Abstract
Krüppel-like factor 5 (KLF5) is an oncogenic factor that is highly expressed in basal-like breast cancer (BLBC) and promotes cell proliferation, survival, migration, stemness, and tumor growth; however, its posttranslational modifications are poorly defined. Protein arginine methyltransferase 5 (PRMT5) is also an oncogene implicated in various carcinomas, including breast cancer. In this study, we found that PRMT5 interacts with KLF5 and catalyzes the di-methylation of KLF5 at Arginine 57 (R57) in a methyltransferase activity-dependent manner in BLBC cells. Depletion or pharmaceutical inhibition (using PJ-68) of PRMT5 decreased the expression of KLF5 and its downstream target genes in vitro and in vivo. PRMT5-induced KLF5R57me2 antagonizes GSK3β-mediated KLF5 phosphorylation and subsequently Fbw7-mediated KLF5 ubiquitination and coupled degradation. Functionally, PRMT5 promotes breast cancer stem cell maintenance and proliferation, at least partially, by stabilizing KLF5. PRMT5 and KLF5 protein levels were positively correlated in clinical BLBCs. Taken together, PRMT5 methylates KLF5 to prevent its phosphorylation, ubiquitination, and degradation, and thus promotes breast cancer stem cell maintenance and proliferation. These findings suggest that PRMT5 is a potential therapeutic target for BLBC.
Similar content being viewed by others
Introduction
Breast cancer is a common malignant tumor that seriously threatens women’s health. According to the latest cancer statistics reported by the American Cancer Society, breast cancer is the leading cause of cancer-related death among women aged 20–59 years [1]. The basal-like breast cancer (BLBC) subtype is characterized by the expression of basal markers, but not estrogen receptor α, progesterone receptor, and human epidermal growth factor 2 [2, 3]. Currently, BLBCs occur more frequently in younger women and BLBC patients display more aggressive clinical outcomes [4].
Cancer stem cells (CSCs) play key roles in intra- and intertumoral heterogeneities, and are responsible for tumor progression through metastasis, drug resistance, and recurrence [5]. Breast CSCs (BCSCs) represent a dynamic subpopulation of breast cancer cells that have the abilities of tumor initiation, self-renewal, and the potential to give rise to a more differentiated progeny upon xenotransplantation into immunocompromised mice [6]. Thus, identification of CSC population in BLBCs may help to identify new targeted therapies.
Knockdown of Krüppel-like factor 5 (KLF5) prevents embryonic stem cells (ESC) differentiation partially by promoting the transcription of Nanog and Oct3/4. KLF5 is a stem cell transcription factor, expressed in mouse ESCs [7]. In our previous studies, we implicated KLF5 in the promotion of breast cancer cell proliferation, survival, stemness, and tumorigenesis [8, 9]. KLF5 expression is also a prognostic marker for poorer survival of patients with breast cancer. Compared to the other breast cancer subtypes, KLF5 is highly expressed in BLBC [10]. KLF5 promotes breast cancer partially by promoting Cyclin D1 [11], FGF-BP1 [12], Slug [13], and Nanog [14]. Our previous studies showed that KLF5 is an unstable protein ubiquitinated by E3 ubiquitin ligases including SCFFBW7 [15], WWP1 [16, 17], and SMURF2 [18]. KLF5 ubiquitination is reversed by deubiquitinases, such as BAP1, ATXN3L, and USP3 [19,20,21]. Whether KLF5 protein is modified by methylation has not been explored.
Protein arginine methyltransferase 5 (PRMT5) catalyzes the symmetric di-methylation of arginine [22]. Targeted deletion of Prmt5 resulted in early embryonic lethality and suppression of pluripotency of ESCs by reprogramming a set of genes that orchestrate stem cell self-renewal and differentiation [23]. In breast cancer, PRMT5 is a key regulator of BCSC proliferation, self-renewal [24], and genome stability [25]. PRMT5 requires its partner methylome protein (MEP50/WDR77) to form an active enzymatic complex [26]. CDK4 phosphorylates MEP50 and increases PRMT5/MEP50 activity, resulting in prolonged survival of tumor cells [27]. Loss of PRMT5 or MEP50 in MDA-MB-231 cells results in the defects of alternative splicing then leads to inhibition of cell proliferation and migration of breast cancer [28].
PRMT5 methylate proteins besides histones and plays an important role in a variety of cellular processes, including transcription, DNA damage repair, RNA metabolism, and signaling [29, 30]. Several important oncoproteins, such as CFLAR, EGFR, and KLF4, are methylated by PRMT5 [25, 31, 32]. PRMT5 methylate KLF4 and inhibits KLF4 ubiquitination by VHL, thereby reducing KLF4 degradation, contributing to breast cancer initiation and invasion [25]. Considering the substantial accumulation of PRMT5 in breast cancer, melanoma, and acute myeloid leukemia, and its role in promoting cell survival following chemotherapy, the clinical effects of targeting PRMT5 have attracted extensive attention [33,34,35].
Here, we identified PRMT5 as a KLF5 arginine methyltransferase, which increased KLF5 protein stability in breast cancer. PRMT5 promoted BLBC cell stemness and proliferation that were inhibited by the PRMT5-specific inhibitor, PJ-68. These findings suggest that PRMT5 may be a novel therapeutic target and PJ-68 is potential compound for BLBC therapy.
Materials and methods
Cell culture
All cell lines were purchased from American Type Culture Collection (Manassas, VA, USA) and validated via short tandem repeat analysis. HCC1806 and HCC1937 were cultured in Roswell Park Memorial Institute-1640 Medium containing 5% fetal bovine serum. All cells were maintained at 37 °C in an incubator with 5% CO2. HEK293T cells were grown in Dulbecco’s modified eagle’s medium supplemented with 5% fetal bovine serum.
Mammosphere assay
Mammospheres were performed using a Mammosphere Culture Kit (no. 05620; STEMCELL Technologies, Vancouver, BC, Canada). HCC1937 cells were cultured in ultra-low attachment plates (no. 3473, Corning Inc., Corning, NY, USA) at a density of 500 cells/well. Mammospheres were counted after 10–14 days culture.
Flow cytometry
Cells were grown in 6-well plates (105 cells/well). HCC1806 and HCC1937 were trypsinized into single cells, washed once using Hanks Balanced Salt Solution with 2% FBS (HF solution), and stained freshly with antibodies against CD24 and CD44 (all antibody information is listed in Supplementary Table S1) or used for the ALDEFLUOR assay (STEMCELL Technologies) according to the manufacturer’s instructions. Briefly, for CD24/CD44 staining, the cells were incubated with anti-CD24 and anti-CD44 antibodies on ice for 25 min. After centrifugation at 500 × g for 5 min, the cells were collected, washed once using HF solution, and subjected to flow cytometric analysis.
In vivo tumorigenesis assay
Nude female mice (aged 7 weeks) were used. In total, 1 × 105 HCC1806 cells (for single injection) suspended in a 1:3 mixture of Matrigel and PBS (total volume of 100 μl) were injected into the mammary fat pads. Tumor size was measured every 3 days. Tumor volume (cm3) = π (length × width2)/6. For PJ-68 experiments, mice were randomly re-divided into two groups for each group when the tumor volume was close to 50 mm3. PJ-68 (20 mg/kg) and (40 mg/kg) were administered through intraperitoneal injection (not in double-blinding manner). The animal experiment was approved by the Animal Ethics Committee of the Kunming Institute of Zoology, CAS.
Plasmids and transfection
All siRNAs were transfected using Lipofectamine 2000 and plasmids were transfected using PEI. All transfection assays were performed according to the manufacturer’s instructions. All siRNA and primer sequences are shown in Supplementary Table S2.
Quantitative RT-PCR
Total RNA was extracted from cells using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. Reverse transcription was performed using the iScript cDNA synthesis kit (Bio-Rad Laboratories Inc., CA), and mRNA levels were quantified using RT Real-Time™ SYBR Green/ROX PCR master mix (SABiosciences, CA) on an ABI-7900 system.
Cell viability assay
Cell viability was measured via sulforhodamine B (SRB) assays. Briefly, cells were plated in 96-well plates at a density of 103/well. The day after cell plating, the cells were fixed using 10% trichloroacetic acid and stained with 0.4% SRB. After dissolving SRB in the cells using 10 mM unbuffered Tris-base, the optical density was read at a single wavelength of 530 nm using an automated spectrophotometric plate reader.
EdU assay
A 5-ethynyl-20-deoxyuridine (EdU) assay kit (RiboBio, Guangzhou, China) was used to determine cell proliferation. Cells were seeded in confocal plates at a density of 1 × 105 cells/well. The cells were incubated with 50 μM EdU buffer at 37 °C for 2 h, fixed with 4% formaldehyde for 0.5 h, and permeabilized with 0.1% Triton X-100 for 20 min. EdU solution was added to the culture followed by staining of nuclei with Hoechst. The results were visualized using a fluorescence microscope.
Co-IP and western blot
Cells were washed with PBS and lysed in ice-cold lysis buffer (containing protease inhibitor) for 30 min. The cell lysates were incubated with the antibodies overnight at 4 °C, followed by incubation with protein A/G beads for 2 h at 4 °C. The beads were washed with RIPA buffer four times. Finally, the beads were boiled in 2× sodium dodecyl sulfate (SDS) buffer for 10 min. The eluents were analyzed using immunoblotting (IB). Lysed samples were separated via SDS-PAGE, transferred onto polyvinylidene difluoride membranes, and blocked with 5% bovine serum albumin for 1 h. The membranes were then incubated with primary antibodies at 4 °C overnight, and with corresponding HRP-conjugated secondary antibody for 1 h, and detected with a chemiluminescent HRP substrate.
Protein–protein interaction
GST pull-down assays were performed using glutathione-sepharose 4B slurry beads by incubating for 2 h at 4 °C. The beads were washed with RIPA buffer gentle shaking for four times and the eluents were analyzed using IB.
In vitro methylation analysis
In vitro methylation analysis was carried out as previously described [36]. In brief, a prokaryotic expression plasmid pGEX-6 × His-GFP-KLF5 was conducted, 6 × His-GFP-KLF5 fusion protein was expressed in E. coli strain BL21 and later purified according to the manufacturer’s directions. Wild-type (WT) PRMT5 protein was obtained from 293T cells and later immunoprecipitated with PRMT5 antibody and 6 × His-GFP-KLF5 fusion protein and immunoprecipitated PRMT5 protein were incubated in reaction buffer containing methyl group donor SAM (1 μM) at 37 °C for 75 min. The reaction stopped with SDS sample buffer was subjected to SDS-PAGE and immunoblot with the antibody SYM10 and KLF5.
Immunohistochemical staining
Paraffin-embedded clinical BLBC specimens were obtained from The First Affiliated Hospital, Zhengzhou University, Zhengzhou, China. Informed consent was obtained from all subjects. PRMT5 antibody was used at a dilution of 1:800. KLF5 staining was performed according to the protocol described in our previous study [37]. Finally, the sections were incubated with avidin–biotin complex reagent for 30 min at room temperature on a shaker. All antibodies and reagents used are listed in Supplementary Table 3.
Statistical analysis
All experiments were performed in triplicates. When appropriate, the data were pooled to generate means ± standard deviation and were analyzed using the t-test. p < 0.05 was considered significant. GraphPad Prism 9 (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses.
Results
PRMT5 interacts with and methylates KLF5
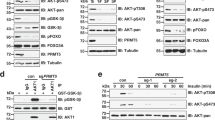
First, we demonstrated that endogenous KLF5 protein interacts with PRMT5 and MEP50 proteins via immunoprecipitation (IP) with anti-KLF5 antibody in HCC1806 and HCC1937 cells, two BLBC cell lines highly expressing KLF5 (Fig. 1a). Using fast protein liquid chromatography, we further demonstrated that KLF5 forms a protein complex with PRMT5 and MEP50 (Fig. 1b). Next, we mapped the interacting motifs of KLF5 and PRMT5 by generating a series of GST-tagged deletion mutants of KLF5 and Myc-tagged truncation mutants of PRMT5. GST pull-down assays revealed that KLF5 zinc-finger domains (373–457 aa) interacted with the PRMT5 N-terminal TIM barrel domain (1-324 aa) (Fig. 1c, d).
a Whole-cell lysate of HCC1806 and HCC1937 cells were collected and subjected to co-immunoprecipitation (Co-IP) assays and immunoblotting (IB). An anti-KLF5 antibody was used for IP. b Protein extracts of HCC1806 cells expressing KLF5 were analyzed via size exclusion chromatography on an FPLC Superdex 200 column. Eluted fractions were resolved using SDS-PAGE for IB analysis. c Identification of the KLF5 region that interacts with PRMT5. KLF5 373–457 aa is necessary for its binding to PRMT5. GST-tagged full-length or deletion mutants of KLF5 from transfected HEK293T cells were precipitated with GST beads. Myc-PRMT5 was blotted with an anti-Myc antibody. XZF zinc finger motifs. d Mapping the PRMT5 domain that interacts with KLF5. PRMT5 1-324 aa is necessary for its binding to KLF5. e Endogenous KLF5 protein is methylated in BLBC cells. Cell lysates from HCC1806 and HCC1937 cells were immunoprecipitated with anti-KLF5 antibody and immunoblotted with anti-SYM10 antibody. f Endogenous KLF5 protein is methylated in HCC1806. Cell lysates from HCC1806 cells were immunoprecipitated with anti-SYM10 antibody and then immunoblotted with anti-KLF5 antibody. g Purified 6 × His-GFP-KLF5 fusion protein, immunoprecipitated PRMT5 protein, PJ-68, and SAM were subjected to in vitro methylation analysis with SYM10 antibody to detect KLF5 methylation. Immunoblotting (two top panels) and Coomassie Brilliant blue staining (bottom panel) were shown. h PRMT5 knockdown decreased KLF5 methylation in HCC1806 and HCC1937 cells. The cells were transfected with either control (siCtrl) or siPRMT5 (pool), treated with MG132 (20 μΜ, 6 h), and whole-cell extracts were collected for IP with the anti-KLF5 antibody, followed by IB analysis with anti-SYM10 antibody. i Flag-PRMT5 overexpression increased KLF5 methylation in HEK293T cells. HEK293T cells were transfected with the indicated plasmids and whole-cell lysates were collected for GST pull-down assays, followed by IB analysis. Flag-PRMT5-R368A is an enzyme activity-dead mutant. Average of three experiments.
Next, we investigated whether PRMT5 methylates KLF5. To test this, we have immunoprecipitated Flag-tagged KLF5 (KLF5-3 × Flag) or endogenous KLF5 proteins and detected KLF5 methylation via western blotting using an antibody against symmetric di-methylated arginine (SYM10). As expected, KLF5 was methylated (Fig. 1e and Supplementary Fig. S1a), which was further confirmed via IP using the SYM10 antibody and IB with KLF5 antibody (Fig. 1f). Moreover, in vitro methylation assay also supports that PRMT5 complex catalyzed methylation of KLF5 protein (Fig. 1g). Most importantly, depletion of PRMT5 resulted in a significant reduction in KLF5 methylation in HCC1806 and HCC1937 cells (Fig. 1h). Consistently, overexpression of WT Flag-PRMT5, but not its catalytically inactive mutant Flag-PRMT5-R368A, increased KLF5 methylation in HEK293T cells (Fig. 1i). These data suggest that PRMT5 interacts with and methylates KLF5 in an enzyme activity-dependent manner.
PRMT5 protects KLF5 protein from ubiquitination and degradation
To determine the mechanism through which PRMT5-mediated KLF5 methylation affects KLF5 expression and function, we knocked down PRMT5 in HCC1806 and HCC1937 cells and measured the expression of KLF5 protein and its downstream targets. We found that depletion of PRMT5 resulted in a marked decrease in KLF5 protein, and its downstream targets, including Slug, FGF-BP1, and Cyclin D1, at both protein and mRNA levels (Fig. 2a, b); conversely, KLF5 knockdown did not affected PRMT5 expression or activity, indicated by the unchanged level of its target E-cadherin (Supplementary Fig. S1b). In addition, depletion of MEP50 also resulted in the significant downregulation of KLF5 expression in these cell lines (Supplementary Fig. S1c). However, PRMT5 knockdown did not affect KLF5 mRNA levels in either cell line (Fig. 2b). To determine whether PRMT5 decreases KLF5 ubiquitin-dependent proteolysis, we knocked down PRMT5 using siRNA and measured the half-life of KLF5 protein using cycloheximide chase assay. Depletion of PRMT5 accelerated KLF5 protein turnover in HCC1806 and HCC1937 cells (Fig. 2c). MG132, a proteasome inhibitor, blocked PRMT5 knockdown-induced KLF5 decrease in both cell lines (Fig. 2d). Furthermore, KLF5 protein half-life was extended by the overexpression of PRMT5, but not PRMT5-R368A, in HEK293T cells (Fig. 2e).
a PRMT5 knockdown led to downregulation of expression of KLF5 protein and its targets (Slug, FGF-BP1, Cyclin D1, and Nanog). b PRMT5 knockdown had no effect on KLF5 mRNA levels in HCC1806 and HCC1937 cells. PRMT5 depletion decreased the mRNA levels of Slug, FGF-BP1, Cyclin D1, and Nanog. c PRMT5 depletion significantly promoted the degradation of KLF5 protein in HCC1806 and HCC1937 cells. HCC1806 and HCC1937 were transfected with siPRMT5 (pool) or siCtrl following treatment with Cycloheximide (CHX, 50 μg/ml) for 0.5, 1, or 2 h. The density of the KLF5 band was quantified and normalized to the control β-actin and plotted on the right. Statistical significance was determined via Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. d The proteasome inhibitor MG132 blocked the decrease in PRMT5 knockdown-induced KLF5 in HCC1806 and HCC1937 cells. siCtrl was used as the negative control. The cells were treated with MG132 (20 μM) for 6 h, and then cell lysates were collected for IB. e PRMT5 overexpression extended the half-life of KLF5. HEK293T cells were transfected with Flag-PRMT5 or Flag-PRMT5-R368A for 48 h and then treated with CHX for the indicated times. The graph shows the quantitative results from the left panel. Statistical significance was determined via Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. f PRMT5 knockdown in HCC1806 cells increased endogenous KLF5 protein ubiquitination. PRMT5 was stably knocked down in HCC1806 cells. g PRMT5 overexpression decreased KLF5 ubiquitination in an arginine methyltransferase activity-dependent manner. GST pull-down was used to detect ubiquitinated KLF5 protein levels. h PRMT5 decreased Fbw7γinduced KLF5 ubiquitination. GST pull-down was used to detect ubiquitinated KLF5 protein levels. PRMT5 decreased GST-KLF5 associated Fbw7γ protein. Average of three experiments.
It is well established that KLF5 is degraded through the ubiquitin-proteasome pathway [38]. We demonstrated that PRMT5 knockdown increased KLF5 ubiquitination in HCC1806 (Fig. 2f). Overexpression of PRMT5, but not PRMT5-R368A, decreased KLF5 ubiquitination in HEK293T cells (Fig. 2g). Given our previous finding that Fbw7γ predominately ubiquitinates KLF5 [39], we sought to determine whether PRMT5-mediated methylation prevents KLF5 ubiquitination by Fbw7γ. We found that PRMT5 overexpression decreased the association of Fbw7γ with KLF5 and Fbw7γ-mediated ubiquitination (Fig. 2h). Collectively, our results indicate that KLF5 methylation by PRMT5 stabilizes KLF5 by antagonizing Fbw7γ-mediated KLF5 ubiquitination.
PRMT5 promotes basal-like breast tumorigenesis partially through KLF5
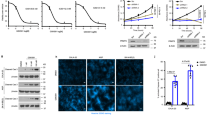
Given that both PRMT5 and KLF5 promote breast cancer stemness [24, 40], we first tested whether PRMT5 knockdown decreased BCSC through downregulation of KLF5. We overexpressed KLF5 in HCC1806 and HCC1937 cells with stable PRMT5 knockdown. As expected, KLF5 overexpression partially restored the decrease in FGF-BP1 and Cyclin D1 protein expression induced by PRMT5 depletion (Fig. 3a). Consistently, ectopic overexpression of KLF5 significantly reduced the loss of cell viability and DNA synthesis (Fig. 3b, c and Supplementary Fig. S2a) and BCSC populations marked by CD24lowCD44+ caused by depletion of PRMT5 (Fig. 3d and Supplementary Fig. S2b, c). These results suggest that endogenous PRMT5 promotes breast cancer cell proliferation and stemness partially through KLF5.
a Stable overexpression of KLF5 partially rescued PRMT5 knockdown-mediated decrease in FGF-BP1 and Cyclin D1 protein expression in HCC1806 and HCC1937 cells, as measured via IB. pCDH was used as a negative control for KLF5. shCtrl was used as a negative control for shPRMT5. b KLF5 overexpression partially restored cell proliferation in PRMT5 knockdown HCC1806 and HCC1937 cells. Cell viability was measured using the SRB assay every day. Error bars represent the standard deviation (SD) of five samples in parallel. Statistical significance was determined via Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001. c KLF5 overexpression partially restored DNA synthesis in PRMT5 knockdown HCC1806 and HCC1937 cells. The percentages of proliferative cells (vs. total cells) from five images were compared using the t-test. Graphs represent the mean ± SD *p < 0.05, **p < 0.01, ***p < 0.001. d KLF5 overexpression significantly restored the percentage of CD24low/CD44+ BCSC population in PRMT5 knockdown HCC1806 and HCC1937 cells. Average of three experiments. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. e Stable overexpression of KLF5 restored PRMT5 knockdown-mediated growth inhibition of HCC1806 xenograft in nude mice. Xenograft tumor growth was measured every 3 days. Data points represent the mean ± SD (N = 6 per group). Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. f Stable overexpression of KLF5 partially rescued PRMT5 knockdown-mediated decrease in tumor weight. *p < 0.05, **p < 0.01, ***p < 0.001, t-test. g Tumor masses collected on day 21.
To test whether PRMT5 promotes basal-like breast tumor growth in vivo, we knocked down PRMT5 in HCC1806 cells that overexpressed KLF5 (Supplementary Fig. S2d). As expected, PRMT5 knockdown cells grew significantly slower than control cells (Fig. 3e). KLF5 overexpression promoted the growth of HCC1806 cell-derived xenografts in nude mice and significantly rescued PRMT5 knockdown-induced growth inhibition (Fig. 3e). Comparison of tumor weights among the different groups further supported this conclusion (Fig. 3f, g). In addition, the primary xenografts showed consistent changes in the expression of KLF5, PRMT5, and FGF-BP1 (Supplementary Fig. S2e). Taken together, these results indicate that PRMT5 promotes BLBC tumorigenesis partially through KLF5.
PRMT5-mediated KLF5 methylation occurs at Arginine 57
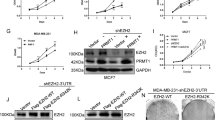
To identify the KLF5 sites that are methylated by PRMT5, we expressed Flag-tagged KLF5 in HEK293T cells and purified KLF5 protein using Flag-M2 beads. The prominent band was excised and subjected to mass spectrometric analysis. Arginine residue 57 (R57), R358, R361, and R362 were identified as candidate di-methylation sites (Fig. 4a and data not shown). To verify this, we mutated these Arginines into Lysines, respectively, and found that the R57K mutation substantially reduced KLF5 methylation (Fig. 4b). In contrast, the R358/361/362K mutantion did not markedly decrease KLF5 methylation (Fig. 4b). Consistently, PRMT5 overexpression substantially increased the methylation level of KLF5-WT, but not that of KLF5-R57K (Fig. 4c). In addition, KLF5-R57K also increased KLF5 ubiquitination and displayed higher affinity for Fbw7γ, compared to KLF5-WT (Fig. 4d). In agreement with the finding that KLF5 methylation decreases Fbw7γ-mediated KLF5 ubiquitination, we found that KLF5-R57K presented a shorter protein half-life than KLF5-WT in the presence of Fbw7γ, but not Fbw7γΔF (an inactive mutant that does not interact with Skp1) (Fig. 4e).
a Identification of R57 as the methylation residue of KLF5 via mass spectrometric analysis. b Validation of R57 as the PRMT5-catalyzed KLF5 methylation site. GST pull-down was used to detect methylated KLF5 protein levels. Mutations in other R residues of KLF5 had no effect on KLF5 methylation. c PRMT5 overexpression increased protein methylation of KLF5-WT, but not KLF5-R57K, in HEK293T cells. d KLF5-R57K increased Fbw7γ-mediated ubiquitination. Myc-Fbw7γ was co-expressed with GST-KLF5 or GST-KLF5-R57K in HEK293T cells. e KLF5-R57K increased Fbw7γ-mediated degradation. Fbw7γΔF served as a negative control. The density of KLF5 bands were quantitated. f Compared with KLF5-WT, the interaction between KLF5-R57K and GSK3β is more obvious. g Joint knockdown of PRMT5 and GSK3β restored KLF5 protein level. h KLF5 R57K/S303A exhibited dramatically higher protein stability than KLF5 R57K. i MG132 blocked degradation of KLF5-R57K protein in HCC1806 and HCC1937 cells. HCC1806 and HCC1937 cells (endogenous KLF5 was stably deleted using Cas9/sgKLF5) were treated with MG132 (20 μM) for 6 h. j Overexpression of KLF5-WT, but not that of methylation-deficient KLF5-R57K, restored FGF-BP1 protein levels in HCC1806 and HCC1937 cells. Endogenous KLF5 was stably deleted using Cas9/sgKLF5. k KLF5-R57K did not completely restore the growth arrest induced by stable KLF5 knockdown in HCC1806 and HCC1937 cells. l KLF5-R57K did not completely restore the decrease in DNA synthesis induced by stable KLF5 knockdown in HCC1806 and HCC1937 cells. Graphs represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. m KLF5-R57K did not completely restore the decrease in BCSCs induced by KLF5 knockdown in HCC1806 and HCC1937 cells. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
It has been established that GSK3β phosphorylates KLF5 on Ser303 (S303) and thus promotes Fbw7-mediated KLF5 ubiquitination in TNBC cells [41]. Thus, we further study the interplay between PRMT5-induced methylation and GSK3β-mediated phosphorylation on KLF5. We first proved that no interfering between the expression of GSK3β and PRMT5 (Supplementary Fig. S3a). Interestingly, we found an enhanced interaction between GSK3β and KLF5 R57K over KLF5 WT (Fig. 4f). Furthermore, methylation and phosphorylation on KLF5 seemed mutually exclusive, which was coordinated by PRMT5 and GSK3β, respectively (Supplementary Fig. S3b). Consistent with that, joint knockdown of PRMT5 and GSK3β rescued endogenous KLF5 expression in both HCC1806 and HCC1937 cells (Fig. 4g), and KLF5 R57K/S303A exhibited dramatically higher protein stability than KLF5 R57K (Fig. 4h). We further demonstrated that MG132 blocked KLF5-R57K degradation in both HCC1806 and HCC1937 cell lines (Fig. 4i).
To test whether KLF5R57me2 increases KLF5-mediated oncogenic functions in breast cancer, we knocked out KLF5 in both HCC1806 and HCC1937 cell lines using the Cas9/sgRNAs targeting non-CDS region. Next, we stably expressed KLF5-WT and KLF5-R57K in KLF5 KO cells (Fig. 4j). Notably, although both KLF5-WT and KLF5-R57K showed similar mRNA levels, KLF5-WT showed higher protein levels than KLF5-R57K (Supplementary Fig. S3c). As shown in Fig. 4k–m, overexpression of KLF5, but not KLF5-R57K, rescued KLF5 KO-induced decreases in FGF-BP1 mRNA expression, cell growth, DNA synthesis, and BCSC population (Supplementary Fig. S3d, e). Taken together, these findings indicate that PRMT5-meditated KLF5R57me2 stabilizes KLF5 protein and promotes BLBC.
PRMT5 inhibitor, PJ-68, suppresses BLBC
PRMT5 is a promising therapeutic target for cancer treatment, and several specific PRMT5 small-molecule inhibitors, such as PJ-68, have been reported [42]. We evaluated whether PJ-68 inhibits KLF5 in BLBC cells. PJ-68 decreased KLF5 protein levels, but not mRNA levels, in both HCC1806 and HCC1937 cells in a dose-dependent manner (Fig. 5a, b). Consistently, PJ-68 also reduced KLF5 methylation (Fig. 5c) and accelerated KLF5 protein turnover in HCC1806 and HCC1937 cells (Fig. 5d). MG132, a proteasome inhibitor, blocked the PJ-68-induced KLF5 decrease in both cell lines (Fig. 5e). We also found that PJ-68 reduced KLF5-WT, but not KLF5-R57K protein levels in HEK293T cells (Fig. 5f), and notably, PJ-68 cannot further decrease KLF5 in PRMT5-knockdown HCC1806 cells (Supplementary Fig. S4a). KLF5 overexpression partially restored PJ-68-induced FGF-BP1 protein levels in HCC1806 and HCC1937 cells (Fig. 5g). Functionally, KLF5 overexpression promoted cell growth and rescued PJ-68-induced cell growth inhibition (Fig. 5h). We further validated this by measuring DNA synthesis (Fig. 5i and Supplementary Fig. S4b). Moreover, KLF5 overexpression significantly rescued the reduction of CD24low/CD44+ BCSC population following PJ-68 treatment (Fig. 5j and Supplementary Fig. S4c).
a PJ-68 inhibited the expression of KLF5 and its downstream target genes in both HCC1806 and HCC1937 cells. Cells were treated with 0, 1.25, 2.5, 5, and 10 μM PJ-68 for 24 h. Whole-cell extracts were collected for IB analysis. b PJ-68 had no effect on the mRNA levels of KLF5 in HCC1806 and HCC1937 cells. In contrast, it decreased the mRNA levels of FGF-BP1. mRNA level of KLF5 was examined via RT-qPCR. *p < 0.05, **p < 0.01, ***p < 0.001, t-test. c PJ-68 decreased KLF5 methylation in HEK293T cells. Cells were transfected with the indicated plasmids and treated with PJ-68 (5 μΜ) for 6 h. Whole-cell lysates were collected for KLF5 methylation analysis. d PJ-68 significantly promoted the degradation of KLF5 protein in HCC1806 and HCC1937 cells. HCC1806 and HCC1937 were treated with PJ-68 or DMSO following treatment with cycloheximide (CHX, 50 μg/ml) for 0.5, 1, or 2 h. The density of the KLF5 band was quantified and normalized to the control β-actin and plotted on the right. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. e The proteasome inhibitor MG132 blocked the decrease in PJ-68-induced KLF5 in HCC1806 and HCC1937 cells. The cells were treated with MG132 (20 μM) for 6 h, and then cell lysates were collected for IB. f PJ-68 decreased the protein levels of KLF5, but not KLF5-R57K, in HEK293T cells. HEK293T cells were transfected with the indicated plasmids and treated with PJ-68 (5 μΜ) for 6 h. KLF5-R57K protein level was lower than that of KLF5-WT in the absence of PJ-68 treatment because of its instability. g Overexpression of KLF5 restored PJ-68-mediated downregulation of FGF-BP1 protein expression in HCC1806 and HCC1937 cells. Cells were treated with PJ-68 (5 μΜ) for 6 h. h KLF5 overexpression partially rescued PJ-68-mediated cell growth inhibition in HCC1806 and HCC1937 as revealed by SRB assays. i KLF5 overexpression partially rescued PJ-68-mediated decrease in DNA synthesis in HCC1806 and HCC1937 cells, as measured via EdU assays. Cells were treated with PJ-68 (0 or 5 μΜ) for 6 h. j KLF5 overexpression partially rescued PJ-68-mediated decrease in CD24low/CD44+ population in HCC1806 and HCC1937. Cells were treated with PJ-68 (0 or 5 μΜ) for 6 h. Average of three experiments. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
Finally, we investigated whether PJ-68 suppresses tumor growth in vivo. We administered the mice that harbored HCC1806 cell-derived xenografts with 20–40 mg/kg/day PJ-68 or vehicle control for 3 weeks (Fig. 6a). As expected, PJ-68 significantly inhibited tumor growth (Fig. 6b–d), and there was no statistical difference in the mouse body weight between the different groups (Fig. 6e). We also analyzed the expression of KLF5, PRMT5, and FGF-BP1 in these primary tumor samples and found that PJ-68 decreased the protein levels of KLF5 and its targets in a dose-dependent manner (Fig. 6f).
a Schematic of the strategy used for testing the efficacy of PJ-68 in vivo. b Pharmacological inhibition of PRMT5 significantly suppressed breast tumor growth in an in vivo mouse xenograft model. Quantification of tumor growth curve based on tumor volume. Statistical significance was determined via Student’s t-test (N = 4 per group). *p < 0.05, **p < 0.01, ***p < 0.001. c PJ-68 significantly decreased tumor weights in a dose-dependent manner as observed in the mice following their sacrifice. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. d Tumor masses collected at the end of the experiment. e No statistical differences were observed in the body weight of mice among the different dose groups. Statistical significance was determined via Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001. f PJ-68 decreased KLF5, Slug, and Cyclin D1 protein levels and increased p21 and cleaved PARP protein levels in primary tumor samples, as measured via IB. g PRMT5 and KLF5 protein expression levels are positively correlated in human BLBC specimens, as measured via IHC. Staining results of the positive and negative controls are shown. h Schematic representation showing that PRMT5-mediated KLF5 protein methylation at R57 decreases GSK3β-mediated KLF5 phosphorylation and Fbw7γ-mediated KLF5 protein ubiquitination and degradation and promotes BLBC tumorigenesis.
Protein expression of PRMT5 and KLF5 are positively correlated in BLBC
To test the clinical relevance of the PRMT5-KLF5 axis in breast cancer, we examined the protein levels of both PRMT5 and KLF5 in breast tumor tissues using immunohistochemistry. A strong positive correlation (R = 0.700, **p < 0.01) was observed between PRMT5 and KLF5 in these BLBC samples (Fig. 6g and Supplementary Fig. S5a, b and Table 1). It is known that high expression of either PRMT5 or KLF5 contributes to the poor survival outcome of breast cancer patients. Despite the universal expression of PRMT5, the high co-expression of PRMT5 and KLF5 in BLBC samples further supported our hypothesis. Our molecular characterization unveiled a novel mechanism in which crosstalk between ubiquitination, phosphorylation, and arginine methylation orchestrates KLF5 protein expression and functions in BLBC (Fig. 6h).
Discussion
As the most malignant subtype of breast cancer, BLBC has the worst prognosis. We previously reported that KLF5 is highly expressed in BLBC and serves as a driver oncogene for BLBC [10] and that Fbw7 targets KLF5 for proteasomal degradation [43]. Here, we identified PRMT5 as a stabilizer of KLF5 that further enhances KLF5 expression in BLBCs. PRMT5 has been suggested as a potential therapeutic target in cancers. Inhibition of PRMT5 in melanoma cells led to decreased MDM4 protein expression and subsequent p53 activation, independent of methylation [32]. Additionally, PRMT5 inhibition decreased the methylation of E2F1 and reduced expression of its target genes, leading to attenuated DNA damage repair, cell cycle arrest, and apoptosis [44]. PRMT5 contributes to gastric cancer, at least in part, by methylating BCL6 at R305, a modification necessary for the full transcriptional repressive effects of BCL6. Inhibition of PRMT5 in B-cell lymphoma lines led to significant upregulation of BCL6 target genes, and concomitant inhibition of both BCL6 and PRMT5 exhibited synergistic killing of BCL6-expressing lymphoma cells [45].
Our previous study suggested that KLF5 is a stem cell factor in BLBC. Interestingly, both KLF5 and PRMT5 play key roles in cell proliferation and stem cell self-renewal [46, 47]. We have previously reported that GSK3β phosphorylates KLF5 at S303 and thus promotes Fbw7-mediated KLF5 degradation [41]. Moreover, PRMT5 epigenetically silenced the expression of the tumor suppressor Fbw7 in cells [48]. Here, we showed that PRMT5 methylates KLF5 and attenuates the interaction of KLF5 with GSK3β and Fbw7γ. Therefore, our findings integrated those clues and reported that the novel crosstalk among methylation, phosphorylation, and ubiquitination coordinates KLF5 expression and function in BLBC, in which PRMT5 plays a vital role. We have noticed that KLF5 only partially rescued tumor growth in the shPRMT5 context (Fig. 3e–g). We guess that KLF5R57me2 has additional roles such as transcriptional regulation of some specific targets, since KLF5 forms a complex with PRMT5/MEP50 (Fig. 1b), which function as important epigenetic regulators. Thus, exogenous KLF5 rescued the expression level in PRMT5-deficient cells, but PRMT5-dependent KLF5R57me2 was still absent, resulting in limited promotion on tumor growth. Additionally, PRMT5 is a well-known epigenetic regulator methylating histones such as H3R8 and H4R3, thus PRMT5 also functions through other genes besides KLF5. Another KLF family protein, KLF4, is also targeted by PRMT5 [25]. Future studies will identify more targets of PRMT5 and reveal their functions in breast cancer. High expression of PRMT5 predicts worse clinical outcome in several cancers, including breast cancer, but its mRNA level showed little difference among the breast cancer subtypes [49]. Thus, it is of interest to study the mechanism regulating PRMT5 expression in BLBC.
In a mouse transgenic study, accumulating physiological evidence revealed that PRMT5 was initially associated with cell proliferation and development [23]. Targeted deletion of Prmt5 results in early embryonic lethality and suppression of pluripotency of ES cells by rebalancing a set of genes that orchestrate stem cell self-renewal and differentiation [50]. Conditional ablation of Prmt5 in progenitors results in hypomyelination and reduced survival and differentiation [51]. In addition, accumulation of PRMT5 in blood [42], lung [52], and liver [53] cancers appears to promote cell survival. It is therefore necessary to study the development of breast and breast cancer in breast-specific Prmt5 KO mice.
Anticancer compounds targeting PRMT5 have attracted considerable attention over the past decade, and several PRMT5 inhibitors have entered phase I/III clinical trials [54]. For example, EPZ015666, also known as GSK3235025, induced the death of mantle-cell lymphoma cells and suppressed tumor growth in xenograft models [55]. Based on its safety and selectivity, JNJ-64619178 was chosen as a clinical candidate [56]. Inhibition of PRMT5-induced cell death and suppressed cell proliferation in solid and hematologic cancers [57]. The small-molecule inhibitor of PRMT5, PJ-68, prolonged survival in a murine model of retroviral BCR-ABL-driven CML and impaired the in vivo self-renewal capacity of transplanted CML LSCs [42]. Here, we tested the anticancer effects of PJ-68 against breast tumors, and our data support its potential therapeutic application in BLBC. Our current study suggests that PRMT5-mediated KLF5 methylation at R57 prevents the E3 ubiquitin ligase, Fbw7γ, from binding to KLF5 protein, and therefore, stabilizes KLF5 protein to promote the transcription of KLF5 downstream target genes, and thereby BLBC cell stemness and proliferation. Thus, PRMT5 may be a potential target for the treatment of KLF5-positive BLBC.
Data availability
All data generated and analyzed during this study are included in this published article. The datasets supporting the conclusions of this article are included within the article and its additional files.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS. 2001;98:10869–74.
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–40.
Brooks MD, Burness ML, Wicha MS. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell. 2015;17:260–71.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, et al. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–34.
Liu R, Zheng HQ, Zhou ZM, Dong JT, Chen CS. KLF5 promotes breast cell survival partially through fibroblast growth factor-binding protein 1-pERK-mediated dual specificity MKP-1 protein phosphorylation and stabilization. J Biol Chem. 2009;284:16791–8.
Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, et al. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–8.
Chen CH, Yang N, Zhang YC, Ding JC, Zhang WJ, Liu R, et al. Inhibition of super enhancer downregulates the expression of KLF5 in basal-like breast cancers. Int J Biol Sci. 2019;15:1733–42.
Chen CH, Benjamin MS, Sun XD, Otto KB, Guo P, Dong XY, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulation in the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–55.
Zheng HQ, Zhou Z, Huang J, Chaudhury L, Dong JT, Chen C. Kruppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of broblast growth factor binding protein 1. Oncogene. 2009;28:3702–13.
Liu R, Shi PG, Zhou ZM, Zhang HL, Li W, Zhang H, et al. Krupple-like factor 5 is essential for mammary gland development and tumorigenesis. J Pathol. 2018;246:497–507.
Long XL, Singla DK. Inactivation of Klf5 by zinc finger nuclease downregulates expression of pluripotent genes and attenuates colony formation in embryonic stem cells. Mol Cell Biochem. 2013;382:113–9.
Yumimoto K, Nakayama KI. Recent insight into the role of FBXW7 as a tumor suppressor. Semin Cancer Biol. 2020;67:1–15.
Zhao D, Zhi X, Zhou ZM, Chen CS. TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis. 2012;33:59–67.
Chen CS, Sun XD, Guo P, Dong XY, Sethi P, Cheng XH, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–61.
Du JX, Hagos EG, Nandan MO, Bialkowska AB, Yu B, Yang VW. The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Kruppel-like factor 5 protein. J Biol Chem. 2011;286:40354–64.
Ge F, Chen WL, Qin JY, Zhou ZM, Liu R, Liu LL, et al. Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Kruppel-like factor 5 (KLF5). Oncotarget. 2015;6:21369–78.
Qin JY, Zhou ZM, Chen WL, Wang CY, Zhang HL, Ge GZ, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471.
Wu YY, Qin JY, Li FB, Yang CY, Li Z, Zhou ZM, et al. USP3 promotes breast cancer cell proliferation by deubiquitinating KLF5. J Biol Chem. 2019;294:17837–47.
Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24.
Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–7.
Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL, et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21:3498–513.
Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419.
Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, et al. Crystal structure of the human PRMT5:MEP50 complex. PNAS. 2012;109:17960–5.
Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–40.
Rengasamy M, Zhang F, Vashisht A, Song WM, Aguilo F, Sun YD, et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017;45:11106–20.
Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13.
Li MY, An WT, Xu LY, Lin YD, Su L, Liu XG. The arginine methyltransferase PRMT5 and PRMT1 distinctly regulate the degradation of anti-apoptotic protein CFLAR(L) in human lung cancer cells. J Exp Clin Canc Res. 2019;38:64.
Ge L, Wang HZ, Xu X, Zhou ZR, He JB, Peng WX, et al. PRMT5 promotes epithelial-mesenchymal transition via EGFR-beta-catenin axis in pancreatic cancer cells. J Cell Mol Med. 2020;24:1969–79.
AbuHammad S, Cullinane C, Martin C, Bacolas Z, Ward T, Chen HQ, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. PNAS. 2019;116:17990–8000.
Vinet M, Suresh S, Maire V, Monchecourt C, Nemati F, Lesage L, et al. Protein arginine methyltransferase 5: a novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019;8:2414–28.
Kim H, Kim H, Feng YM, Li Y, Tamiya H, Tocci S, et al. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci Transl Med. 2020;12:551.
Radzisheuskaya A, Shliaha PV, Grinev V, Lorenzini E, Kovalchuk S, Shlyueva D, et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat Struct Mol Biol. 2019;26:999–1012.
Song HW, Feng X, Zhang M, Jin X, Xu XD, Wang L, et al. Crosstalk between lysine methylation and phosphorylation of ATG16L1 dictates the apoptosis of hypoxia/reoxygenation-induced cardiomyocytes. Autophagy. 2018;14:825–44.
Shi PG, Liu WJ, Tala, Wang HX, Li FB, Zhang HL, et al. Metformin suppresses triple-negative breast cancer stem cells by targeting KLF5 for degradation. Cell Discov. 2017;3:17010.
Chen CS, Sun XD, Ran QM, Wilkinson KD, Murphy TJ, Simons JW, et al. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–27.
Liu N, Li H, Li SX, Shen MY, Xiao N, Chen YF, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–67.
Liu N, Yang R, Shi Y, Chen L, Liu YT, Wang ZL, et al. The cross-talk between methylation and phosphorylation in lymphoid-specific helicase drives cancer stem-like properties. Signal Transduct Tar. 2020;5:197.
Zhao D, Zheng HQ, Zhou ZM, Chen CS. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–38.
Jin YL, Zhou JF, Xu F, Jin B, Cui LJ, Wang Y, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126:3961–80.
Luan Y, Wang P. FBW7-mediated ubiquitination and degradation of KLF5. World J Biol Chem. 2014;5:216–23.
Pastore F, Bhagwat N, Pastore A, Radzisheuskaya A, Karzai A, Krishnan A, et al. PRMT5 inhibition modulates E2F1 methylation and gene-regulatory networks leading to therapeutic efficacy in JAK2(V617F)-mutant MPN. Cancer Discov. 2020;10:1742–57.
Lu XQ, Femando TM, Lossos C, Yusufova N, Liu F, Fontan L, et al. PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood. 2018;132:2026–39.
Tan DQ, Li Y, Yang C, Li J, Tan SH, Chin DWL, et al. PRMT5 modulates splicing for genome integrity and preserves proteostasis of hematopoietic stem cells. Cell Rep. 2019;26:2316–28.
Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6:533–44.
Qin Y, Hu QS, Xu J, Ji SR, Dai WX, Liu WS, et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun Signal. 2019;17:30.
Wu Y, Wang Z, Zhang J, Ling R. Elevated expression of protein arginine methyltransferase 5 predicts the poor prognosis of breast cancer. Tumor Biol. 2017;39:10104283117695917.
He JP, Fu XL, Zhang M, He FF, Li WJ, Abdul MM, et al. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat Commun. 2019;10:34.
Scaglione A, Patzig J, Liang JL, Frawley R, Bok J, Mela A, et al. PRMT5-mediated regulation of developmental myelination. Nat Commun. 2018;9:2840.
Sheng XM, Bowen N, Wang ZX. GLI pathogenesis-related 1 functions as a tumor-suppressor in lung cancer. Mol Cancer. 2016;15:25.
Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9:1572.
Desterro J, Bak-Gordon P, Carmo-Fonseca M. Targeting mRNA processing as an anticancer strategy. Nat Rev Drug Disco. 2020;19:112–29.
Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–7.
Wu TF, Millar H, Gaffney D, Beke L, Mannens G, Vinken P, et al. JNJ-64619178, a selective and pseudo-irreversible PRMT5 inhibitor with potent in vitro and in vivo activity, demonstrated in several lung cancer models. Abstract of Annual Meeting of the American-Association-for-Cancer-Research (AACR). Cancer Res. 2018;78:4859.
Wang YX, Hu WH, Yuan YQ. Protein arginine methyltransferase 5 (PRMT5) as an anticancer target and its inhibitor discovery. J Med Chem. 2018;61:9429–41.
Acknowledgements
We thank all members of the laboratory for their help.
Funding
This work was supported by the National Key Research and Development Program of China (2020YFA0112300 and 2018YFC2000400 to CC), National Natural Science Foundation of China (31771516 and 81830087 to CC; 81802671 and 81872414 to DJ), Yunnan Fundamental Research Projects (2019FB112 and 202001AW070018 to DJ), and Project of Innovative Research Team of Yunnan Province (2019HC005).
Author information
Authors and Affiliations
Contributions
CC and DJ were responsible for the conception and design and study supervision. XW and TQ were responsible for the development of the methodology, analysis and interpretation of the data, and experiments in vivo. CY, YW, and YS were responsible for the development of IHC. XW, TQ, CC, GD, YH, WL, and RL were responsible for the experiments in vitro. XW, TQ, DJ, and CC wrote and discussed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Sibilia
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, X., Qiu, T., Wu, Y. et al. Arginine methyltransferase PRMT5 methylates and stabilizes KLF5 via decreasing its phosphorylation and ubiquitination to promote basal-like breast cancer. Cell Death Differ 28, 2931–2945 (2021). https://doi.org/10.1038/s41418-021-00793-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00793-0
This article is cited by
-
GlPRMT5 inhibits GlPP2C1 via symmetric dimethylation and regulates the biosynthesis of secondary metabolites in Ganoderma lucidum
Communications Biology (2024)
-
Histone modifications in drug-resistant cancers: From a cancer stem cell and immune evasion perspective
Experimental & Molecular Medicine (2023)
-
Arginine methylation of MTHFD1 by PRMT5 enhances anoikis resistance and cancer metastasis
Oncogene (2022)
-
BAP1 in cancer: epigenetic stability and genome integrity
Discover Oncology (2022)