutant (mSIM3 His-Trim39). The reaction samples were then treated or not with the SUMO-specific protease SENP1 for 15 min at 37 °C. The three last samples were not subjected to in vitro ubiquitination to assess the results of SUMOylation and SENP1 reactions. Poly-ubiquitinated and multi-SUMOylated forms of NFATc3 were detected by immunoblotting using an anti-HA antibody (left panel). The same membrane was immunoblotted with an anti-Trim39 antibody to assess the auto-ubiquitination of the two forms of Trim39. I Purified recombinant SUMO-2 chains were incubated in the in vitro ubiquitination reaction mix, in the absence or the presence of ubiquitin, with purified recombinant His-Trim39 (WT) or its SIM3 mutant (mSIM3). The reaction samples were analyzed by immunoblotting with an antibody against SUMO-2. The poly-ubiquitinated forms of SUMO-2 chains appear as a more intense smear above 100 kDa. In a separate SDS-PAGE, the same volumes of WT and mSIM3 His-Trim39 as used in the in vitro ubiquitination experiment were analyzed with antibodies against Trim39.
Proteins interacting non-covalently with SUMO generally harbor SUMO-interacting motifs (SIMs), which typically consist of three hydrophobic residues in a sequence of four amino acids, sometimes flanked by acidic or phosphorylated residues [35]. Using GPS-SUMO [36] and JASSA [37] web-based tools, we identified three putative SIMs in the Trim39 sequence, which are conserved from mouse to human and which we named SIM1 (39-PVII-42), SIM2 (125-VCLI-128) and SIM3 (211-LLSRL-215) (Fig. 7D). Trim39 constructs were generated in which most residues of the three SIMs were mutated to Ala (mSIM1: 39-PAAA-42, mSIM2: 125-AAAA-128 and mSIM3: 211-AAARA-215) and GST pull-down experiments were performed using purified recombinant proteins. Interestingly, GST-Trim39 could bind di-, tri-, tetra- and higher-order SUMO-2 chains but not free SUMO-2, whereas GST alone showed no interaction (Fig. 7E). Mutation of SIM1 and SIM2 had no significant effect but mutation of SIM3 strongly reduced the SUMO-binding ability of Trim39 (Fig. 7E, Supplementary Fig. S5). Consistently, mutation of SIM3 reduced the ability of Trim39 to bind NFATc3 in co-immunoprecipitation experiments (Fig. 7F). Indeed, the quantification of band intensities from three independent experiments indicates that the amount of Trim39-mSIM3 co-immunoprecipitated with NFATc3 is 35.6 ± 7.1% compared with WT Trim39 (average ± SD). Moreover, SIM3 mutation reduced the ability of Trim39 to ubiquitinate NFATc3 in Neuro2A cells (Fig. 7G).
To further characterize Trim39 as a STUbL, in vitro translated HA-NFATc3 was first subjected to in vitro SUMOylation, then immunopurified and finally subjected to in vitro ubiquitination using WT Trim39 or mSIM3-Trim39. In some samples, the SUMO-specific protease SENP1 was then used to remove SUMO chains from NFATc3 and better visualize the ubiquitinated forms. Importantly, ubiquitination of NFATc3 by WT Trim39 was strongly increased by its prior SUMOylation and the resulting higher molecular weight forms of NFATc3 were not drastically changed by SENP1 (Fig. 7H). In contrast, mSIM3-Trim39 did not significantly modify NFATc3 and the higher bands disappeared following SENP1 treatment (Fig. 7H). Yet, the E3 ubiquitin-ligase activity of mSIM3-Trim39 assessed by its auto-ubiquitination was not significantly different from that of WT Trim39 (Fig. 7H). Furthermore, as described for other STUbLs [19], recombinant purified WT Trim39 was able to ubiquitinate SUMO chains in vitro, whereas its mSIM3 mutant was not (Fig. 7I). Collectively, these data indicate that Trim39 acts as a STUbL for NFATc3 by preferentially binding the SUMOylated forms of NFATc3 through its SIM3, to mediate their ubiquitination.
SUMOylation and Trim39 modulate the pro-apoptotic effect of NFATc3 in neurons
We have previously shown that NFATc3 in involved in serum and KCl deprivation-induced apoptosis, in primary cultures of cerebellar granule neurons (CGNs) [24]. We used this well characterized in vitro models of neuronal apoptosis [38] to examine the impact of NFATc3 SUMOylation on its pro-apoptotic effect. As shown previously [24], apoptosis was significantly increased in CGNs transfected with WT GFP-NFATc3 compared to GFP, as shown by the increased number of condensed nuclei (Fig. 8A, B). Interestingly, neuronal apoptosis was further increased in neurons overexpressing GFP-NFATc3-EallA (Fig. 8A,B), suggesting that stabilization of NFATc3 by preventing its SUMOylation increases its pro-apoptotic effect. Consistently, efficient silencing of endogenous Trim39 (Supplementary Fig. S6A) aggravated neuronal apoptosis. Indeed, the number of apoptotic nuclei (Fig. 8C, D) and the level of active caspase 3 (Fig. 8E, F) were significantly increased by a specific shRNA against Trim39 compared to a control shRNA. Importantly, efficient knock-down of endogenous NFATc3 using two different shRNAs (Supplementary Fig. S6B) completely abrogated the increase in apoptosis induced by the shRNA-Trim39 (Fig. 8E, F), indicating that the pro-apoptotic effect of Trim39 silencing depends on NFATc3. Our data therefore strongly suggest that SUMOylation and Trim39 negatively regulate the pro-apoptotic function of NFATc3, most likely by reducing its protein level and thereby its transcription factor activity.
A CGN primary cultures were transfected after 5 days in vitro (DIV) with GFP, WT GFP-NFATc3 or GFP-NFATc3-EallA for 16 h. Then, neurons were switched to serum-free medium containing 5 mM KCl (K5) for 7 h or were left untreated (control). Indeed, CGNs survive in the presence of serum and depolarizing concentrations of KCl (25 mM) that mimic the neuronal activity required for their survival in vivo. They undergo apoptosis following withdrawal of serum and lowering of KCl to 5 mM (K5), which recapitulates the programmed cell death naturally occurring in the cerebellum during post-natal development. Nuclei were visualized by DAPI staining and proteins fused to GFP were detected by fluorescent microscopy. Arrows indicate GFP-positive neurons with thick arrows for neurons undergoing apoptosis and thin arrows for healthy neurons. B The percentage of transfected, GFP-positive neurons undergoing apoptosis was assessed by examining cell morphology and nuclear condensation. Data are the means ± SEM of four independent experiments performed as in (A). *P < 0.05; ***P < 0.001 significantly different (two-way ANOVA followed by Sidak’s multiple comparisons test). Apoptosis was highly significantly different in K5 versus Ctrl in all conditions (not shown). C CGNs were transduced with lentiviral particles expressing a control shRNA (directed against Luciferase) or an shRNA specifically targeting Trim39 one day after plating. At DIV 6, they were incubated for 8 h in K5 medium, fixed and stained with DAPI. D The percentage of apoptotic neurons was estimated by examining nuclear condensation. Data are the means ± SEM of four independent experiments performed as in (C). ****P < 0.0001 significantly different (two-way ANOVA followed by Sidak’s multiple comparisons test). Apoptosis was highly significantly different in K5 versus Ctrl in both conditions (not shown). E CGNs were transduced with lentiviral particles expressing a control shRNA, or an shRNA specifically targeting Trim39, or one of two different shRNAs against NFATc3, or a combination of shRNAs, one day after plating. At DIV 6 transduced neurons were incubated for 8 h in K5 medium, and total cell extracts were analyzed by western blot using antibodies against the cleaved (active) form of caspase 3 and actin. F The intensity of the bands of different experiments performed as in E was quantified. For each experiment, the amount of active caspase 3 was normalized by the level of actin, only in K5 conditions, and expressed as the percentage of shRNA control. Data are the mean ± SEM of four independent experiments. *P < 0.05, **P < 0.01 significantly different (one-way ANOVA followed by Tukey’s multiple comparisons test). G CGNs were left untreated (ctrl), or incubated in K5 medium for 6 h and total cell extracts were analyzed by western blot using antibodies against NFATc3 and actin. The intensity of the NFATc3 bands was normalized by the intensity of the actin bands. Relative values are indicated in red. In parallel, total RNA was extracted and the mRNA level of NFATc3 was estimated by quantitative PCR. Data are the mean ± SEM of triplicate determinations. H CGNs were left untreated (control), switched to K5 medium for 8 h or treated with 0.3 μM ML-792 for 8 h. Then neurons were fixed and subjected to in situ PLA using rabbit anti-NFATc3 and mouse anti-SUMO-2 antibodies. Each bright red spot indicates that NFATc3 is SUMOylated. Negative control was obtained by omitting anti-NFATc3 antibody. Images were analyzed by confocal microscopy. To better visualize the differences in PLA intensity, maximum intensity projection was applied to the z-stacks of images. Nuclear staining was performed using DAPI and visualized in merged pictures with PLA signal (merge). The number of dots per cell was determined in one slice of each image using Fiji. The graph shows the mean ± SEM of 5 images per condition, including a total of more than 100 cells, from one experiment representative of three independent experiments. ****P < 0.0001 significantly different from control (one way ANOVA followed by Dunnet’s multiple comparison test).
Since Trim17 expression is highly induced in early apoptotic CGN [31] and Trim17 inhibits Trim39 (Fig. 4), Trim39-mediated degradation of NFATc3 is expected to be decreased following apoptosis induction. Indeed, in serum and KCl-deprived neurons, the protein level of NFATc3 was slightly increased although its mRNA level was unchanged (Fig. 8G). To further study the underlying mechanisms, we estimated the level of NFATc3 SUMOylation by PLA, using antibodies against NFATc3 and SUMO-2. This type of assay has been set up to measure the SUMOylation level of a given protein [39] and indeed, the SUMOylation inhibitor ML-792 led to a strong decrease of the PLA signal (Fig. 8H). Interestingly, the SUMOylation of endogenous NFATc3 was strongly decreased following serum and KCl withdrawal (Fig. 8H), which should also participate in the stabilization of NFATc3.
Discussion
The regulation of NFAT protein stability by the ubiquitin-proteasome system has been poorly studied. Independent studies have suggested that certain E3 ubiquitin-ligases may be responsible for ubiquitination and proteasomal degradation of different NFAT members: HDM2 for NFATc2 in breast and pancreatic cancers [6, 7]; Cbl-b, c-Cbl, VHL or KBTBD11/Cullin3 for NFATc1 during osteoclastogenesis [8,9,10, 40]; CHIP for NFATc3 in LPS-induced cardiomyopathies [11]. However, these proteins have not been shown to be genuine NFAT E3 ubiquitin-ligases, with the exception of HDM2 for NFATc2 [6]. In the present study, we show several lines of evidence demonstrating that Trim39 is an E3 ubiquitin-ligase for NFATc3 which targets it for proteasomal degradation. First, we found a physical interaction between Trim39 and NFATc3 proteins. Second, Trim39 ubiquitinated NFATc3 in vitro, predominantly forming K48-ubiquitin chains. Third, overexpression of WT Trim39, but not its inactive mutant, increased the ubiquitination of NFATc3 and decreased its protein level in cells, whereas silencing of Trim39 had opposite effects. As a physiological consequence, overexpression of Trim39 decreased the transcription factor activity of NFATc3 without affecting its nuclear translocation. Conversely, silencing of endogenous Trim39 increased both the transcriptional activity of NFATc3 and its pro-apoptotic effect in neurons. Taken together, our data strongly suggest that Trim39 modulates neuronal apoptosis by acting as a physiological E3 ubiquitin-ligase for NFATc3.
We also show that Trim17 inhibits Trim39-mediated ubiquitination of NFATc3 in vitro and in cells. Trim39 and Trim17 belong to the family of TRIM proteins which forms one of the largest classes of RING-containing E3 ubiquitin-ligases [41]. TRIM proteins are characterized by their N-terminal tripartite motif that consists of a RING domain, one or two B-box domains and a coiled-coil domain [42, 43]. While the RING domain confers an E3 ubiquitin-ligase activity by binding ubiquitin-loaded E2 ubiquitin-conjugating enzymes, the B-box and coiled-coil domain are generally involved in the formation of homo- or hetero-dimers or multimers [44,45,46,47]. As homo-multimerization seems to be necessary for the E3 ubiquitin-ligase activity of TRIM proteins [47,48,49,50], Trim17 may inhibit Trim39 by forming inactive hetero-multimers with Trim39 at the expense of Trim39 homo-multimers. Similarly, we have previously shown that TRIM17 inhibits two other TRIM proteins to which it binds: TRIM41 [26] and TRIM28 [27]. The inhibitory effect of Trim17 might result from two mechanisms that are not mutually exclusive. First, the binding of Trim17 may inhibit the E3 ubiquitin-ligase activity of its TRIM partner, possibly by preventing the binding of the E2 enzyme. Indeed, TRIM17 prevented the auto-ubiquitination of TRIM39 in vitro, as previously shown for TRIM41 inhibition by TRIM17 [26]. Second, Trim17 may prevent the binding of the substrate to its TRIM partner. Indeed, Trim17 reduced the interaction between Trim39 and NFATc3. This is reminiscent of the effect of TRIM17 on the interaction between TRIM41 or TRIM28 and their respective substrates [51]. Our current data indicate that the RING domain of TRIM17 is required to inhibit TRIM39, in line with our previous data showing that Trim17, but not its RING-deletion mutant, inhibits basal ubiquitination of NFATc3 [24]. This does not necessarily imply that the E3 ubiquitin-ligase activity of TRIM17 is involved in its inhibitory effect. In particular, it is clear that Trim17 does not inhibit Trim39 by inducing its ubiquitination and subsequent degradation because Trim17 rather decreased Trim39 ubiquitination. Instead, its RING domain may be required for TRIM17 to bind TRIM39, and to prevent the interaction between TRIM39 and the E2 enzyme or NFATc3.
SUMOylation has recently emerged as an important regulator of protein stability [18]. Consistently, we show here that SUMOylation of NFATc3 promotes its ubiquitination and degradation. Indeed, the E/A mutation of the SUMOylation consensus sites of NFATc3 decreased its ubiquitination level, increased its half-life and aggravated its pro-apoptotic effect. This mechanism could be conserved in other members of the NFAT family. Indeed, the double K/R mutation of two SUMOylation sites in NFATc1 and NFATc2 has been shown to reduce their ubiquitination and degradation [7, 14]. Although it cannot be excluded that these Lys residues are directly ubiquitinated, it is also possible that SUMOylation of NFATc1 and NFATc2 on these consensus sites is necessary for their ubiquitination.
A few SUMO-targeted E3 ubiquitin-ligases (STUbLs) have been described [19,20,21]. These proteins generally combine a RING domain that confers them an E3 ubiquitin-ligase activity and SIMs that mediate their preference for SUMOylated substrates. Proteasome inhibition leads to an important accumulation of SUMO-modified proteins in yeast and human cells [52, 53], suggesting that ubiquitination and degradation of SUMOylated proteins mediated by STUbLs play an important role in proteostasis. However, only two STUbLs have been identified so far in mammals: RNF4 and Arkadia/RNF111 [54], which may not be sufficient to account for the regulation of all SUMOylated proteins. Therefore, additional STUbLs probably remain to be discovered. Here we provide evidence indicating that Trim39 acts as a STUbL for NFATc3. First, Trim39 is able to bind and ubiquitinate SUMO-2 chains in vitro. Second, mutation of a putative SIM motif in Trim39 (SIM3) strongly decreases its capacity to bind and ubiquitinate both SUMO-2 chains in vitro, and NFATc3 in cells. Third, in vitro ubiquitination of NFATc3 by Trim39 is increased by prior in vitro SUMOylation of NFATc3. Finally, mutation of SUMOylation consensus sites in NFATc3 and treatment of cells with the SUMOylation inhibitor ML-792, reduce Trim39-mediated ubiquitination of NFATc3. Most of the STUbLs studied so far bear multiple SIMs, which mediate cooperative binding to multiple SUMO units [21]. Of the three SIMs predicted in Trim39 with a high score, only one is instrumental for the binding of Trim39 to SUMO chains. SIMs are characterized by a loose consensus sequence and some non-canonical SIMs have been described [21, 35]. Therefore, it is possible that another SIM in Trim39, that was not identified by GPS-SUMO or JASSA, may participate in the binding of Trim39 to SUMO. It is also possible that a single SIM is sufficient to fulfill this function, as reported for other STUbLs [55, 56]. Moreover, it is possible that in its multimerized form, Trim39 harbors several SIMs in close proximity. Indeed, SIM3 is located in the coiled-coil domain of Trim39 that is expected to form antiparallel dimers or multimers [44,45,46,47]. Alternatively, another binding site, such as the RING domain, may cooperate with the SUMO-SIM interaction to bind the substrate, as reported for the Drosophila STUbL Degringolade [57]. Trim39 may also bind NFATc3 by a dual mechanism, involving both SUMO-dependent and SUMO-independent recognition, as shown for viral STUbLs [58, 59].
We show here that silencing of Trim39 enhances apoptosis triggered by serum and KCl deprivation in primary CGNs. Since Trim39 induces NFATc3 degradation and NFATc3 aggravates neuronal apoptosis, it can be assumed that Trim39 silencing favors neuronal apoptosis by stabilizing NFATc3. Additional effects of Trim39 on apoptosis regulation cannot be excluded. For example, silencing of Trim39 has been reported to aggravate apoptosis following a genotoxic stress in HCT116 cells [60] and to increase nutlin3a-induced apoptosis, presumably by stabilizing p53 [61]. Conversely, in p53 null cell lines, silencing of Trim39 reduces DNA damage-induced apoptosis [61, 62], probably because Trim39 inhibits APC/CCdh1-mediated degradation of the pro-apoptotic protein MOAP-1 [62, 63]. However, in the present study, the increase in apoptosis triggered by Trim39 knock-down is completely abrogated by NFATc3 silencing, strongly suggesting that Trim39 regulates neuronal apoptosis by controlling the stability of NFATc3. We and others have reported that Trim17 also plays an important role in apoptosis regulation [24, 27, 31, 64, 65]. Notably, we have shown that Trim17 is highly induced in CGNs following serum and KCl deprivation, and that Trim17 is both sufficient and necessary to trigger neuronal apoptosis [31]. This pro-apoptotic effect is in part mediated by the ubiquitination and degradation of the anti-apoptotic protein Mcl-1 [64]. In addition, Trim17 can modulate neuronal apoptosis by acting on NFATc3 through antagonistic mechanisms. On the one hand, we have previously shown that Trim17 prevents the nuclear translocation and activation of NFATc3 [24] and should therefore inhibit its pro-apoptotic effect. On the other hand, we show here that Trim17 can inhibit Trim39-mediated ubiquitination and degradation of NFATc3 and should therefore aggravate its pro-apoptotic effect. Moreover, we have identified Trim17 as a target gene of NFATc3 [24]. The effects of Trim17 on NFATc3 should therefore influence its own expression, creating both negative and positive feedback loops and eventually resulting in fine tuning of neuronal apoptosis. Moreover, we show here that NFATc3 SUMOylation is reduced after serum and KCl deprivation in CGNs. This, combined with the induction of Trim17, should therefore contribute to the initiation of neuronal apoptosis by inhibiting Trim39-mediated degradation of NFATc3.
Methods and materials
Materials
Culture media were from Thermo Fisher Scientific. Foetal calf serum, other culture reagents, protease inhibitor cocktail, DAPI, PMA, A23187, cycloheximide, N-ethylmaleimide (NEM), MG-132, puromycin, anti-Flag M2 affinity gel beads (#A2220), ML-792 (#HY-108702) and other chemicals were from Sigma-Aldrich. Protein G-agarose and protein A-agarose beads were from Roche. GFP-Trap®-A beads were from Chromotek (Planneg-Martinsried, Germany). Rat monoclonal anti-HA antibody (clone 3F10; #11867432001), mouse monoclonal anti-Flag antibody (clone M2, #F3165), mouse monoclonal anti-tubulin antibody (clone DM1A, #T6199), rabbit antibody against human TRIM17 (#AV34547) and rabbit IgG (#I5006) were from Sigma-Aldrich. Rabbit anti-GFP antibody (#TP401) was from Torrey Pines Biolabs Inc. (Houston, TX USA). Mouse monoclonal antibody against actin (clone C4) was from Millipore (#MAB1501). Rabbit polyclonal antibody against NFATc3 was from Proteintech (18222-1-AP). Mouse monoclonal antibody against Trim39 was from Origene (#TA505761). Rabbit polyclonal antibody against Trim17 was generated by Eurogentec, as described previously [31]. Mouse monoclonal antibody against SUMO-2 (clone #8A2) was purified from hybridomas obtained from the Developmental Studies Hybridoma Bank. Fluorescent and horseradish peroxidase-conjugated goat anti-rabbit, anti-rat and anti-mouse secondary antibodies were from Thermo Fisher Scientific and Jackson ImmunoResearch Laboratories Inc. (West Grove, PA, USA), respectively.
Cell culture and transient transfection
Lenti-X 293T (Clontech), Neuro2A (mouse neuroblastoma) and Baby Hamster Kidney (BHK) cell lines were grown in Dulbecco’s modified Eagle’s medium containing 4.5 g/l glucose supplemented with 10% foetal bovine serum and penicillin-streptomycin 100 IU/ml-100 μg/ml. Cells were regularly tested for mycoplasma contamination. Cells were transfected with plasmids using GenJet™ in vitro transfection reagent (Ver. II) pre-optimized and conditioned for transfecting Neuro2A and BHK-21 cells respectively (SignaGen laboratories, Ijamsville, MD) according to the manufacturer’s instructions. Neuro2A cells were transfected with siRNAs using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) following the manufacturer’s instruction. For one 35 mm dish, 2.5 μl of transfection reagent was used with 25 pmoles of siRNA. The sequences of the siRNAs used were as follows: siTrim39#1 5′ CCAAGCGGGTAGGCATATT 3′; siTrim39#2 5′ GCGTCAAGTTTGTGGAGACAA3′; siRNA ctrl (targeting Luciferase gene): 5′ CGTACGCGGAATACTTCGA 3′.
Primary cultures of CGNs were prepared from 7-day-old murine pups (C57Bl/6J mice) as described previously and cultured in Basal Medium Eagle (BME) medium supplemented with 10% fetal bovine serum, 2 mM L-Gln, 10 mM HEPES, penicillin-streptomycin 100 IU/ml–100 μg/ml and 20 mM KCl to reach a final concentration of 25 mM [31]. Primary CGNs, grown on glass coverslips in 24-well plates, were transfected at DIV 5 with 2 μg of plasmids using a calcium phosphate protocol optimized for neuronal cultures as previously described [31].
Silencing of Trim39 using shRNA-expressing lentiviruses
The HIV-derived lentiviral vectors pLKO.1 containing control shRNAs respectively against eGFP and Luciferase (SHC005, SHC007), shRNAs against Trim39: TRCN0000037281 (shRNA Trim39#1), TRCN0000037282 (shRNA Trim39#2) and TRCN0000438509 (shRNA Trim39#3), and shRNAs against NFATc3: TRCN0000377122 (shRNA NFATC3#1) and TRCN0000097129 (shRNA NFATC3#2) were from Sigma-Aldrich. Lentiviral particles were produced as previously described [26]. Neuro2A cells and CGNs were transduced one day after plating. The lentiviral preparations were added directly to the culture medium for 8 h. Cells were then replaced in fresh medium. Culture was continued until 6 days in vitro for CGNs. Neuro2A cells were kept in culture for 24 h after transduction and then selected using 2 μg/ml puromycin for an additional 48 h.
Expression vectors
The following plasmids were described previously: pCI-GFP, pCS2-3×HA-NFATc3, pCS2-3×HA-NFATc3-KallR, pCS2-GFP-NFATc3 and pCI-Trim17-GFP [24]. Site directed mutagenesis and new expression constructs used in the present study are described in supplementary information.
Protein extraction and western blot analysis
Cells were harvested in lysis buffer A (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM NaF, 5 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 5 mM EDTA, 10 mM NEM, 20 μM MG-132, and protease inhibitor cocktail) supplemented with 1% NP-40 and homogenized by thorough vortexing. Cell debris were removed by centrifugation at 1000 × g for 5 min at 4 °C and the protein concentration of the resulting supernatant was estimated using the BCA protein assay kit (Thermo Fisher Scientific) with bovine serum albumin as the standard. Total lysates were diluted in 3× Laemmli sample buffer and incubated at 95 °C for 5 min. Proteins were separated by 8–12% SDS–PAGE and transferred to Immobilon-P PVDF membrane (Millipore). Blocking and probing with antibodies were performed as previously described [26]. Visualization of immunoreactive proteins was performed using horseradish peroxidase-linked secondary antibodies and Covalight enhanced chemiluminescent substrate (Covalab, Bron, France) or Immobilon® Western (Millipore). Membranes were revealed using films or Amersham Imager 680 (GE Healthcare). When necessary, membranes were stripped using Restore™ PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific) and re-probed with additional antibodies.
Co-immunoprecipitation
Following transfection with the indicated plasmids for 24 h, Neuro2A or BHK cells were incubated for 5 h with 10–20 μM MG-132. They were then homogenized in lysis buffer A, supplemented with 1% NP-40 for immunoprecipitation with anti-HA, 0.5% NP-40 for immunoprecipitation with GFP-Trap-A and 1% Triton X-100 for immunoprecipitation with anti-Flag. For anti-HA and anti-Flag immunoprecipitation, cell lysates (500 μl) were centrifuged at 300 × g for 5 min at 4 °C. The resulting supernatants were pre-cleared by rotation for 1–3 h at 4 °C with 15 μl protein G-agarose beads and then rotated overnight at 4 °C with 25 μl protein G agarose beads together with 7 μl anti-HA antibody or with 30 μl anti-Flag M2 affinity gel beads. The beads were recovered by centrifugation and washed four times with 1 ml of lysis buffer A without detergent and containing 0.3 M NaCl for anti-HA or 0.5 M NaCl for anti-Flag (instead of 150 mM NaCl). For GFP-Trap precipitation, cell lysates (200 μl) were diluted with 300 μl dilution buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM NaF, 5 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 0.5 mM EDTA, 20 μM MG-132, 10 mM NEM and protease inhibitor cocktail) and cell debris were removed by centrifugation. Resulting supernatants were rotated for 2 h at 4 °C with 10–25 μl GFP-Trap®-A beads to immunoprecipitate proteins fused to GFP. Beads were recovered by centrifugation and washed four times with dilution buffer. Material bound to the protein G agarose, anti-Flag M2 affinity gel or GFP-Trap beads was eluted by the addition of 3× Laemmli sample buffer and incubation at 95 °C for 5 min. Precipitated proteins were analyzed by western blot.
In situ proximity ligation assay
Neuro2A cells seeded onto glass coverslips were left untreated or transfected with indicated plasmids for 24 h, and then treated with 10 μM MG-132 for 4 h. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, washed with 0.1 M Gly (pH = 7.11), permeabilized with 0.2% Triton X-100 in PBS for 10 min and washed with PBS. The close proximity between proteins was detected using the Duolink® In Situ kit (Olink® Bioscience, Uppsala, Sweden), according to the manufacturer’s instructions, as described previously [24]. Cells were analyzed using a confocal Leica SP5-SMD microscope, with a LEICA 63x/1.4 OIL HCX PL APO CS objectives. Images were acquired by the Confocal head TCS SP5 II using the Leica Application Suite X software. Images were processed using Image J. When indicated, z-stacks of images were submitted to maximum intensity projection. The number of dots per transfected cell was estimated in one slice, in around 100 cells in each condition, with an automated procedure using plugins from the Image J software.
Immunofluorescence
BHK and Neuro2A cells were seeded onto glass coverslips. Primary CGNs were cultured on coverslips previously coated with laminin (16.67 μg/ml) and poly-D-lysine (33.3 μg/ml). Cells and neurons were treated as described in the figure legends and then fixed with 4% paraformaldehyde. Overexpressed HA-NFATc3 and endogenous Trim39 were detected using anti-HA (1:500) and anti-Trim39 (1:400) antibodies respectively, as described previously [26]. GFP and mCherry-fused proteins were visualized by fluorescence and nuclei were stained with DAPI. Coverslips were analyzed by conventional or confocal microscopy, as mentioned in the figure legends. Image acquisition and analysis were performed on work stations of the Montpellier imaging facility (Leica DM600 for conventional microscopy, Leica SP5-SMD for confocal microscopy). For quantification of NFATc3 nuclear localization, BHK cells with predominant cytoplasmic or nuclear localization of NFATc3 were counted, in a blinded manner, among double GFP/HA positive cells. At least 100 double positive cells were scored for each experiment and each condition.
Ubiquitination of NFATc3 in cells
Neuro2A or BHK cells cultured in 60 mm or 100 mm dishes were transfected with pCI-HA-NFATc3 or NFATc3 E/A mutants together with a plasmid expressing eight His6-tagged ubiquitin (His-Ub), or empty pCI, in the presence or the absence of pCI-Flag-Trim39, pCI-Flag-Trim39-ΔRING, pCI-Flag-Trim39-mSIM3, pCI-Trim17-GFP or a combination of these plasmids. Following 24 h transfection, the medium was supplemented with 20 μM MG-132, in the presence or the absence of 0.3 μM ML-792, for 6–8 h. Then, cells were harvested in 1 ml PBS without Ca2+ and Mg2+ supplemented with 10 μM MG-132. A 100 μl sample of the homogenous cell suspension was taken for input analysis and transfection efficiency control. After centrifugation, the pellet from the remaining 900 μl cell suspension was homogenized in 1 ml lysis buffer B (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl [pH = 8.0]) supplemented with 5 mM imidazole, 510 mM β-mercaptoethanol, 0.5 M NaCl and 10 mM NEM. The lysate was sonicated, cleared by centrifugation at 1500 × g for 5 min at room temperature and added to 40 μl magnetic nickel beads (MagneHis™ Ni-Particles, Promega). Beads were rotated for 2 h at room temperature to purify ubiquitinated proteins and washed once with lysis buffer B supplemented with 20 mM imidazole, 0.5 M NaCl and 10 mM NEM; once with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH = 8.0), 20 mM imidazole,0.5 M NaCl and 10 mM NEM; three times with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH = 6.3), 20 mM imidazole,0.5 M NaCl, 10 mM NEM, 0.2% Triton X-100; once with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH = 6.3), 20 mM imidazole,0.5 M NaCl, 10 mM NEM, 0.1% Triton X-100 and once with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH = 6.3), 10 mM imidazole, 10 mM NEM. Materials bound to the beads were eluted by the addition of 3× Laemmli sample buffer and boiling for 5 min. These purification products and initial total lysates were resolved by SDS-PAGE and analyzed by immunoblotting.
Production of recombinant TRIM proteins
BL21-CodonPlus®(DE3)-RP competent cells (Agilent) were transformed with the expression vectors pGEX-4T1 expressing GST-Trim39 fusion proteins. Protein expression was induced by the addition of 500 μM IPTG and was carried out at 20 °C overnight in the presence of 100 μM ZnCl2 and 200 μM MgSO4. Bacteria were harvested by centrifugation and resuspended in BugBuster® Protein Extraction Reagent (Millipore #70584-4) supplemented with protease inhibitor cocktail (cOmplete EDTA-free, Sigma-Aldrich). Bacterial suspensions were incubated for 30 min at room temperature with 1 mg/ml lysozyme (Fluka) and further lysed by sonication. The soluble protein fraction was recovered by centrifugation. GST fusion proteins were isolated by binding to glutathione magnetic beads (MagneGST™ Glutathione Particles, Promega) for 30 min at room temperature on a rotating wheel. The beads were then washed three times with PBS.
ArcticExpress (DE3) competent cells (Agilent) were transformed with the expression vector pET-15-HIS-TRIM39 and pMAL-c2-TRIM17 (expressing MBP-TRIM17 fusion protein). Bacteria were grown overnight in LB medium supplemented with Ampicillin and Gentamycin (20 µg/ml). Recombinant protein expression was induced by the addition of 1 mM IPTG and was carried out at 12 °C overnight in the presence of 100 μM ZnCl2 and 200 μM MgSO4. Bacteria were harvested by centrifugation. Pellets were resuspended in bacterial lysis buffer (50 mM Tris-HCl [pH = 8.6], 0.5 M NaCl, 50 mM MgSO4) supplemented with lysozyme and protease inhibitors, and frozen in liquid nitrogen to lyse bacteria. Lysates were then cleared by centrifugation. MBP-TRIM17 proteins were purified on amylose resin (New England BioLabs, #E8021L) and then eluted in a buffer containing 20 mM maltose before dialysis in PBS. His-TRIM39 proteins were purified on Ni-NTA agarose beads (Qiagen, #1018244) and then eluted in a buffer containing 0.5 M imidazole before dialysis in PBS.
In vitro ubiquitination assay
NFATc3 was first transcribed and translated in vitro. For this, 1 μg of pCS2-HA-NFATc3 was incubated for 2 h at 30 °C in 50 μl of the TNT® SP6 coupled wheat germ extract system (Promega, #L5030), according to the instructions of the manufacturer. For each ubiquitination condition, the equivalent of 3 μl of the in vitro translation reaction was immunopurified using 1 μl rat anti-HA antibody and 10 μl protein G-agarose beads in a buffer containing 50 mM Tris-HCl (pH = 7.5) and 50 mM NaCl for 1 h. Beads were washed 3 times in the same buffer, as described above for co-immunoprecipitation. Then, beads carrying NFATc3 were incubated in 20 μl of ubiquitination reaction buffer (50 mM Tris-HCl [pH = 7.5], 50 mM NaCl, 4 mM ATP, 4 mM MgCl2, 2 mM DTT, 10 mM phosphocreatine, 0.5 U creatine kinase, 20 μM ZnCl2), in the presence of 50 ng human recombinant His-tagged ubiquitin-activating enzyme E1 (from BostonBiochem, #E-304), 500 ng human recombinant His-tagged ubiquitin-conjugating enzyme (E2) Ube2d3 (from BIOMOL International, #U0880), in the presence or the absence of 10 μg N-terminal-His-tagged ubiquitin (Sigma-Aldrich, #U5507), or N-terminal-His-tagged ubiquitin mutants (R&D Systems, UM-K630, UM-K480, UM-K270, UM-K290, UM-K330, UM-K60), and ≈2 μg of purified recombinant mouse GST-Trim39 or His-Trim39 (WT) or GST-Trim39-C49S/C52S or mSIM3 His-Trim39, or ≈2 μg of purified recombinant human His-TRIM39 in the presence or the absence of ≈2 μg MBP-TRIM17 or MBP-TRIM17-ΔRING. Reactions were incubated at 37 °C for 1 h. Beads were washed once and the reaction was stopped by adding 10 μl of 3× Laemmli sample buffer and by heating at 95 °C for 5 min. The samples were analyzed western bot.
In vitro SUMOylation assay
WT NFATc3 and its KallR and EallA mutants were first transcribed and translated in vitro as described above. Equivalent amounts of the different forms of NFATc3 (between 1,5 and 6 μl of the in vitro translation reaction) were incubated for 1 h to 1 h 30 min at 37 °C in the presence of 3 μg recombinant SUMO-1, 150 ng recombinant His-tagged Aos1/Uba2 (E1 enzyme), 100 ng recombinant Ubc9 (E2 enzyme) and 300 ng recombinant GST-PIASxα (E3 enzyme) in 20 μl shift-assay buffer (20 mM Hepes [pH = 7.3], 110 mM KOAc, 2 mM Mg(OAc)2, 0.5 mM EGTA, 1 mM DTT, 0.05% Tween 20, 0.2 mg/ml ovalbumin, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin) supplemented with 1 mM ATP. Negative controls (time 0) were obtained by mixing all reagents directly into loading buffer. When SUMOylation was followed by in vitro ubiquitination, the SUMO-specific protease GST-SENP1 was added at the end of the reaction, for 15 min at 37 °C, to remove SUMO chains from NFATc3. Recombinant proteins used in this assay were produced and purified as previously described [66]. Reaction products were analyzed by western blot.
RNA preparation and quantitative RT-PCR
Total RNA was extracted using the RNeasy kit (Qiagen) and treated with the DNase I from the DNA-free™ kit (Thermo Fisher Scientific) according to manufacturer’s instructions. RNA was used to perform a two-step reverse-transcription quantitative polymerase chain reaction (RT-qPCR) as described previously [26]. Specific primers used to amplify mouse Trim39 and mouse Trim17 are listed in Supplementary Table 1. Data were analyzed and relative amounts of specifically amplified cDNA were calculated with MxPro software (Agilent), using the mouse Gapdh amplicon as a reference.
Protein sequence analysis
The sequence of mouse Trim39 (GenBank: NM_024468) was analyzed by using the prediction web-based tools JASSA (Joined Advanced SUMOylation site and SIM analyzer, (http://www.jassa.fr/) and GPS SUMO (group-based prediction system, http://sumosp.biocuckoo.org/online.php) to identify putative SIMs.
Production of SUMO chains and GST-pull down
Recombinant free SUMO-2 and poly-SUMO-2 chains were produced in bacteria co-expressing His-SUMO-2, Aos1/Uba2 (SUMO conjugating E1 enzyme) and Ubc9 (SUMO E2 conjugating enzyme). For this, BL21 competent cells were transformed with the plasmid pE1-E2-His-Su2 (described in [67]). The transformed bacteria were grown with strong agitation (210 rpm) at 37 °C until the OD reaches 0.4–0.6. Protein expression was induced by adding 1 mM IPTG for 6 h at 25 °C. The bacteria were resuspended in 30 ml of bacterial lysis buffer, frozen in liquid nitrogen and stored at −80 °C. The resuspended bacteria were thawed and supplemented with lyzozyme (1 mg/ml), 8 mM ß-mercaptoethanol, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin and incubated for 1 h on ice before centrifugation at 100,000 × g for 1 h at 4 °C. The supernatant was loaded on a Ni-NTA column equilibrated in bacterial lysis buffer supplemented with 8 mM ß-mercaptoethanol, 0.5% Triton X-100, 10 mM imidazole and protease inhibitors. The column was washed 3 times with 10 ml of the same buffer and eluted with 15 ml of Ni-NTA elution buffer (bacterial lysis buffer supplemented with 250 mM imidazole and protease inhibitors).
For GST-pull down, GST-Trim39 and its different SIM mutants were produced in bacteria and purified as described above. The quantity and the quality of the different forms of GST-Trim39 bound to glutathione magnetic beads was first estimated on a poly-acrylamide gel using Coomassie blue staining, by reference to known amounts of BSA. Beads binding approximately 1 μg of each form of GST-Trim39 were incubated with ≈1 μg SUMO-2 chains for 1 h at room temperature in 200 μl shift assay buffer. Beads were recovered by centrifugation and washed four times with PBS. Material bound to the beads was eluted by the addition of 3× Laemmli sample buffer and incubation at 95 °C for 5 min. Both GST-Trim39 proteins and bound SUMO-2 chains were analyzed by western blot.
Assessment of neuronal apoptosis
After 6 days in vitro, transfected or transduced CGNs were maintained in initial culture medium (control) or were washed once and incubated in serum-free BME supplemented with L-Gln, HEPES, antibiotics and 1 μM (+)-MK-801, and containing 5 mM KCl (K5 medium) for the indicated times. Neurons were then stained with DAPI and mounted on glass slides in Mowiol. In experiments in which the CGNs were transfected with GFP, GFP-NFATc3 or GFP-NFATc3-EallA, apoptosis was assessed among GFP-positive neurons, by examining neuronal morphology and nuclear condensation. For each experiment and each condition, at least 100 GFP-positive neurons were scored in a blinded manner (the experimenter did not know which conditions he was counting). In experiments in which CGNs were transduced with shRNA-expressing lentiviruses, apoptosis was estimated by counting the percentage of condensed nuclei in five random fields for each condition (more than 500 cells) in a blinded manner.
Statistics
Data plotting and statistical analyses were performed using GraphPad Prism version 7.0c for Mac OS X (GraphPad Software, San Diego California USA, www.graphpad.com). Statistical analyses are described in the legends of the figures. Normal distribution with similar variance between groups was assumed. None of the samples were excluded except when a technical problem occurred. Unless stated, data are representative of at least three independent experiments.
Data availability
The data used during the current study are available from the corresponding author on reasonable request.
References
Fric J, Zelante T, Wong AYW, Mertes A, Yu H-B, Ricciardi-Castagnoli P. NFAT control of innate immunity. Blood. 2012;120:1380–9.
Kipanyula MJ, Kimaro WH, Seke Etet PF. The emerging roles of the calcineurin-nuclear factor of activated T-lymphocytes pathway in nervous system functions and diseases. J Aging Res. 2016;2016:5081021.
Mognol GP, Carneiro FRG, Robbs BK, Faget DV, Viola JPB. Cell cycle and apoptosis regulation by NFAT transcription factors: new roles for an old player. Cell Death Dis. 2016;7:e2199.
Wu H, Peisley A, Graef IA, Crabtree GR. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007;17:251–60.
Müller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–56.
Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–50.
Singh SK, Baumgart S, Singh G, Konig AO, Reutlinger K, Hofbauer LC, et al. Disruption of a nuclear NFATc2 protein stabilization loop confers breast and pancreatic cancer growth suppression by zoledronic acid. J Biol Chem. 2011;286:28761–71.
Youn M-Y, Yokoyama A, Fujiyama-Nakamura S, Ohtake F, Minehata K, Yasuda H, et al. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J Biol Chem. 2012;287:12994–3004.
Li X, Wei W, Huynh H, Zuo H, Wang X, Wan Y. Nur77 prevents excessive osteoclastogenesis by inducing ubiquitin ligase Cbl-b to mediate NFATc1 self-limitation. Elife. 2015;4:e07217.
Narahara S, Sakai E, Kadowaki T, Yamaguchi Y, Narahara H, Okamoto K, et al. KBTBD11, a novel BTB-Kelch protein, is a negative regulator of osteoclastogenesis through controlling Cullin3-mediated ubiquitination of NFATc1. Sci Rep. 2019;9:3523.
Chao C-N, Lai C-H, Badrealam KF, Lo J-F, Shen C-Y, Chen C-H, et al. CHIP attenuates lipopolysaccharide-induced cardiac hypertrophy and apoptosis by promoting NFATc3 proteasomal degradation. J Cell Physiol. 2019;234:20128–38.
Terui Y, Saad N, Jia S, McKeon F, Yuan J. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J Biol Chem. 2004;279:28257–65.
Nayak A, Glockner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T, et al. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J Biol Chem. 2009;284:10935–46.
Vihma H, Timmusk T. Sumoylation regulates the transcriptional activity of different human NFAT isoforms in neurons. Neurosci Lett. 2017;653:302–7.
Kim ET, Kwon KM, Lee MK, Park J, Ahn J-H. Sumoylation of a small isoform of NFATc1 is promoted by PIAS proteins and inhibits transactivation activity. Biochem Biophys Res Commun. 2019;513:172–8.
Zhao X. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell. 2018;71:409–18.
Henley JM, Carmichael RE, Wilkinson KA. Extranuclear SUMOylation in neurons. Trends Neurosci. 2018;41:198–210.
Liebelt F, Vertegaal ACO. Ubiquitin-dependent and independent roles of SUMO in proteostasis. Am J Physiol Cell Physiol. 2016;311:C284–296.
Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–8.
Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJP, Tainer JA, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101.
Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. 2014;1843:75–85.
Ulrich JD, Kim MS, Houlihan PR, Shutov LP, Mohapatra DP, Strack S, et al. Distinct activation properties of the nuclear factor of activated T-cells (NFAT) isoforms NFATc3 and NFATc4 in neurons. J Biol Chem. 2012;287:37594–609.
Luo J, Sun L, Lin X, Liu G, Yu J, Parisiadou L, et al. A calcineurin- and NFAT-dependent pathway is involved in α-synuclein-induced degeneration of midbrain dopaminergic neurons. Hum Mol Genet. 2014;23:6567–74.
Mojsa B, Mora S, Bossowski JP, Lassot I, Desagher S. Control of neuronal apoptosis by reciprocal regulation of NFATc3 and Trim17. Cell Death Differ. 2015;22:274–86.
Caraveo G, Auluck PK, Whitesell L, Chung CY, Baru V, Mosharov EV, et al. Calcineurin determines toxic versus beneficial responses to α-synuclein. PNAS. 2014;111:E3544–52.
Lassot I, Mora S, Lesage S, Zieba BA, Coque E, Condroyer C, et al. The E3 ubiquitin ligases TRIM17 and TRIM41 modulate α-synuclein expression by regulating ZSCAN21. Cell Rep. 2018;25:2484–96. e9
Lionnard L, Duc P, Brennan MS, Kueh AJ, Pal M, Guardia F, et al. TRIM17 and TRIM28 antagonistically regulate the ubiquitination and anti-apoptotic activity of BCL2A1. Cell Death Differ. 2019;26:902–17.
Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–26.
Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8.
Woodsmith J, Jenn RC, Sanderson CM. Systematic analysis of dimeric E3-RING interactions reveals increased combinatorial complexity in human ubiquitination networks. Mol Cell Proteom. 2012;11:M111.016162.
Lassot I, Robbins I, Kristiansen M, Rahmeh R, Jaudon F, Magiera MM, et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 2010;17:1928–41.
Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33.
Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–9.
He X, Riceberg J, Soucy T, Koenig E, Minissale J, Gallery M, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol. 2017;13:1164–71.
Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5.
Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–330.
Beauclair G, Bridier-Nahmias A, Zagury J-F, Saïb A, Zamborlini A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics. 2015;31:3483–91.
Contestabile A. Cerebellar granule cells as a model to study mechanisms of neuronal apoptosis or survival in vivo and in vitro. Cerebellum. 2002;1:41–55.
Ristic M, Brockly F, Piechaczyk M, Bossis G. Detection of protein-protein interactions and posttranslational modifications using the proximity ligation assay: application to the study of the SUMO pathway. Methods Mol Biol. 2016;1449:279–90.
Kim JH, Kim K, Jin HM, Song I, Youn BU, Lee SH, et al. Negative feedback control of osteoclast formation through ubiquitin-mediated down-regulation of NFATc1. J Biol Chem. 2010;285:5224–31.
Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–57.
Esposito D, Koliopoulos MG, Rittinger K. Structural determinants of TRIM protein function. Biochem Soc Trans. 2017;45:183–91.
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–51.
Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y, et al. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24:762–5.
Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71.
Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci USA. 2014;111:2494–9.
Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35:1204–18.
Fiorentini F, Esposito D, Rittinger K. Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochem Soc Trans. 2020;48:2615–24.
Streich FC, Ronchi VP, Connick JP, Haas AL. Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J Biol Chem. 2013;288:8209–21.
Yudina Z, Roa A, Johnson R, Biris N, de Souza Aranha Vieira DA, Tsiperson V, et al. RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 2015;12:788–97.
Basu-Shrivastava M, Kozoriz A, Desagher S, Lassot I. To ubiquitinate or not to ubiquitinate: TRIM17 in cell life and death. Cells. 2021;10:1235.
Bailey D, O’Hare P. Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem J. 2005;392:271–81.
Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–75.
Jansen NS, Vertegaal ACO. A chain of events: regulating target proteins by SUMO polymers. Trends Biochem Sci. 2021;46:113–23.
Parker JL, Ulrich HD. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 2012;40:11380–8.
Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, Levy L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol. 2013;33:2163–77.
Abed M, Barry KC, Kenyagin D, Koltun B, Phippen TM, Delrow JJ, et al. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011;30:1289–301.
Boutell C, Cuchet-Lourenço D, Vanni E, Orr A, Glass M, McFarlane S, et al. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011;7:e1002245.
Wang L, Oliver SL, Sommer M, Rajamani J, Reichelt M, Arvin AM. Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 2011;7:e1002157.
Zhang L, Mei Y, Fu NY, Guan L, Xie W, Liu HH, et al. TRIM39 regulates cell cycle progression and DNA damage responses via stabilizing p21. Proc Natl Acad Sci USA. 2012;109:20937–42.
Zhang L, Huang NJ, Chen C, Tang W, Kornbluth S. Ubiquitylation of p53 by the APC/C inhibitor Trim39. Proc Natl Acad Sci USA. 2012;109:20931–6.
Lee SS, Fu NY, Sukumaran SK, Wan KF, Wan Q, Yu VC. TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp Cell Res. 2009;315:1313–25.
Huang NJ, Zhang L, Tang W, Chen C, Yang CS, Kornbluth S. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J Cell Biol. 2012;197:361–7.
Magiera MM, Mora S, Mojsa B, Robbins I, Lassot I, Desagher S. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 2013;20:281–92.
Song K-H, Choi CH, Lee H-J, Oh SJ, Woo SR, Hong S-O, et al. HDAC1 upregulation by NANOG promotes multidrug resistance and a stem-like phenotype in immune edited tumor cells. Cancer Res. 2017;77:5039–53.
Bossis G, Chmielarska K, Gartner U, Pichler A, Stieger E, Melchior F. A fluorescence resonance energy transfer-based assay to study SUMO modification in solution. Methods Enzymol. 2005;398:20–32.
Brockly F, Piechaczyk M, Bossis G. Production and purification of recombinant SUMOylated proteins using engineered bacteria. Methods Mol Biol. 2016;1475:55–65.
Acknowledgements
This article is based upon work from COST Action (PROTEOSTASIS BM1307), supported by COST (European Cooperation in Science and Technology). We would like to thank the staff of the Montpellier Genomic Collection platform for providing human TRIM39 and human TRIM17 cDNA clones. We acknowledge the imaging facility MRI (Montpellier Ressources Imagerie), member of the national infrastructure France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, “Investments for the future”). We are grateful to Frédérique Brockly for the production and purification of recombinant proteins and Dr Olivier Coux for providing ubiquitin mutants. We thank Drs Dimitris Liakopoulos and Manuel Rodriguez for interesting discussions.
Funding
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Université de Montpellier, La Fondation de l’Association pour la Recherche contre le Cancer (ARC, grant number PJA 20141201882 to SD), La Ligue contre le Cancer (grant number TDUM13665 to BM) and La Fondation pour la Recherche Médicale (grant number FDT201904008340 to MBS).
Author information
Authors and Affiliations
Contributions
MBS, BM and IL designed and performed experiments, and analyzed the data; SM performed experiments; SD and BM conceived the study; SD performed the statistical analysis, wrote and revised the manuscript, and prepared the figures; IR cloned Trim39 and reviewed the manuscript; GB provided material support and contributed in the design of experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M Piacentini
Supplementary information
Rights and permissions
About this article
Cite this article
Basu-Shrivastava, M., Mojsa, B., Mora, S. et al. Trim39 regulates neuronal apoptosis by acting as a SUMO-targeted E3 ubiquitin-ligase for the transcription factor NFATc3. Cell Death Differ 29, 2107–2122 (2022). https://doi.org/10.1038/s41418-022-01002-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-01002-2